Structural Characterization of Canine Minute Virus, Rat and Porcine Bocavirus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus-like Production and Purification
2.2. Cryo-EM Data Collection
2.3. Three-Dimensional Particle Reconstruction
2.4. Model Building and Structure Refinement
2.5. Structural Comparison
3. Results and Discussion
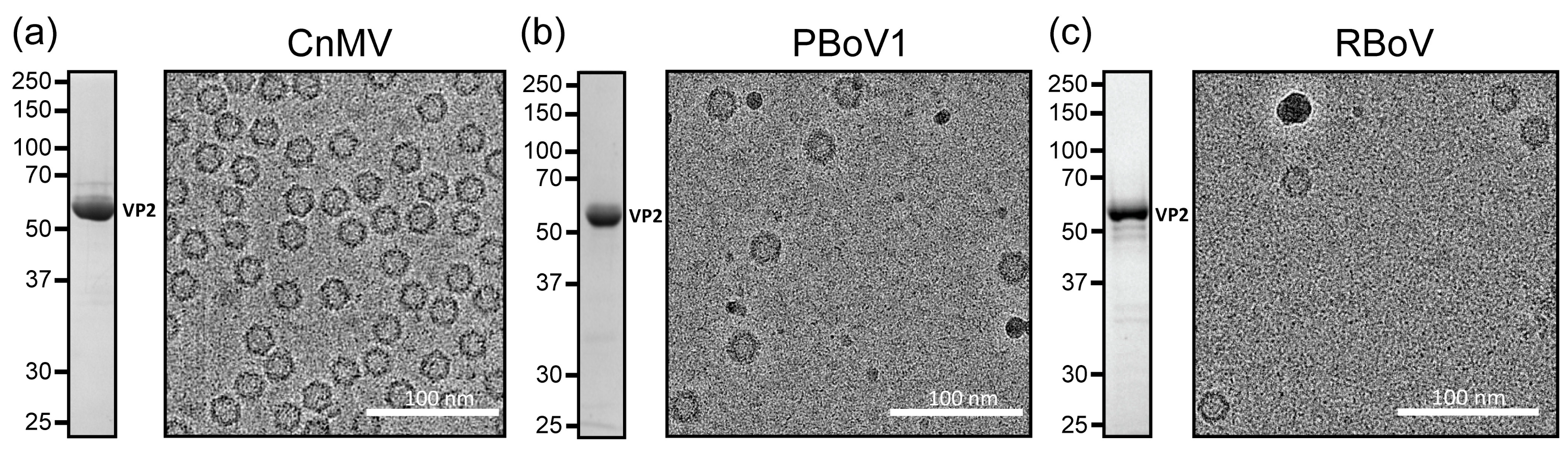
3.1. Expression and Purification of VLPs of CnMV, PBoV, and RBoV
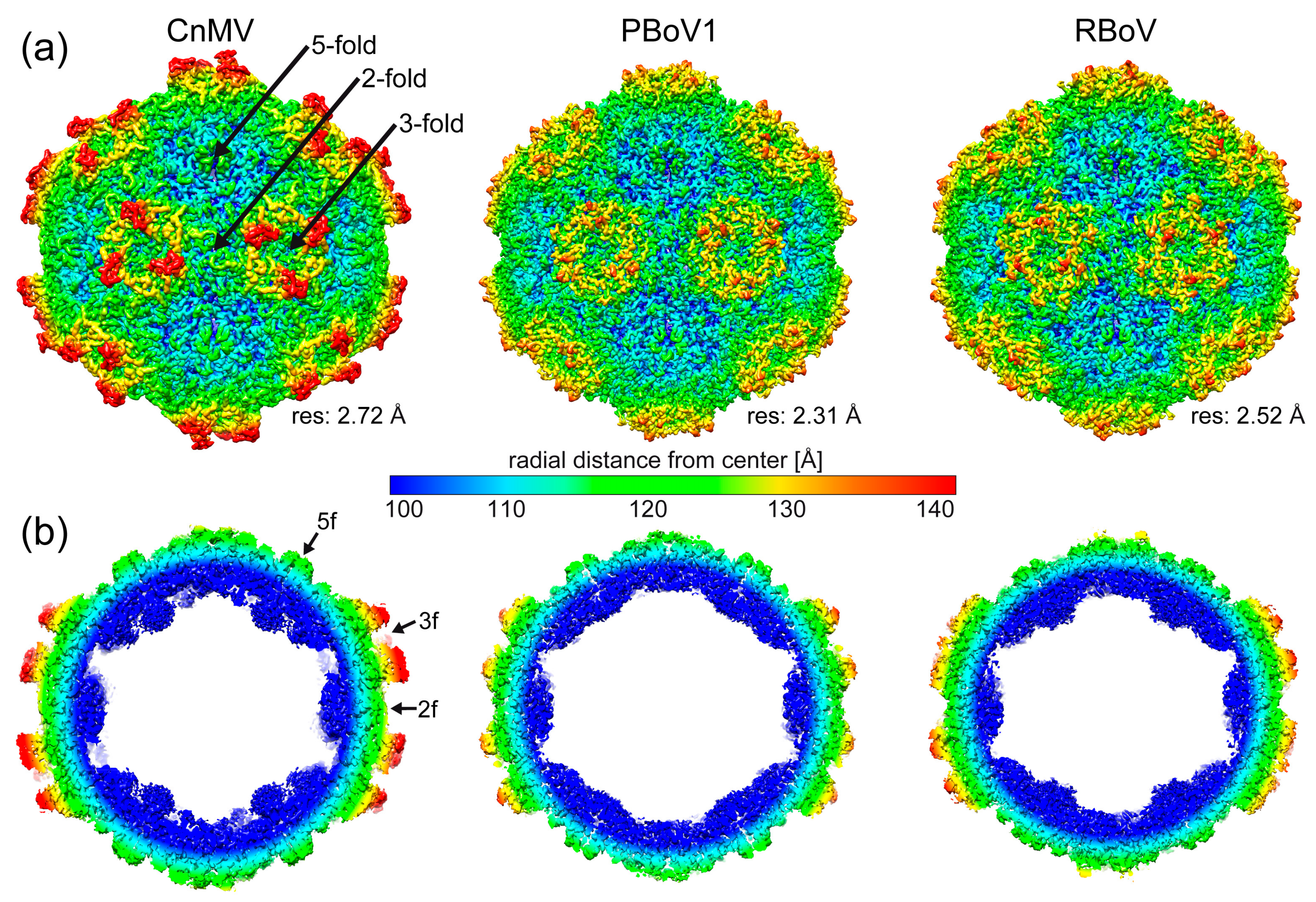
3.2. Determination of the CnMV, PBoV1, and RBoV Capsid Structures
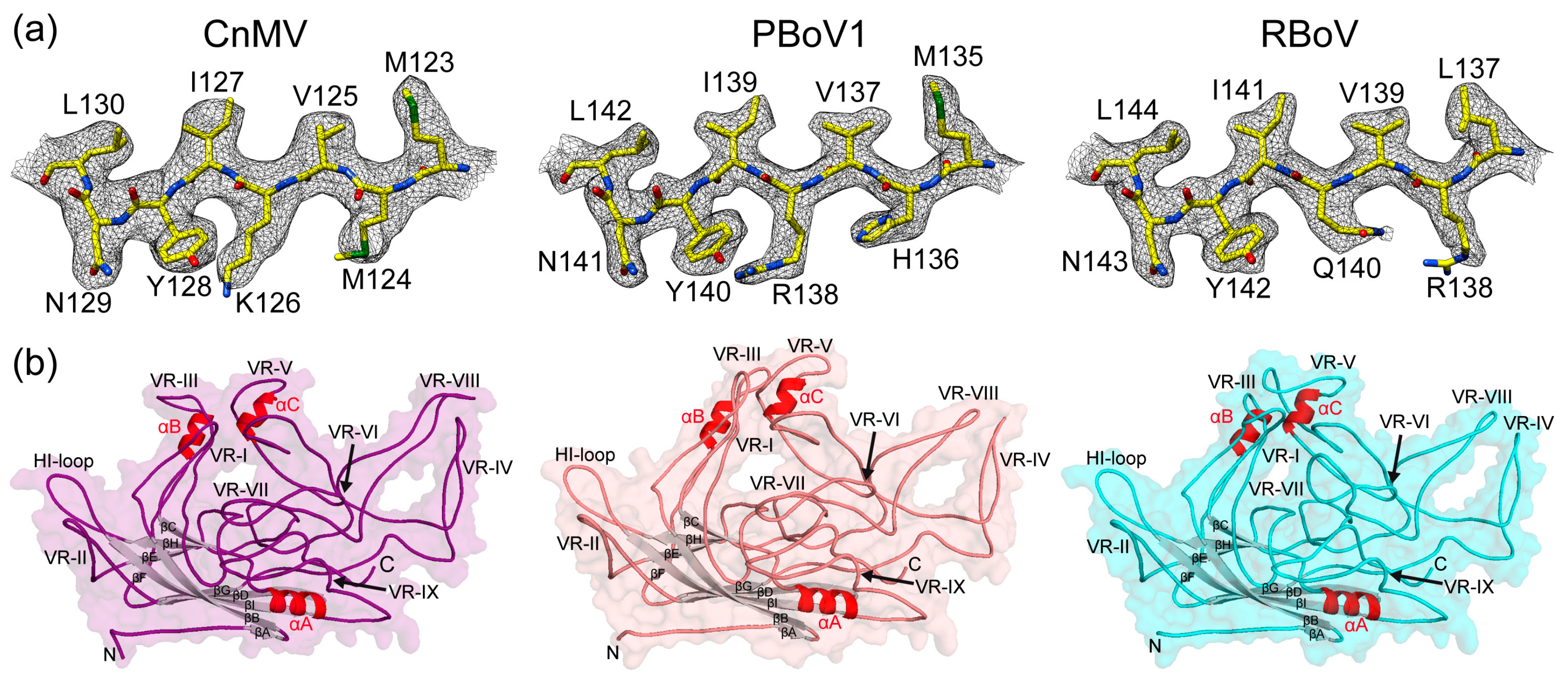
3.3. CnMV, PBoV1, and RBoV Possess an Additional Surface α-Helix
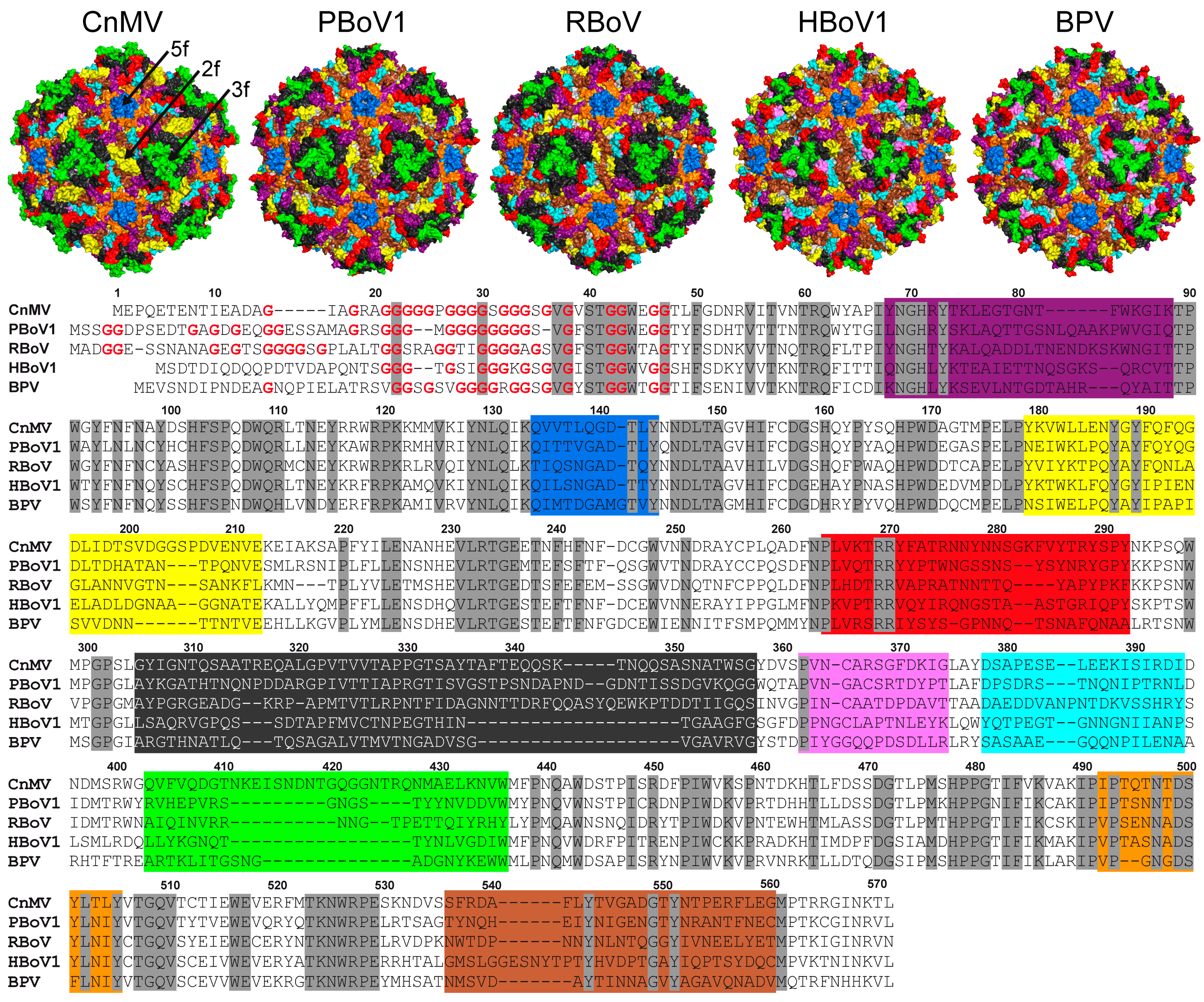
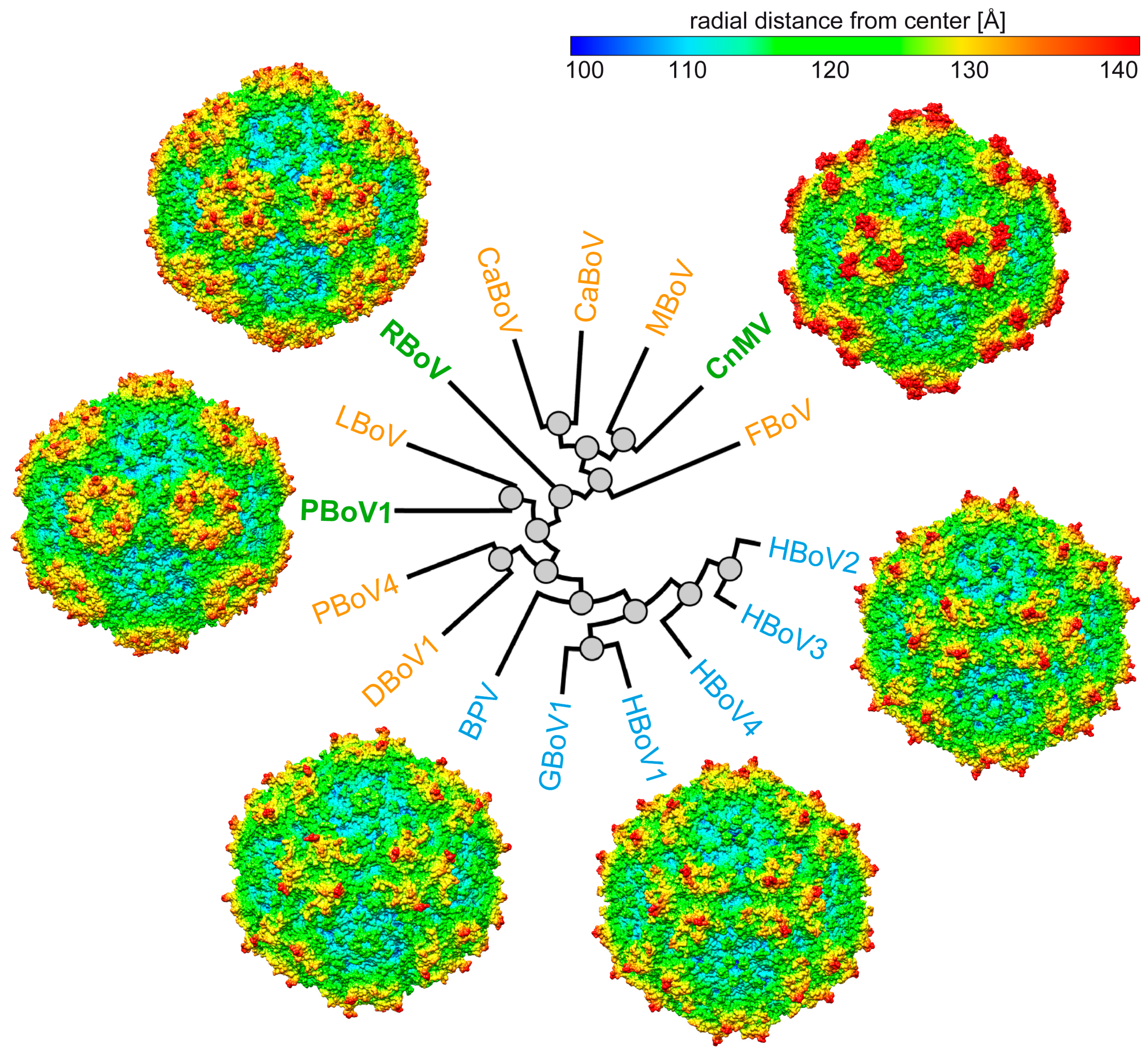
3.4. Structural Differences among the Bocaviruses Are Localized to the Variable Regions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Penzes, J.J.; et al. ICTV Virus Taxonomy Profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef] [PubMed]
- Penzes, J.J.; Soderlund-Venermo, M.; Canuti, M.; Eis-Hubinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. Reorganizing the family Parvoviridae: A revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020, 165, 2133–2146. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.C.; Tomlinson, J.E.; Lopez-Astacio, R.A.; Parrish, C.R.; Van de Walle, G.R. Small but mighty: Old and new parvoviruses of veterinary significance. Virol. J. 2021, 18, 210. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Shen, W.; Wang, S.; Qiu, J. Recent Advances in Molecular Biology of Human Bocavirus 1 and Its Applications. Front. Microbiol. 2021, 12, 696604. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Cheng, F.; Shen, W.; Engelhardt, J.F.; Yan, Z.; Qiu, J. Nonstructural Protein NP1 of Human Bocavirus 1 Plays a Critical Role in the Expression of Viral Capsid Proteins. J. Virol. 2016, 90, 4658–4669. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Deng, X.; Zou, W.; Cheng, F.; Engelhardt, J.F.; Yan, Z.; Qiu, J. Identification and Functional Analysis of Novel Nonstructural Proteins of Human Bocavirus 1. J. Virol. 2015, 89, 10097–10109. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Soderlund-Venermo, M.; Young, N.S. Human Parvoviruses. Clin. Microbiol. Rev. 2017, 30, 43–113. [Google Scholar] [CrossRef]
- Qu, X.W.; Liu, W.P.; Qi, Z.Y.; Duan, Z.J.; Zheng, L.S.; Kuang, Z.Z.; Zhang, W.J.; Hou, Y.D. Phospholipase A2-like activity of human bocavirus VP1 unique region. Biochem. Biophys. Res. Commun. 2008, 365, 158–163. [Google Scholar] [CrossRef]
- Mietzsch, M.; Penzes, J.J.; Agbandje-McKenna, M. Twenty-Five Years of Structural Parvovirology. Viruses 2019, 11, 362. [Google Scholar] [CrossRef]
- Kailasan, S.; Halder, S.; Gurda, B.; Bladek, H.; Chipman, P.R.; McKenna, R.; Brown, K.; Agbandje-McKenna, M. Structure of an enteric pathogen, bovine parvovirus. J. Virol. 2015, 89, 2603–2614. [Google Scholar] [CrossRef]
- Mietzsch, M.; Kailasan, S.; Garrison, J.; Ilyas, M.; Chipman, P.; Kantola, K.; Janssen, M.E.; Spear, J.; Sousa, D.; McKenna, R.; et al. Structural Insights into Human Bocaparvoviruses. J. Virol. 2017, 91, e00261-17. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.C.; Mietzsch, M.; Singh, A.; Jimenez Ybargollin, A.; Kailasan, S.; Chipman, P.; Bhattacharya, N.; Fakhiri, J.; Grimm, D.; Kapoor, A.; et al. Characterization of the GBoV1 Capsid and Its Antibody Interactions. Viruses 2021, 13, 330. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Mietzsch, M.; Chipman, P.; Song, K.; Xu, C.; Spear, J.; Sousa, D.; McKenna, R.; Söderlund-Venermo, M.; Agbandje-McKenna, M. pH-Induced Conformational Changes of Human Bocavirus Capsids. J. Virol. 2021, 95, e02329-20. [Google Scholar] [CrossRef] [PubMed]
- Gurda, B.L.; Parent, K.N.; Bladek, H.; Sinkovits, R.S.; DiMattia, M.A.; Rence, C.; Castro, A.; McKenna, R.; Olson, N.; Brown, K.; et al. Human bocavirus capsid structure: Insights into the structural repertoire of the parvoviridae. J. Virol. 2010, 84, 5880–5889. [Google Scholar] [CrossRef] [PubMed]
- Binn, L.N.; Lazar, E.C.; Eddy, G.A.; Kajima, M. Recovery and characterization of a minute virus of canines. Infect. Immun. 1970, 1, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Harrison, L.R.; Styer, E.L.; Pursell, A.R.; Carmichael, L.E.; Nietfeld, J.C. Fatal disease in nursing puppies associated with minute virus of canines. J. Vet. Diagn. Investig. 1992, 4, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, L.E.; Schlafer, D.H.; Hashimoto, A. Pathogenicity of minute virus of canines (MVC) for the canine fetus. Cornell Vet. 1991, 81, 151–171. [Google Scholar] [PubMed]
- Choi, J.W.; Jung, J.Y.; Lee, J.I.; Lee, K.K.; Oem, J.K. Molecular characteristics of a novel strain of canine minute virus associated with hepatitis in a dog. Arch. Virol. 2016, 161, 2299–2304. [Google Scholar] [CrossRef]
- Blomström, A.L.; Belák, S.; Fossum, C.; McKillen, J.; Allan, G.; Wallgren, P.; Berg, M. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 2009, 146, 125–129. [Google Scholar] [CrossRef]
- Zhai, S.; Yue, C.; Wei, Z.; Long, J.; Ran, D.; Lin, T.; Deng, Y.; Huang, L.; Sun, L.; Zheng, H.; et al. High prevalence of a novel porcine bocavirus in weanling piglets with respiratory tract symptoms in China. Arch. Virol. 2010, 155, 1313–1317. [Google Scholar] [CrossRef]
- Zhang, H.B.; Huang, L.; Liu, Y.J.; Lin, T.; Sun, C.Q.; Deng, Y.; Wei, Z.Z.; Cheung, A.K.; Long, J.X.; Yuan, S.S. Porcine bocaviruses: Genetic analysis and prevalence in Chinese swine population. Epidemiol. Infect. 2011, 139, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, R.; Tang, X.; Wu, C.; He, Q.; Zhao, Z.; Chen, H.; Wu, B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 2013, 158, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Cságola, A.; Lőrincz, M.; Cadar, D.; Tombácz, K.; Biksi, I.; Tuboly, T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 2012, 157, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Pfankuche, V.M.; Bodewes, R.; Hahn, K.; Puff, C.; Beineke, A.; Habierski, A.; Osterhaus, A.D.; Baumgärtner, W. Porcine Bocavirus Infection Associated with Encephalomyelitis in a Pig, Germany(1). Emerg. Infect. Dis. 2016, 22, 1310–1312. [Google Scholar] [CrossRef] [PubMed]
- Safamanesh, S.; Azimian, A.; Shakeri, A.; Ghazvini, K.; Jamehdar, S.A.; Khosrojerdi, M.; Youssefi, M. Detection of Porcine Bocavirus From a Child With Acute Respiratory Tract Infection. Pediatr. Infect. Dis. J. 2018, 37, e338–e339. [Google Scholar] [CrossRef] [PubMed]
- Aryal, M.; Liu, G. Porcine Bocavirus: A 10-Year History since Its Discovery. Virol. Sin. 2021, 36, 1261–1272. [Google Scholar] [CrossRef]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef]
- Lau, S.K.; Yeung, H.C.; Li, K.S.; Lam, C.S.; Cai, J.P.; Yuen, M.C.; Wang, M.; Zheng, B.J.; Woo, P.C.; Yuen, K.Y. Identification and genomic characterization of a novel rat bocavirus from brown rats in China. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2017, 47, 68–76. [Google Scholar] [CrossRef]
- Berger, I.; Poterszman, A. Baculovirus expression: Old dog, new tricks. Bioengineered 2015, 6, 316–322. [Google Scholar] [CrossRef]
- Ilyas, M.; Mietzsch, M.; Kailasan, S.; Vaisanen, E.; Luo, M.; Chipman, P.; Smith, J.K.; Kurian, J.; Sousa, D.; McKenna, R.; et al. Atomic Resolution Structures of Human Bufaviruses Determined by Cryo-Electron Microscopy. Viruses 2018, 10, 22. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, R.; Mietzsch, M.; Jimenez Ybargollin, A.; Chipman, P.; Fu, X.; Qiu, J.; Söderlund-Venermo, M.; McKenna, R. Capsid Structure of Aleutian Mink Disease Virus and Human Parvovirus 4: New Faces in the Parvovirus Family Portrait. Viruses 2022, 14, 2219. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Ho, P.T.; Montiel-Garcia, D.J.; Wong, J.J.; Carrillo-Tripp, M.; Brooks, C.L., 3rd; Johnson, J.E.; Reddy, V.S. VIPERdb: A Tool for Virus Research. Annu. Rev. Virol. 2018, 5, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kleywegt, G.J.; Jones, T.A. xdlMAPMAN and xdlDATAMAN—Programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. D Biol. Crystallogr. 1996, 52 Pt 4, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60 Pt 12 Pt 1, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 213–221. [Google Scholar] [CrossRef]
- Bleker, S.; Sonntag, F.; Kleinschmidt, J.A. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 2005, 79, 2528–2540. [Google Scholar] [CrossRef]
- Stahnke, S.; Lux, K.; Uhrig, S.; Kreppel, F.; Hosel, M.; Coutelle, O.; Ogris, M.; Hallek, M.; Buning, H. Intrinsic phospholipase A2 activity of adeno-associated virus is involved in endosomal escape of incoming particles. Virology 2011, 409, 77–83. [Google Scholar] [CrossRef]
- Mietzsch, M.; Jose, A.; Chipman, P.; Bhattacharya, N.; Daneshparvar, N.; McKenna, R.; Agbandje-McKenna, M. Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features. Viruses 2021, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- Mietzsch, M.; McKenna, R.; Vaisanen, E.; Yu, J.C.; Ilyas, M.; Hull, J.A.; Kurian, J.; Smith, J.K.; Chipman, P.; Lasanajak, Y.; et al. Structural Characterization of Cuta- and Tusavirus: Insight into Protoparvoviruses Capsid Morphology. Viruses 2020, 12, 653. [Google Scholar] [CrossRef] [PubMed]
- Kailasan, S.; Garrison, J.; Ilyas, M.; Chipman, P.; McKenna, R.; Kantola, K.; Soderlund-Venermo, M.; Kucinskaite-Kodze, I.; Zvirbliene, A.; Agbandje-McKenna, M. Mapping Antigenic Epitopes on the Human Bocavirus Capsid. J. Virol. 2016, 90, 4670–4680. [Google Scholar] [CrossRef]
- Meyer, N.L.; Chapman, M.S. Adeno-associated virus (AAV) cell entry: Structural insights. Trends Microbiol. 2022, 30, 432–451. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, S.D.; Cline, S.E.; Hemming, J.P.; Johnson, F.B. Attachment of bovine parvovirus to O-linked alpha 2,3 neuraminic acid on glycophorin A. Arch. Virol. 2005, 150, 1477–1484. [Google Scholar] [CrossRef]
- Johnson, F.B.; Fenn, L.B.; Owens, T.J.; Faucheux, L.J.; Blackburn, S.D. Attachment of bovine parvovirus to sialic acids on bovine cell membranes. J. Gen. Virol. 2004, 85 Pt 8, 2199–2207. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Fakhiri, J.; Linse, K.P.; Mietzsch, M.; Xu, M.; Schneider, M.A.; Meister, M.; Schildgen, O.; Schnitzler, P.; Soderlund-Venermo, M.; Agbandje-McKenna, M.; et al. Impact of Natural or Synthetic Singletons in the Capsid of Human Bocavirus 1 on Particle Infectivity and Immunoreactivity. J. Virol. 2020, 94, e00170-20. [Google Scholar] [CrossRef] [PubMed]
- Parrish, C.R.; Kawaoka, Y. The origins of new pandemic viruses: The acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 2005, 59, 553–586. [Google Scholar] [CrossRef]

| Parameter | CnMV | PBoV1 | RBoV |
|---|---|---|---|
| Total no. of micrographs | 653 | 7949 | 7616 |
| Defocus range (µm) | 0.5–2.0 | 0.8–2.1 | 0.8–2.1 |
| Electron dose (e−/Å2) | 59 | 50 | 50 |
| No. of frames/micrograph | 52 | 50 | 50 |
| Pixel size (Å/pixel) | 0.92 | 0.95 | 0.95 |
| No. of particles used for final map | 50,866 | 98,388 | 9199 |
| Resolution of final map (Å) | 2.72 | 2.31 | 2.52 |
| PHENIX model refinement statistics | |||
| Map CC | 0.84 | 0.88 | 0.88 |
| RMSD Bonds (Å) | 0.01 | 0.01 | 0.01 |
| RMSD Angles (°) | 0.83 | 0.90 | 0.82 |
| All-atom clash score | 9.34 | 7.81 | 7.95 |
| Ramachandran plot (%) | |||
| Favored | 97.8 | 97.9 | 98.5 |
| Allowed | 2.2 | 2.1 | 1.5 |
| Outliers | 0.0 | 0.0 | 0.0 |
| Rotamer outliers | 0.0 | 0.0 | 0.0 |
| No. of Cβ deviations | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velez, M.; Mietzsch, M.; Hsi, J.; Bell, L.; Chipman, P.; Fu, X.; McKenna, R. Structural Characterization of Canine Minute Virus, Rat and Porcine Bocavirus. Viruses 2023, 15, 1799. https://doi.org/10.3390/v15091799
Velez M, Mietzsch M, Hsi J, Bell L, Chipman P, Fu X, McKenna R. Structural Characterization of Canine Minute Virus, Rat and Porcine Bocavirus. Viruses. 2023; 15(9):1799. https://doi.org/10.3390/v15091799
Chicago/Turabian StyleVelez, Michael, Mario Mietzsch, Jane Hsi, Logan Bell, Paul Chipman, Xiaofeng Fu, and Robert McKenna. 2023. "Structural Characterization of Canine Minute Virus, Rat and Porcine Bocavirus" Viruses 15, no. 9: 1799. https://doi.org/10.3390/v15091799
APA StyleVelez, M., Mietzsch, M., Hsi, J., Bell, L., Chipman, P., Fu, X., & McKenna, R. (2023). Structural Characterization of Canine Minute Virus, Rat and Porcine Bocavirus. Viruses, 15(9), 1799. https://doi.org/10.3390/v15091799





