Enhancement of IL-6 Production Induced by SARS-CoV-2 Nucleocapsid Protein and Bangladeshi COVID-19 Patients’ Sera
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Population and Data Collection
2.3. Molecular Testing for SARS-CoV-2 Detection
2.4. Hematological Parameter Detection
2.5. Biochemical Parameter Detection
2.6. K-ML2 Cell Culture and Induction with SARS-CoV-2 N Protein
2.7. IL-6 Measurement from Myeloid Cell Culture and Directly from Untreated Serum
2.8. In-House ELISA Measurement of Anti-N and Anti-N Protein Epitope (Ep9) Antibodies from Heat-Inactivated Serum
3. Data Processing and Analysis
4. Results
4.1. Demographics and Clinical Characteristics
4.2. Vaccination Status and Clinical Outcome
4.3. Laboratory Findings
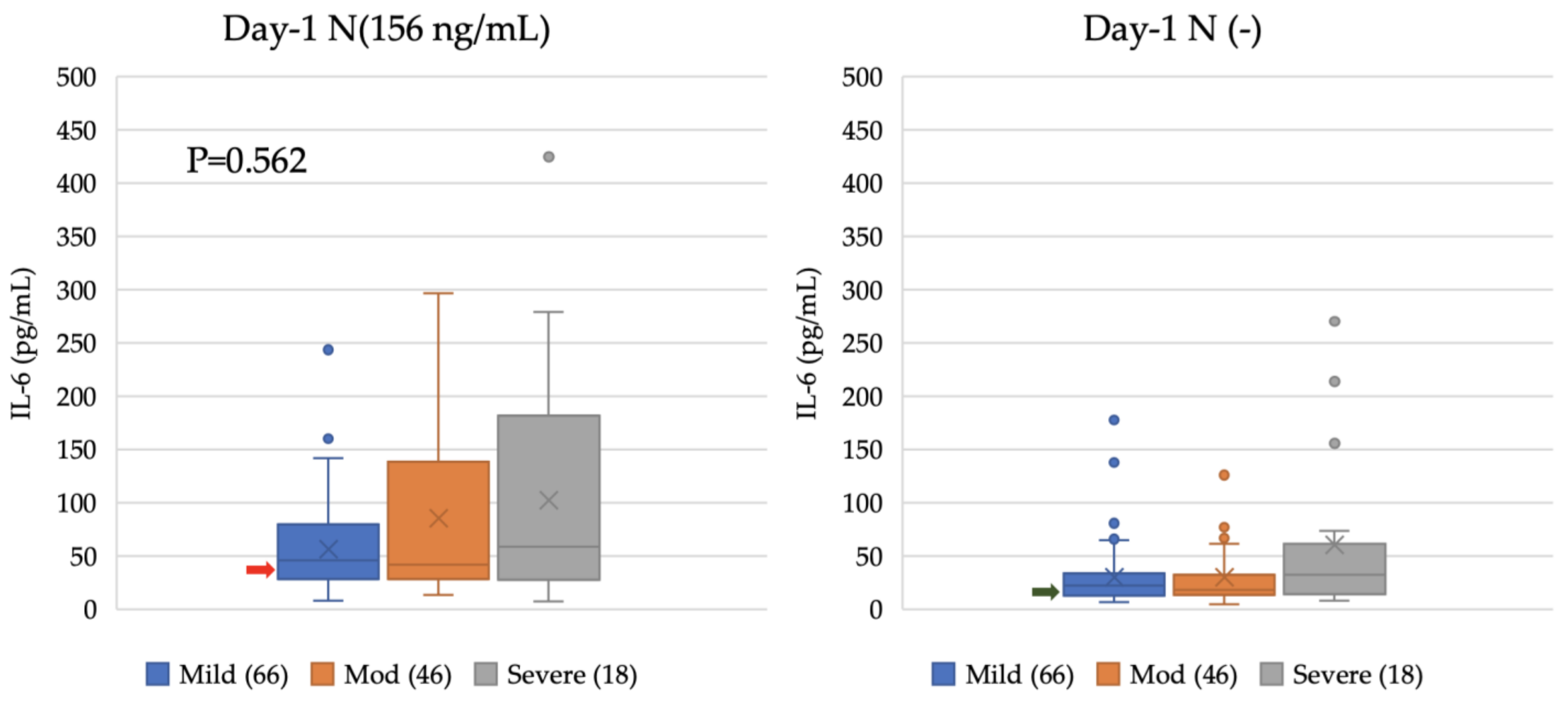
4.4. Enhancement of IL-6 Production from Myeloid Cell Line Induced by COVID-19 Patients’ Sera at Different Time Points
4.5. Longitudinal Analysis of Potency of IL-6 Induction via Patient Samples
4.6. Comparison between Anti-N Antibodies and Induced IL-6 Levels: A Longitudinal Analysis of Anti-N Antibodies
4.7. Lack of Correlation between Epitope Ep9 in N Protein and IL-6-Inducing Capability
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Ciaccio, M.; Agnello, L. Biochemical biomarkers alterations in Coronavirus Disease 2019 (COVID-19). Diagnosis 2020, 7, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Sahin, A.R.; Erdogan, A.; Agaoglu, P.M.; Dineri, Y.; Cakirci, A.Y.; Senel, M.E.; Okyay, R.A.; Tasdogan, A.M. 2019 novel coronavirus (COVID-19) outbreak: A review of the current literature. EJMO 2020, 4, 1–7. [Google Scholar] [CrossRef]
- Satarker, S.; Nampoothiri, M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch. Med. Res. 2020, 51, 482–491. [Google Scholar] [CrossRef]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Zhang, Z.; Nomura, N.; Muramoto, Y.; Ekimoto, T.; Uemura, T.; Liu, K.; Yui, M.; Kono, N.; Aoki, J.; Ikeguchi, M.; et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat. Commun. 2022, 13, 4399. [Google Scholar] [CrossRef]
- McBride, R.; Fielding, B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses 2012, 4, 2902–2923. [Google Scholar] [CrossRef]
- Driggin, E.; Madhavan, M.V.; Bikdeli, B.; Chuich, T.; Laracy, J.; Biondi-Zoccai, G.; Brown, T.S.; Der Nigoghossian, C.; Zidar, D.A.; Haythe, J.; et al. Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems during the COVID-19 Pandemic. J. Am. Coll. Cardiol. 2020, 75, 2352–2371. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef]
- Medetalibeyoglu, A.; Catma, Y.; Senkal, N.; Ormeci, A.; Cavus, B.; Kose, M.; Bayramlar, O.F.; Yildiz, G.; Akyuz, F.; Kaymakoglu, S.; et al. The effect of liver test abnormalities on the prognosis of COVID-19. Ann. Hepatol. 2020, 19, 614–621. [Google Scholar] [CrossRef]
- Adapa, S.; Chenna, A.; Balla, M.; Merugu, G.P.; Koduri, N.M.; Daggubati, S.R.; Gayam, V.; Naramala, S.; Konala, V.M. COVID-19 Pandemic Causing Acute Kidney Injury and Impact on Patients with Chronic Kidney Disease and Renal Transplantation. J. Clin. Med. Res. 2020, 12, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Ellul, M.A.; Benjamin, L.; Singh, B.; Lant, S.; Michael, B.D.; Easton, A.; Kneen, R.; Defres, S.; Sejvar, J.; Solomon, T. Neurological associations of COVID-19. Lancet Neurol. 2020, 19, 767–783. [Google Scholar] [CrossRef] [PubMed]
- Broxmeyer, H.E.; Parker, G.C. Impact of COVID-19 and Future Emerging Viruses on Hematopoietic Cell Transplantation and Other Cellular Therapies. Stem. Cells Dev. 2020, 29, 625–626. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Schultze, J.L.; Aschenbrenner, A.C. COVID-19 and the human innate immune system. Cell 2021, 184, 1671–1692. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Renia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Burke, H.; Freeman, A.; Cellura, D.C.; Stuart, B.L.; Brendish, N.J.; Poole, S.; Borca, F.; Phan, H.T.T.; Sheard, N.; Williams, S.; et al. Inflammatory phenotyping predicts clinical outcome in COVID-19. Respir. Res. 2020, 21, 245. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Manik, M.; Singh, R.K. Role of toll-like receptors in modulation of cytokine storm signaling in SARS-CoV-2-induced COVID-19. J. Med. Virol. 2022, 94, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Lin, Y.X.; Zha, Y.; Sun, Y.; Tian, J.; Yang, Z.; Lin, S.W.; Yu, F.; Chen, Z.S.; Kuang, B.H.; et al. A Low-Producing Haplotype of Interleukin-6 Disrupting CTCF Binding Is Protective against Severe COVID-19. mBio 2021, 12, e0137221. [Google Scholar] [CrossRef] [PubMed]
- Elahi, R.; Karami, P.; Heidary, A.H.; Esmaeilzadeh, A. An updated overview of recent advances, challenges, and clinical considerations of IL-6 signaling blockade in severe coronavirus disease 2019 (COVID-19). Int. Immunopharmacol. 2022, 105, 108536. [Google Scholar] [CrossRef] [PubMed]
- Ashrafzadeh-Kian, S.; Campbell, M.R.; Jara Aguirre, J.C.; Walsh, J.; Kumanovics, A.; Jenkinson, G.; Rinaldo, P.; Snyder, M.R.; Algeciras-Schimnich, A. Role of immune mediators in predicting hospitalization of SARS-CoV-2 positive patients. Cytokine 2022, 150, 155790. [Google Scholar] [CrossRef] [PubMed]
- Jose, R.J.; Manuel, A. COVID-19 cytokine storm: The interplay between inflammation and coagulation. Lancet Respir. Med. 2020, 8, e46–e47. [Google Scholar] [CrossRef]
- Darif, D.; Hammi, I.; Kihel, A.; El Idrissi Saik, I.; Guessous, F.; Akarid, K. The pro-inflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021, 153, 104799. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Hirano, T. Interleukin 6 in autoimmune and inflammatory diseases: A personal memoir. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 717–730. [Google Scholar] [CrossRef]
- Testing.com. Interleukin-6: OneCare Media, 2023 [updated Nov 9, 2021]. Available online: https://www.testing.com/tests/interleukin-6 (accessed on 15 April 2023).
- Ulhaq, Z.S.; Soraya, G.V. Interleukin-6 as a potential biomarker of COVID-19 progression. Med. Mal. Infect 2020, 50, 382–383. [Google Scholar] [CrossRef]
- Cifaldi, L.; Prencipe, G.; Caiello, I.; Bracaglia, C.; Locatelli, F.; De Benedetti, F.; Strippoli, R. Inhibition of natural killer cell cytotoxicity by interleukin-6: Implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol. 2015, 67, 3037–3046. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Yu, H.; Chen, H.; Qi, W.; Chen, L.; Chen, G.; Yan, W.; Chen, T.; Ning, Q.; Han, M.; et al. Longitudinal changes of inflammatory parameters and their correlation with disease severity and outcomes in patients with COVID-19 from Wuhan, China. Crit. Care 2020, 24, 525. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Xu, X.; Wei, H. Why tocilizumab could be an effective treatment for severe COVID-19? J. Transl. Med. 2020, 18, 164. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lu, J.; Tang, Y.; Dai, Y.; Zhou, J.; Wu, Y. Tocilizumab for treating COVID-19: A systemic review and meta-analysis of retrospective studies. Eur. J. Clin. Pharmacol. 2021, 77, 311–319. [Google Scholar] [CrossRef]
- Michot, J.M.; Albiges, L.; Chaput, N.; Saada, V.; Pommeret, F.; Griscelli, F.; Balleyguier, C.; Besse, B.; Marabelle, A.; Netzer, F.; et al. Tocilizumab, an anti-IL-6 receptor antibody, to treat COVID-19-related respiratory failure: A case report. Ann. Oncol. 2020, 31, 961–964. [Google Scholar] [CrossRef]
- Galvan-Roman, J.M.; Rodriguez-Garcia, S.C.; Roy-Vallejo, E.; Marcos-Jimenez, A.; Sanchez-Alonso, S.; Fernandez-Diaz, C.; Alcaraz-Serna, A.; Mateu-Albero, T.; Rodriguez-Cortes, P.; Sanchez-Cerrillo, I.; et al. IL-6 serum levels predict severity and response to tocilizumab in COVID-19: An observational study. J. Allergy Clin. Immunol. 2021, 147, 72–80.e8. [Google Scholar] [CrossRef]
- Shimizu, J.; Sasaki, T.; Yamanaka, A.; Ichihara, Y.; Koketsu, R.; Samune, Y.; Cruz, P.; Sato, K.; Tanga, N.; Yoshimura, Y.; et al. The potential of COVID-19 patients’ sera to cause antibody-dependent enhancement of infection and IL-6 production. Sci. Rep. 2021, 11, 23713. [Google Scholar] [CrossRef]
- Karwaciak, I.; Salkowska, A.; Karas, K.; Dastych, J.; Ratajewski, M. Nucleocapsid and Spike Proteins of the Coronavirus SARS-CoV-2 Induce IL6 in Monocytes and Macrophages-Potential Implications for Cytokine Storm Syndrome. Vaccines 2021, 9, 54. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, K.; Wang, D.; Yue, X.; Song, D.; Zhu, Y.; Wu, J. Nucleocapsid protein of SARS-CoV activates interleukin-6 expression through cellular transcription factor NF-kappaB. Virology 2007, 365, 324–335. [Google Scholar] [CrossRef]
- Pan, P.; Shen, M.; Yu, Z.; Ge, W.; Chen, K.; Tian, M.; Xiao, F.; Wang, Z.; Wang, J.; Jia, Y.; et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021, 12, 4664. [Google Scholar] [CrossRef]
- Nakayama, E.E.; Kubota-Koketsu, R.; Sasaki, T.; Suzuki, K.; Uno, K.; Shimizu, J.; Okamoto, T.; Matsumoto, H.; Matsuura, H.; Hashimoto, S.; et al. Anti-nucleocapsid antibodies enhance the production of IL-6 induced by SARS-CoV-2 N protein. Sci. Rep. 2022, 12, 8108. [Google Scholar] [CrossRef]
- Sen, S.R.; Sanders, E.C.; Gabriel, K.N.; Miller, B.M.; Isoda, H.M.; Salcedo, G.S.; Garrido, J.E.; Dyer, R.P.; Nakajima, R.; Jain, A.; et al. Predicting COVID-19 Severity with a Specific Nucleocapsid Antibody plus Disease Risk Factor Score. mSphere 2021, 6, 00203–00221. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, G.; Greco, M.; Zanella, A.; Albano, G.; Antonelli, M.; Bellani, G.; Bonanomi, E.; Cabrini, L.; Carlesso, E.; Castelli, G.; et al. Risk Factors Associated with Mortality among Patients with COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020, 180, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Bwire, G.M. Coronavirus: Why Men are More Vulnerable to COVID-19 Than Women? SN Compr. Clin. Med. 2020, 2, 874–876. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Bai, H.; Chen, X.; Gong, J.; Li, D.; Sun, Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020, 18, 1094–1099. [Google Scholar] [CrossRef]
- Lippi, G.; Plebani, M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin. Chem. Lab. Med. 2020, 58, 1063–1069. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated with Acute Respiratory Distress Syndrome and Death in Patients with Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef]
- Liu, Y.; Du, X.; Chen, J.; Jin, Y.; Peng, L.; Wang, H.H.X.; Luo, M.; Chen, L.; Zhao, Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020, 81, e6–e12. [Google Scholar] [CrossRef]
- Toori, K.U.; Qureshi, M.A.; Chaudhry, A.; Safdar, M.F. Neutrophil to lymphocyte ratio (NLR) in COVID-19: A cheap prognostic marker in a resource constraint setting. Pak. J. Med. Sci. 2021, 37, 1435–1439. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.; Mao, Z.; Xiao, M.; Wang, L.; Qi, S.; Zhou, F. Predictive values of neutrophil-to-lymphocyte ratio on disease severity and mortality in COVID-19 patients: A systematic review and meta-analysis. Crit. Care 2020, 24, 647. [Google Scholar] [CrossRef]
- Yang, A.P.; Liu, J.P.; Tao, W.Q.; Li, H.M. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020, 84, 106504. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef] [PubMed]
- El Hajj, S.; Canabady-Rochelle, L.; Gaucher, C. Nature-Inspired Bioactive Compounds: A Promising Approach for Ferroptosis-Linked Human Diseases? Molecules 2023, 28, 2636. [Google Scholar] [CrossRef] [PubMed]
- Hirschhorn, T.; Stockwell, B.R. The development of the concept of ferroptosis. Free Radic. Biol. Med. 2019, 133, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Cavezzi, A.; Troiani, E.; Corrao, S. COVID-19: Hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020, 10, 1271. [Google Scholar] [CrossRef]
- Hasan, A.; Rahim, R.; Rahman, M. Alteration of biomarkers of expired and cured COVID-19 ICU patients in a tertiary care hospital. Bioresearch Commun. 2021, 7, 1031–1037. [Google Scholar] [CrossRef]
- Batra, M.; Tian, R.; Zhang, C.; Clarence, E.; Sacher, C.S.; Miranda, J.N.; De La Fuente, J.R.O.; Mathew, M.; Green, D.; Patel, S.; et al. Role of IgG against N-protein of SARS-CoV2 in COVID19 clinical outcomes. Sci. Rep. 2021, 11, 3455. [Google Scholar] [CrossRef]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef]
- Li, K.; Huang, B.; Wu, M.; Zhong, A.; Li, L.; Cai, Y.; Wang, Z.; Wu, L.; Zhu, M.; Li, J.; et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat. Commun. 2020, 11, 6044. [Google Scholar] [CrossRef]
- Hashem, A.M.; Algaissi, A.; Almahboub, S.A.; Alfaleh, M.A.; Abujamel, T.S.; Alamri, S.S.; Alluhaybi, K.A.; Hobani, H.I.; AlHarbi, R.H.; Alsulaiman, R.M.; et al. Early Humoral Response Correlates with Disease Severity and Outcomes in COVID-19 Patients. Viruses 2020, 12, 1390. [Google Scholar] [CrossRef]
- Santa Cruz, A.; Mendes-Frias, A.; Oliveira, A.I.; Dias, L.; Matos, A.R.; Carvalho, A.; Capela, C.; Pedrosa, J.; Castro, A.G.; Silvestre, R. Interleukin-6 Is a Biomarker for the Development of Fatal Severe Acute Respiratory Syndrome Coronavirus 2 Pneumonia. Front. Immunol. 2021, 12, 613422. [Google Scholar] [CrossRef]
- Avila-Nava, A.; Cortes-Telles, A.; Torres-Erazo, D.; Lopez-Romero, S.; Chim Ake, R.; Gutierrez Solis, A.L. Serum IL-6: A potential biomarker of mortality among SARS-CoV-2 infected patients in Mexico. Cytokine 2021, 143, 155543. [Google Scholar] [CrossRef]
- Lee, J.H.; Jang, J.H.; Park, J.H.; Jang, H.J.; Park, C.S.; Lee, S.; Kim, S.H.; Kim, J.Y.; Kim, H.K. The role of interleukin-6 as a prognostic biomarker for predicting acute exacerbation in interstitial lung diseases. PLoS ONE 2021, 16, e0255365. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.T.; Garcia-Carreras, B.; Hitchings, M.D.T.; Yang, B.; Katzelnick, L.C.; Rattigan, S.M.; Borgert, B.A.; Moreno, C.A.; Solomon, B.D.; Trimmer-Smith, L.; et al. A systematic review of antibody mediated immunity to coronaviruses: Kinetics, correlates of protection, and association with severity. Nat. Commun. 2020, 11, 4704. [Google Scholar] [CrossRef] [PubMed]
- Gerhards, C.; Thiaucourt, M.; Kittel, M.; Becker, C.; Ast, V.; Hetjens, M.; Neumaier, M.; Haselmann, V. Longitudinal assessment of anti-SARS-CoV-2 antibody dynamics and clinical features following convalescence from a COVID-19 infection. Int. J. Infect. Dis. 2021, 107, 221–227. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, L.M.; Wan, L.; Xiang, T.X.; Le, A.; Liu, J.M.; Peiris, M.; Poon, L.L.M.; Zhang, W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020, 20, 656–657. [Google Scholar] [CrossRef]
- Manjili, R.H.; Zarei, M.; Habibi, M.; Manjili, M.H. COVID-19 as an Acute Inflammatory Disease. J. Immunol. 2020, 205, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Zanza, C.; Romenskaya, T.; Manetti, A.C.; Franceschi, F.; La Russa, R.; Bertozzi, G.; Maiese, A.; Savioli, G.; Volonnino, G.; Longhitano, Y. Cytokine Storm in COVID-19: Immunopathogenesis and Therapy. Medicina 2022, 58, 144. [Google Scholar] [CrossRef]
- Group, A.-T.S.; Rogers, A.J.; Wentworth, D.; Phillips, A.; Shaw-Saliba, K.; Dewar, R.L.; Aggarwal, N.R.; Babiker, A.G.; Chang, W.; Dharan, N.J.; et al. The Association of Baseline Plasma SARS-CoV-2 Nucleocapsid Antigen Level and Outcomes in Patients Hospitalized with COVID-19. Ann. Intern. Med. 2022, 175, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Bowe, B.; Xie, Y.; Al-Aly, Z. Acute and postacute sequelae associated with SARS-CoV-2 reinfection. Nat. Med. 2022, 28, 2398–2405. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Chen, X.; Xu, Y.; Wang, P.; Wang, J.; Yu, L.; et al. Antigenicity and infectivity characterization of SARS-CoV-2 BA.2.86. bioRxiv 2023. [Google Scholar] [CrossRef]
- Nakayama, E.E.; Shioda, T. SARS-CoV-2 Related Antibody-Dependent Enhancement Phenomena In Vitro and In Vivo. Microorganisms 2023, 11, 1015. [Google Scholar] [CrossRef]
- Huang, W.; Li, M.; Luo, G.; Wu, X.; Su, B.; Zhao, L.; Zhang, S.; Chen, X.; Jia, M.; Zhu, J.; et al. The Inflammatory Factors Associated with Disease Severity to Predict COVID-19 Progression. J. Immunol. 2021, 206, 1597–1608. [Google Scholar] [CrossRef]
- Munipalli, B.; Seim, L.; Dawson, N.L.; Knight, D.; Dabrh, A.M.A. Post-acute sequelae of COVID-19 (PASC): A meta-narrative review of pathophysiology, prevalence, and management. SN Compr. Clin. Med. 2022, 4, 90. [Google Scholar] [CrossRef] [PubMed]
- Lam, I.C.H.; Wong, C.K.H.; Zhang, R.; Chui, C.S.L.; Lai, F.T.T.; Li, X.; Chan, E.W.Y.; Luo, H.; Zhang, Q.; Man, K.K.C.; et al. Long-term post-acute sequelae of COVID-19 infection: A retrospective, multi-database cohort study in Hong Kong and the UK. EClinicalMedicine 2023, 60, 102000. [Google Scholar] [CrossRef] [PubMed]
- Thaweethai, T.; Jolley, S.E.; Karlson, E.W.; Levitan, E.B.; Levy, B.; McComsey, G.A.; McCorkell, L.; Nadkarni, G.N.; Parthasarathy, S.; Singh, U.; et al. Development of a Definition of Postacute Sequelae of SARS-CoV-2 Infection. JAMA 2023, 329, 1934–1946. [Google Scholar] [CrossRef]
- Woodruff, M.C.; Bonham, K.S.; Anam, F.A.; Walker, T.A.; Faliti, C.E.; Ishii, Y.; Kaminski, C.Y.; Ruunstrom, M.C.; Cooper, K.R.; Truong, A.D.; et al. Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID. Nat. Commun. 2023, 14, 4201. [Google Scholar] [CrossRef]
- Orban, Z.S.; Visvabharathy, L.; Perez Giraldo, G.S.; Jimenez, M.; Koralnik, I.J. SARS-CoV-2–Specific Immune Responses in Patients with Postviral Syndrome After Suspected COVID-19. Neurol. Neuroimmunol. Neuroinflamm. 2023, 10, e200159. [Google Scholar] [CrossRef]
- Tsuji, S.; Minami, S.; Hashimoto, R.; Konishi, Y.; Suzuki, T.; Kondo, T.; Sasai, M.; Torii, S.; Ono, C.; Shichinohe, S.; et al. SARS-CoV-2 infection triggers paracrine senescence and leads to a sustained senescence-associated inflammatory response. Nat. Aging 2022, 2, 115–124. [Google Scholar] [CrossRef]



| Characteristics | Mild, % | Moderate, % | Severe, % | Total, % | p-Value | |
|---|---|---|---|---|---|---|
| (n = 66) | (n = 46) | (n = 18) | (n = 130) | |||
| Age—median year (IQR) | 43 (22–58) | 68 (51–75) | 76 (61–81) | 57 (31–72) | <0.001 | |
| Male | 45 (68.2) | 22 (47.8) | 11 (61.1) | 78 (60) | 0.182 | |
| Baseline symptoms | ||||||
| Fever | 60 (90.9) | 38 (82.6) | 18 (100) | 116 (89.2) | 0.106 | |
| Cough | 38 (57.6) | 30 (65.2) | 15 (83.3) | 83 (63.8) | 0.126 | |
| Runny nose | 13 (19.7) | 7 (15.2) | 1 (5.6) | 21 (16.2) | 0.367 | |
| Shortness of breath | 0 | 15 (32.6) | 15 (83.3) | 30 (23.1) | <0.001 | |
| Nausea or vomiting | 13 (19.7) | 9 (19.6) | 1 (5.6) | 23 (17.7) | 0.374 | |
| Diarrhea | 2 (3) | 2 (4.3) | 0 | 4 (3.1) | 0.844 | |
| Fatigue | 18 (27.3) | 17 (37) | 4 (22.2) | 39 (30) | 0.419 | |
| Headache | 14 (21.2) | 6 (13) | 1 (5.6) | 21 (16.2) | 0.218 | |
| Chest pain | 4 (6.1) | 8 (17.4) | 1 (5.6) | 13 (10) | 0.119 | |
| Abdominal pain | 1 (1.5) | 1 (2.2) | 0 | 2 (1.5) | 1 | |
| Myalgia or arthralgia | 9 (13.6) | 6 (13) | 3 (16.7) | 18 (13.8) | 1 | |
| Treatment | ||||||
| Antiviral treatment | ||||||
| Remdesivir | 3 (4.5) | 22 (47.8) | 13 (72.2) | 38 (29.2) | <0.001 | |
| Molnupiravir | 9 (13.6) | 4 (8.7) | 0 | 13 (10) | 0.221 | |
| Paxlovid | 4 (6.1) | 2 (4.3) | 0 | 6 (4.6) | 0.659 | |
| Acyclovir | 1 (1.5) | 1 (2.2) | 0 | 2 (1.5) | 1 | |
| Antibiotic treatment | 31 (47) | 39 (84.8) | 18 (100) | 88 (67.7) | <0.001 | |
| No antiviral/antibiotic treatment | 26 (39.4) | 0 | 0 | 26 (20) | <0.001 | |
| Convalescent plasma therapy | 0 | 0 | 3 (16.7) | 3 (2.3) | 0.002 | |
| Oxygen support | 0 | 17 (37) | 18 (100) | 35 (26.9) | <0.001 | |
| High-flow nasal cannula | 0 | 0 | 15 (83.3) | 15 (11.5) | <0.001 | |
| Mechanical ventilation | 0 | 0 | 3 (16.7) | 3 (2.3) | 0.002 | |
| Vaccinated | 49 (74.2) | 45 (97.8) | 18 (100) | 112 (86.2) | <0.001 | |
| Clinical outcome | ||||||
| Outpatients | 35 (53) | 4 (8.7) | 0 | 39 (30) | <0.001 | |
| Inpatients | 31 (47) | 42 (91.3) | 18 (100) | 91 (70) | <0.001 | |
| Hospital stays (days)—median (IQR) | 2 (2–3) | 5 (2–8) | 10 (6–18) | 4 (2–8) | <0.001 | |
| Death | 0 | 0 | 11 (61.1) | 11 (8.5) | <0.001 | |
| Laboratory Parameters | Mild | Moderate | Severe | p-Value | |||
|---|---|---|---|---|---|---|---|
| Blood Routine Biomarkers | Median (IQR) | N | Median (IQR) | N | Median (IQR) | N | |
| White blood cells, ×109/L | 6.68 (5.26–8.47) | 66 | 8.28 (6.40–9.99) | 46 | 11.53 (8.47–16.86) | 18 | <0.001 |
| Neutrophils, ×109/L | 4.44 (3.32–5.70) | 66 | 6.28 (4.53–8.09) | 46 | 10.45 (7.41–15.32) | 18 | <0.001 |
| Lymphocytes, ×109/L | 1.69 (1.21–2.25) | 66 | 1.35 (1.01–1.80) | 46 | 1.11 (0.86–1.67) | 18 | 0.04 |
| Hemoglobin, g/dL | 12.95 (12–13.90) | 66 | 11.5 (10.05–12.58) | 46 | 10.2 (9.65–11.3) | 18 | <0.001 |
| Platelet, ×109/L | 220 (177.75–284.50) | 66 | 191 (160–249.50) | 46 | 205.5 (156.25–256.75) | 18 | 0.185 |
| Hematocrit, % | 38.7 (35.85–41.4) | 66 | 33.15 (29.08–38.6) | 46 | 28.85 (26.7–30.75) | 18 | <0.001 |
| Neutrophil-to-lymphocyte ratio (NLR) | 2.66 (1.84–4.03) | 66 | 4.67 (2.92–6.42) | 46 | 7.47 (6.14–15.62) | 18 | <0.001 |
| Inflammatory biomarkers | |||||||
| C-reactive protein, mg/dL | 0.68 (0.33–1.22) | 66 | 3.88 (1.79–5.52) | 46 | 13.25 (6.53–19.50) | 18 | <0.001 |
| Ferritin, µg/L | 97 (60–119.25) | 12 | 342.50 (143.25–702) | 16 | 961 (271–3,037) | 13 | <0.001 |
| Procalcitonin, ng/mL | 0.055 (0.05–0.07) | 10 | 0.15 (0.08–6.82) | 23 | 3.39 (1.44–8.13) | 17 | 0.001 |
| Interleukin-6, pg/mL | 2.87 (1.71–12.95) | 66 | 10.74 (3.87–19.98) | 45 | 108.65 (42.17–231.62) | 18 | <0.001 |
| Biochemistry biomarkers | |||||||
| Creatinine, mg/dL | 0.88 (0.79–1.09) | 53 | 1.25 (1.03–1.95) | 43 | 2.15 (1.50–2.65) | 17 | <0.001 |
| ALT/SGPT, U/L | 27 (22–36) | 37 | 34.5 (20.75–49.25) | 40 | 35 (24–41) | 17 | 0.346 |
| AST/SGOT, U/L | 25 (22.50–37) | 23 | 40 (24.50–53.50) | 35 | 54.5 (38.50–96.25) | 16 | 0.002 |
| Albumin, g/dL | 3.60 (3.35–4) | 19 | 3.10 (2.75–3.30) | 31 | 2.60 (2.40–2.60) | 15 | <0.001 |
| Coagulation biomarker | |||||||
| D-dimer, ng/mL | 317.50 (231.25–492.50) | 34 | 930 (374.5–1708.5) | 40 | 3929.5 (1960–12,858.25) | 18 | <0.001 |
| Cardiac biomarkers | |||||||
| High-sensitivity troponin I, ng/L | 7.70 (5.55–32.68) | 8 | 32.50 (8.95–112.48) | 26 | 113 (37–594) | 17 | 0.01 |
| NT-proBNP, pg/mL | 13 (11.50–2165) | 3 | 426 (118–4776) | 21 | 8898 (972–30,000) | 17 | 0.005 |
| IL-6 | Mild Cases (%) | Moderate Cases (%) | Severe Cases (%) |
|---|---|---|---|
| >126 pg/mL | 4 (6.1) | 13 (28.3) | 5 (27.8) |
| <126 pg/mL | 62 (93.9) | 33 (71.7) | 13 (72.2) |
| Total | 66 (100) | 46 (100) | 18 (100) |
| Serum IL-6 | ||||
|---|---|---|---|---|
| IL-6 Induction | <7 pg/mL | >7 pg/mL | ||
| Category | with N Protein | Cases (%) | Cases (%) | Cases |
| Mild | <126 pg/mL | 42 (74) | 12 (21) | 57 |
| >126 pg/mL | 3 (5) | 0 (0) | ||
| Moderate | <126 pg/mL | 17 (40) | 15 (35) | 43 |
| >126 pg/mL | 4 (9) | 7 (16) | ||
| Severe | <126 pg/mL | 1 (6) | 11 (65) | 17 |
| >126 pg/mL | 1 (6) | 4 (24) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, A.; Rahim, R.; Nakayama, E.E.; Uno, K.; Hasan, N.; Rahman, M.; Shioda, T. Enhancement of IL-6 Production Induced by SARS-CoV-2 Nucleocapsid Protein and Bangladeshi COVID-19 Patients’ Sera. Viruses 2023, 15, 2018. https://doi.org/10.3390/v15102018
Hasan A, Rahim R, Nakayama EE, Uno K, Hasan N, Rahman M, Shioda T. Enhancement of IL-6 Production Induced by SARS-CoV-2 Nucleocapsid Protein and Bangladeshi COVID-19 Patients’ Sera. Viruses. 2023; 15(10):2018. https://doi.org/10.3390/v15102018
Chicago/Turabian StyleHasan, Abu, Rummana Rahim, Emi E. Nakayama, Kazuko Uno, Nazmul Hasan, Mizanur Rahman, and Tatsuo Shioda. 2023. "Enhancement of IL-6 Production Induced by SARS-CoV-2 Nucleocapsid Protein and Bangladeshi COVID-19 Patients’ Sera" Viruses 15, no. 10: 2018. https://doi.org/10.3390/v15102018





