Phylogenomics and Floristic Origin of Endiandra R.Br (Lauraceae) from New Caledonia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling, DNA Extraction, and Genomic Sequencing
2.2. Genome Assembly and Annotation
2.3. Phylogenomic Analyses
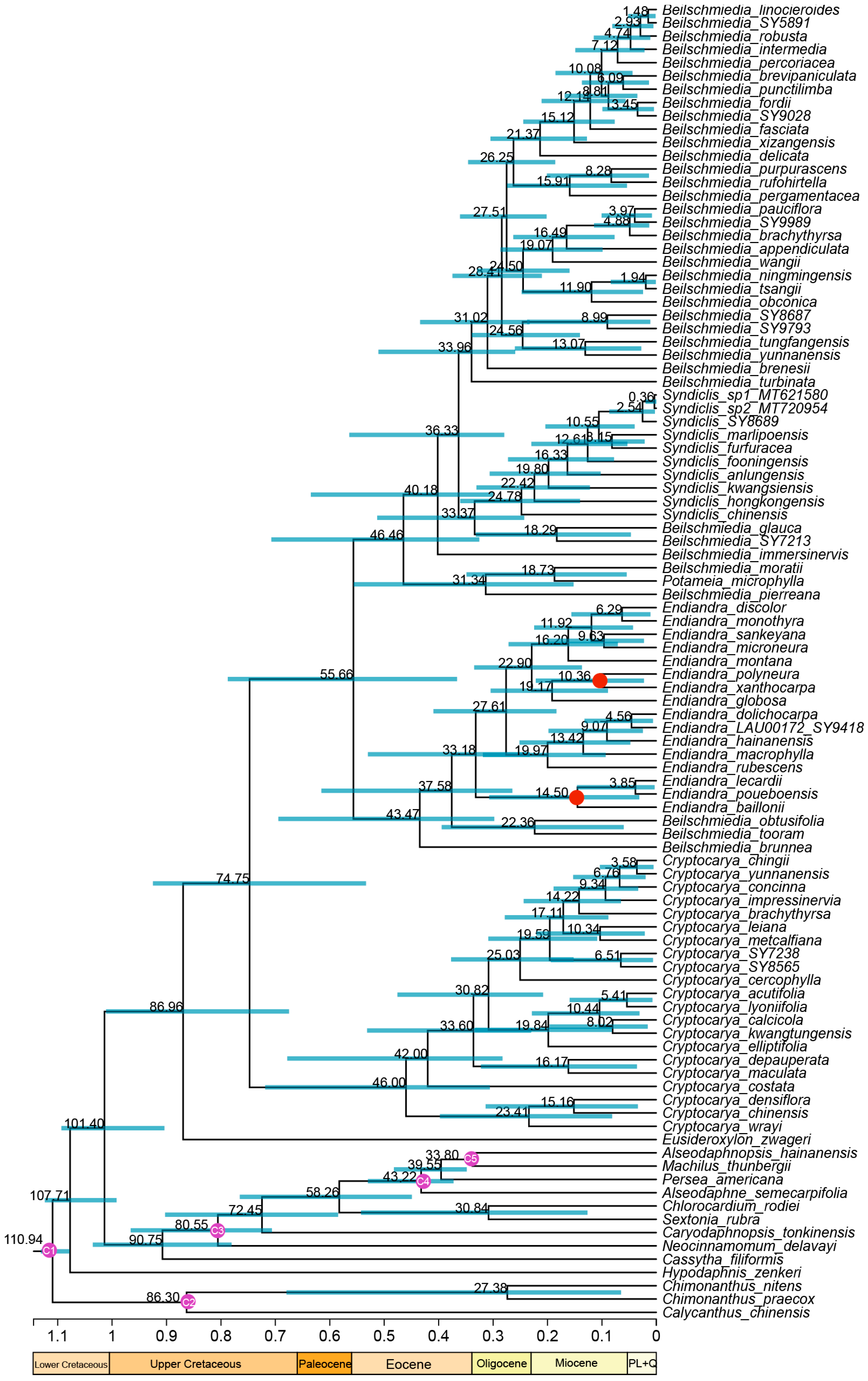
2.4. Divergence Time Estimation
2.5. Analysis of Simple Sequence Repeats and Functional Duplicated Sequences
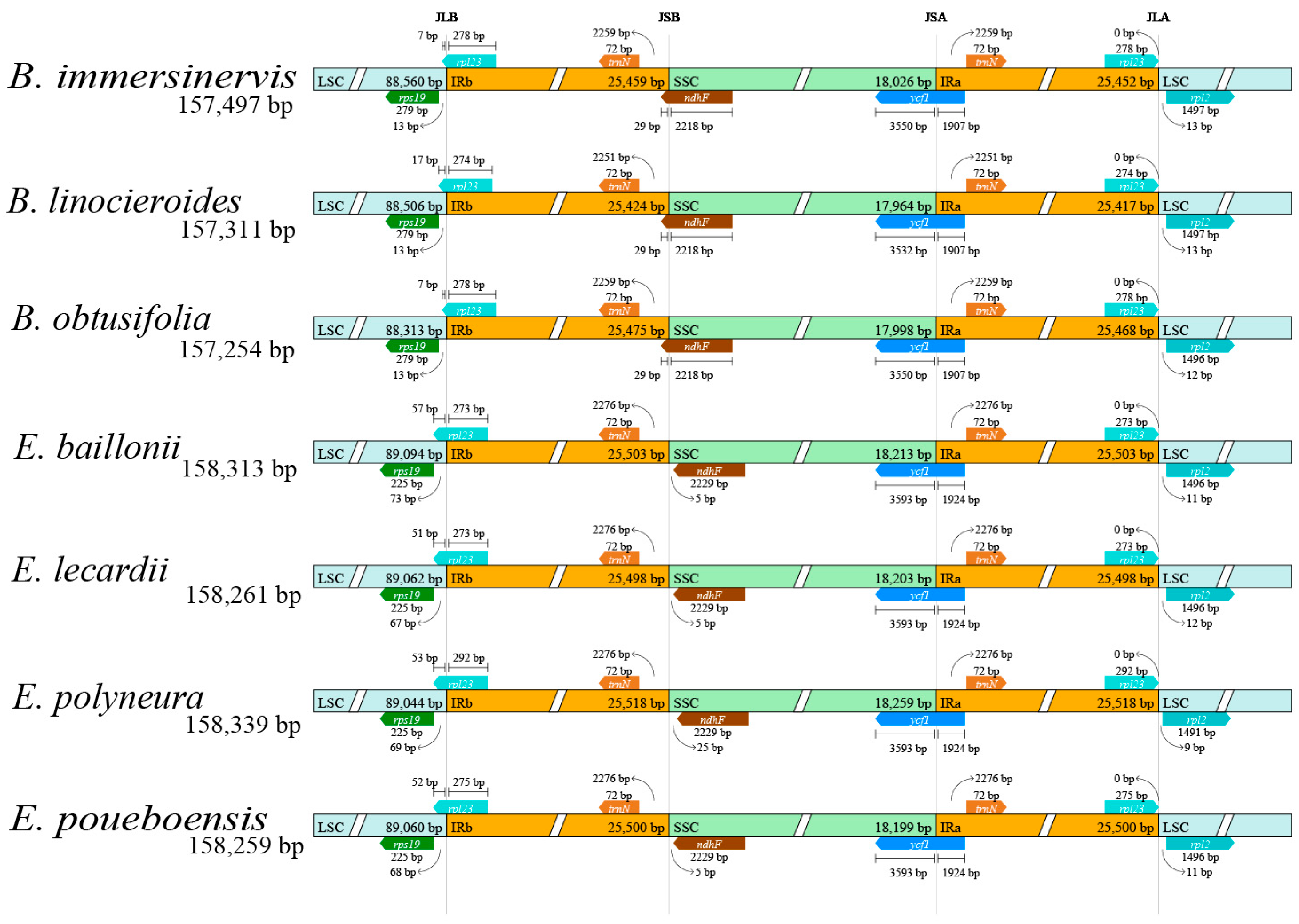
2.6. Analysis of IR Boundaries and Nucleotide Polymorphism
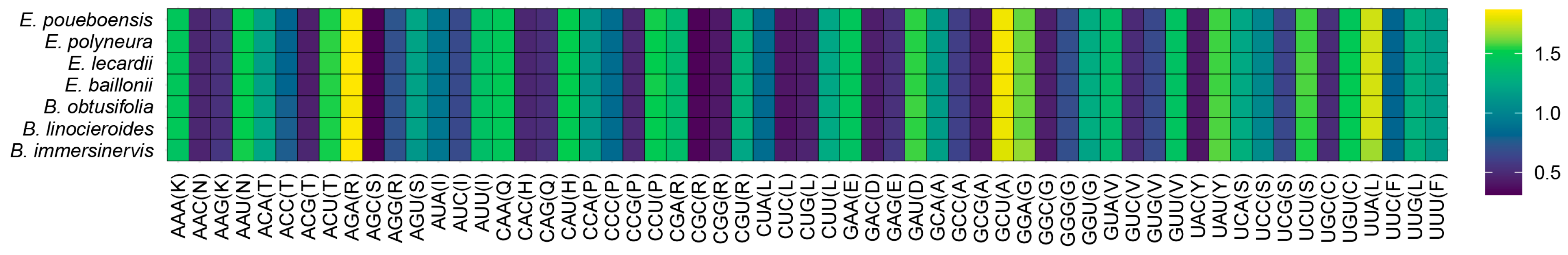
2.7. Codon Usage Bias Analysis
3. Results
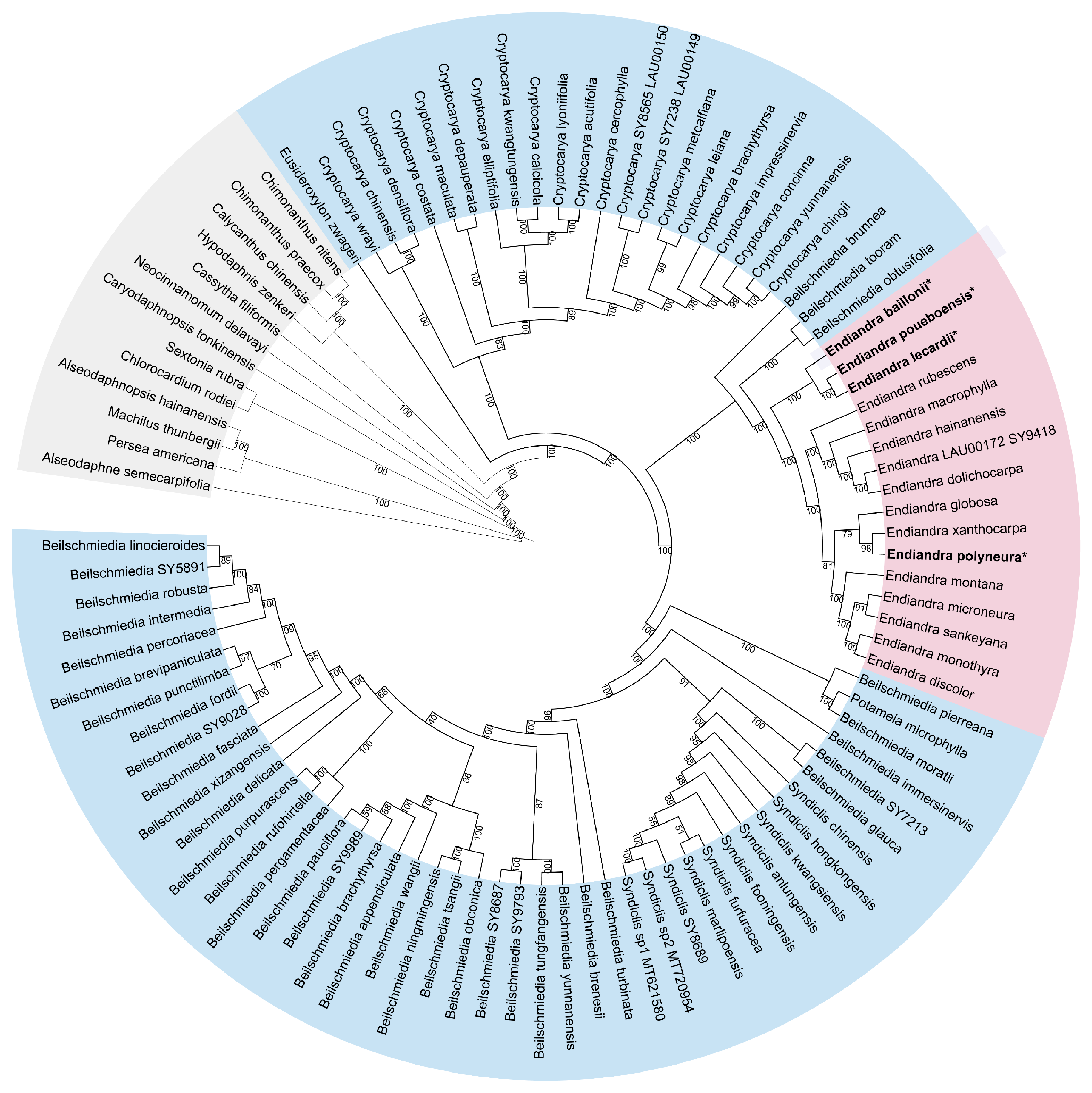
3.1. Phylogenetic Analysis and Diversification Times
3.2. Characteristics of the New Caledonian Endiandra
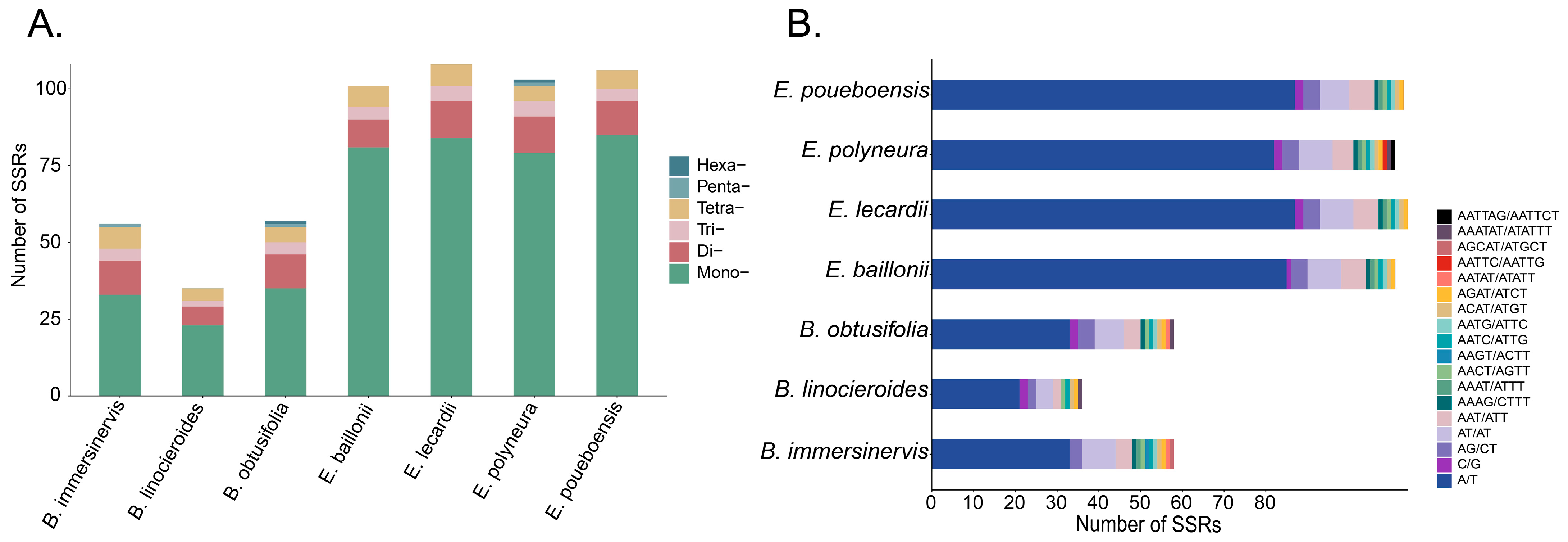
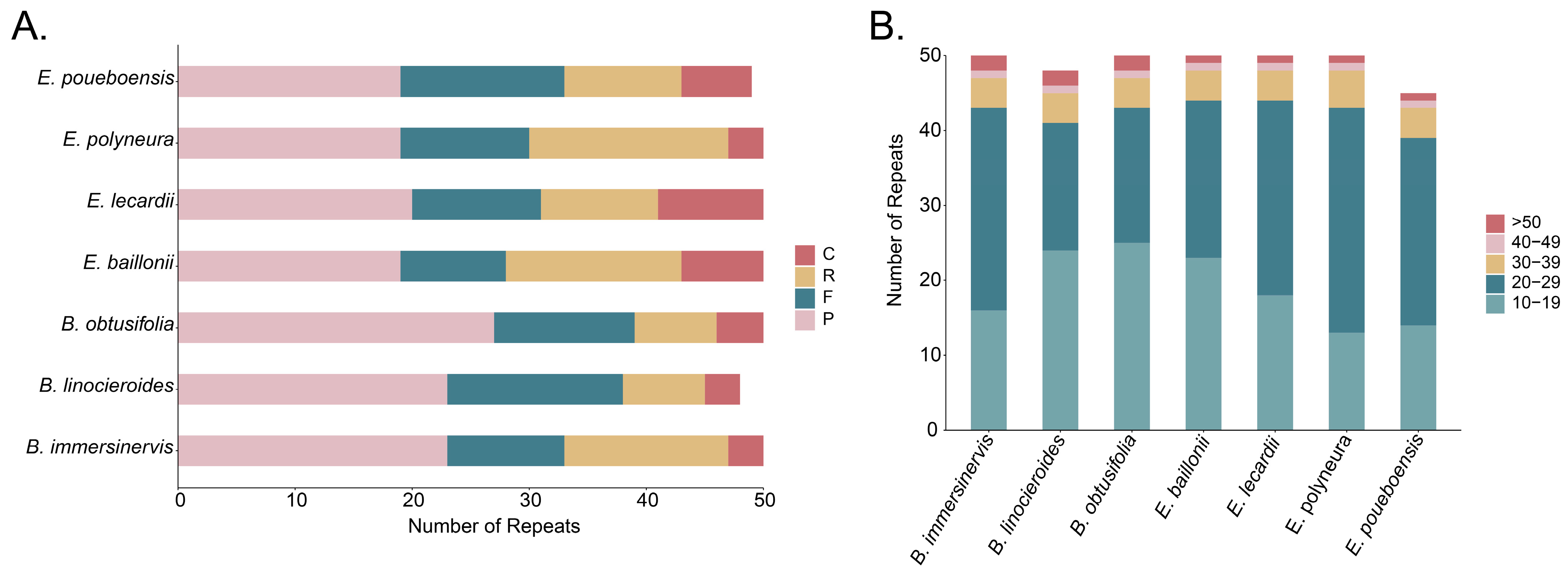
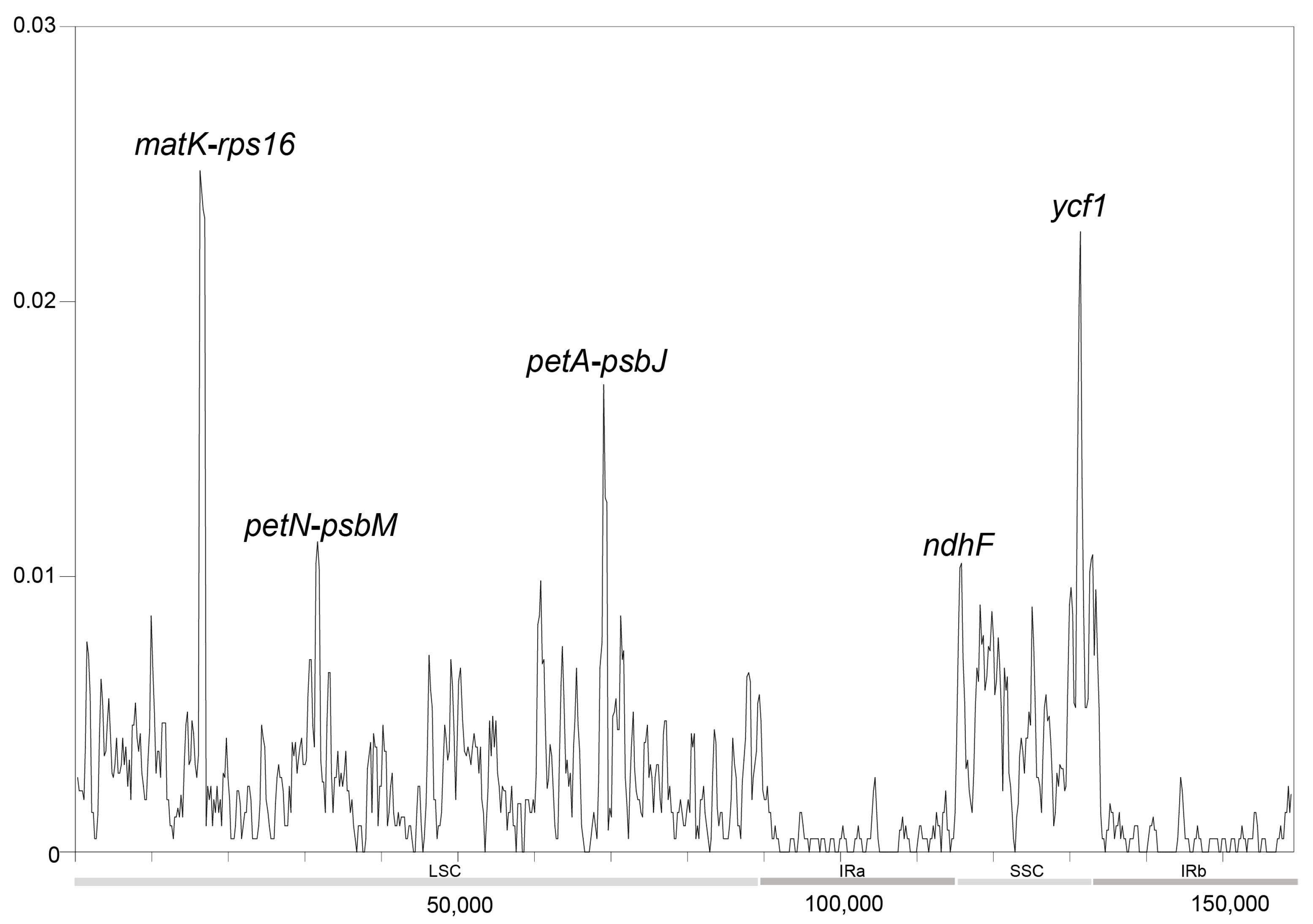
3.3. Analysis of Repeated Sequences
3.4. Analysis of IR Boundaries
3.5. Nucleotide Polymorphism and Codon Usage Bias Analysis
4. Discussion
4.1. Phylogenomics of the Cryptocaryeae and the New Caledonian Species of Endiandra
4.2. Plastome Characteristics of the New Caledonian Species of Endiandra
4.3. The Floristic Origin of Endiandra in New Caledonia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Heads, M. Biogeographical affinities of the New Caledonian biota: A puzzle with 24 pieces. J. Biogeogr. 2010, 37, 1179–1201. [Google Scholar] [CrossRef]
- Pillon, Y. Time and tempo of diversification in the flora of New Caledonia. Bot. J. Linn. Soc. 2012, 170, 288–298. [Google Scholar] [CrossRef]
- Pouteau, R.; Trueba, S.; Field, T.S.; Isnard, S. New Caledonia: A Pleistocene refugium for rain forest lineages of relict angiosperms. J. Biogeogr. 2015, 42, 2062–2077. [Google Scholar] [CrossRef]
- He, T.; Lamont, B.B.; Fogliani, B. Pre-Gondwanan-breakup origin of Beauprea (Proteaceae) explains its historical presence in New Caledonia and New Zealand. Sci. Adv. 2016, 2, 1501648. [Google Scholar] [CrossRef]
- Schellart, W.P.; Lister, G.S.; Toy, V.G. A Late Cretaceous and Cenozoic reconstruction of the Southwest Pacific region: Tectonics controlled by subduction and slab rollback processes. Earth Sci. Rev. 2006, 76, 191–233. [Google Scholar] [CrossRef]
- Rowher, J.G. Toward a phylogenetic classification of the Lauraceae: Evidence from matK sequences. Syst. Bot. 2000, 25, 60–71. [Google Scholar]
- Munzinger, J.; McPherson, G.; Bruy, D. Novitates neocaledonicae XV: Two new species of Endiandra from New Caledonia. Adansonia 2024, 46, 19–28. [Google Scholar] [CrossRef]
- Munzinger, J.; Gemmill, C.E.C. Molecular insights into apecies selimitations of Cryptocarya, Endiandra, and Beilschmiedia (Lauraceae) in New Caledonia and neighbouring islands. Phytotaxa 2025, 681, 233–264. [Google Scholar] [CrossRef]
- Song, Y.; Xia, S.-W.; Tan, Y.-H.; Yu, W.-B.; Yao, X.; Xing, Y.-W.; Corlett, R.T. Phylogeny and biogeography of the Cryptocaryeae (Lauraceae). Taxon 2023, 72, 1244–1261. [Google Scholar] [CrossRef]
- Li, J.-L.; Wang, S.; Yu, J.; Wang, L.; Zhou, S.-L. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 2013, 48, 72–78. [Google Scholar]
- Kates, H.R.; Doby, J.R.; Siniscalchi, C.M.; LaFrance, R.; Soltis, D.E.; Soltis, P.S.; Guralnick, R.P.; Folk, R.A. The effects of herbarium specimen characteristics on short-read NGS sequencing success in nearly 8000 specimens: Old, degraded samples have lower DNA yields but consistent sequencing success. Front. Plant Sci. 2021, 12, 669064. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ferguson, D.K.; Yang, Y. New insights into the plastome evolution of Lauraceae using herbariomics. BMC Plant Biol. 2023, 23, 387. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M. Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.S.; Haeseler, A.V.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2024, 32, 268–274. [Google Scholar] [CrossRef]
- Massoni, J.; Doyle, J.; Sauquet, H. Fossil calibration of Magnoliidae, an ancient lineage of angiosperms. Palaeontol. Electron. 2015, 18, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Madriñán, S.; Li, J. Phylogeny and biogeography of Caryodaphnopsis (Lauraceae) inferred from low-copy nuclear gene and ITS sequences. Taxon 2016, 65, 433–443. [Google Scholar] [CrossRef]
- Eklund, H. Lauraceae flowers from the late cretaceous of North Carolina, U.S.A. Bot. J. Linn. Soc. 2000, 132, 397–428. [Google Scholar] [CrossRef]
- Li, J.-Z.; Qiu, J.; Liao, W.-B.; Jin, J.-H. Eocene fossil Alseodaphne from Hainan Island of China and its paleoclimatic implications. Sci. China Ser. D-Earth Sci. 2009, 52, 1537–1542. [Google Scholar] [CrossRef]
- Puttick, M.N. MCMCtreeR: Functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics 2019, 35, 5321–5322. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Sucnard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Bio. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Xie, J.-M.; Chen, Y.-R.; Cai, G.-J.; Cai, R.-L.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org (accessed on 27 February 2025).
- Li, H.-E.; Guo, Q.-Q.; Xu, L.; Gao, H.-D.; Liu, L.; Zhou, X.-Y. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e14500. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Wall, L.; Christiansen, T.; Orwant, J. Programming Perl, 3rd ed.; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2000. [Google Scholar]
- Li, H.-W.; Liu, B.; Davis, C.C.; Yang, Y. Plastome phylogenomics, systematics, and divergence time estimation of the Beilschmiedia group (Lauraceae). Mol. Phylogenet. Evol. 2020, 151, 106901. [Google Scholar] [CrossRef] [PubMed]
- Proctor, J. Vegetation and soil and plant chemistry on ultramafic rocks in the tropical Far East. Perspect. Plant Ecol. Evol. Syst. 2003, 6, 105–124. [Google Scholar] [CrossRef]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Jin, J.J.; Wang, B.; Yang, J.B.; Corlett, R.T. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J. Syst. Evol. 2020, 58, 423–439. [Google Scholar] [CrossRef]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Yang, J.-P.; Zhang, F.-W.; Ge, Y.-J.; Yu, W.-H.; Xue, Q.-Q.; Wang, M.-T.; Wang, H.-M.; Xue, Q.-Y.; Liu, W.; Niu, Z.-T.; et al. Effects of geographic isolation on the Bulbophyllum chloroplast genomes. BMC Plant Biol. 2022, 22, 201. [Google Scholar] [CrossRef]
- Hu, Q.-D.; Qian, R.-J.; Zhang, Y.-J.; Ma, X.-H.; Ye, Y.-J.; Zhang, X.-L.; Lin, L.; Liu, H.-J.; Zheng, J. Complete chloroplast genome molecular structure, comparative and phylogenetic analyses of Sphaeropteris lepifera of Cyatheaceae family: A tree fern from China. Sci Rep. 2023, 13, 1356. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.-H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef]
- Smith, D.R. Mutation rates in plastid genomes: They are lower than you might think. Genome Biol. Evol. 2015, 7, 1227–1234. [Google Scholar] [CrossRef]
- Wu, C.S.; Chaw, S.M. Evolutionary stasis in cycad plastomes and the first case of plastome gc-biased gene conversion. Genome Biol. Evol. 2015, 7, 2000–2009. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhang, H.-R.; Li, Q.-S.; Cao, Z.-Y.; Song, Y.; Xin, P.-Y. Complete plastid genome sequences of three tropical African Beilschmiediineae trees (Lauraceae: Crytocaryeae). Forests 2024, 15, 832. [Google Scholar] [CrossRef]
- Carpenter, R.J.; Read, J.; Jaffré, T. Reproductive traits of tropical rain-forest trees in New Caledonia. J. Trop. Ecol. 2003, 19, 351–365. [Google Scholar] [CrossRef]
- Grandcolas, P.; Murienne, J.; Robillard, T.; Desutter-Grandcolas, L.; Jourdan, H.; Guilbert, E.; Deharveng, L. New Caledonia: A very old Darwinian island? Phil. Trans. R. Soc. B. 2008, 363, 3309–3317. [Google Scholar] [CrossRef]
- Nattier, R.; Pellens, R.; Robillard, T.; Jourdan, H.; Legendre, F.; Caesar, M.; Nel, A.; Grandcolas, P. Updating the phylogenetic dating of New Caledonian biodiversity with a meta-analysis of the available evidence. Sci. Rep. 2017, 7, 3705. [Google Scholar] [CrossRef]


| Species | Collection | Locality | Herbarium |
|---|---|---|---|
| Endiandra baillonii | Guillaum A & Baumann-Bodenheim MG. 6659 | New Caledonia | A |
| Endiandra lecardii | Lowry II & Prescott P. 6807 | New Caledonia (Province du Nord) | MO |
| Endiandra polyneura | Guillaum A & Baumann-Bodenheim MG. 7213 | New Caledonia | A |
| Endiandra poueboensis | McPherson G. 18988 | New Caledonia (Province du Nord) | MO |
| E. baillonii | E. lecardii | E. polyneura | E. poueboensis | B. immersinervis | B. linocieroides | B. obtusifolia | |
|---|---|---|---|---|---|---|---|
| Size (bp) | 158,313 | 158,261 | 158,339 | 158,259 | 157,497 | 157,311 | 157,254 |
| GC (%) | 39.0 | 39.0 | 39.0 | 39.0 | 39.2 | 39.2 | 39.3 |
| Lsc size in bp | 89,094 | 89,062 | 89,044 | 89,060 | 88,561 | 88,506 | 88,313 |
| Ssc size in bp | 18,213 | 18,203 | 18,213 | 18,199 | 18,026 | 17,964 | 17,998 |
| IRs size in bp | 25,498 | 25,498 | 25,518 | 25,500 | 25,459 | 25,424 | 25,475 |
| Number of coding sequences | 86 | 86 | 86 | 86 | 84 | 84 | 84 |
| Number of rRNA | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Number of tRNA | 37 | 37 | 37 | 37 | 37 | 37 | 37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Shao, C.; Yang, Z.; Yang, Y. Phylogenomics and Floristic Origin of Endiandra R.Br (Lauraceae) from New Caledonia. Forests 2025, 16, 705. https://doi.org/10.3390/f16040705
Song J, Shao C, Yang Z, Yang Y. Phylogenomics and Floristic Origin of Endiandra R.Br (Lauraceae) from New Caledonia. Forests. 2025; 16(4):705. https://doi.org/10.3390/f16040705
Chicago/Turabian StyleSong, Jiayi, Chengyan Shao, Zhi Yang, and Yong Yang. 2025. "Phylogenomics and Floristic Origin of Endiandra R.Br (Lauraceae) from New Caledonia" Forests 16, no. 4: 705. https://doi.org/10.3390/f16040705
APA StyleSong, J., Shao, C., Yang, Z., & Yang, Y. (2025). Phylogenomics and Floristic Origin of Endiandra R.Br (Lauraceae) from New Caledonia. Forests, 16(4), 705. https://doi.org/10.3390/f16040705






