Protective versus Pathogenic Type I Interferon Responses during Virus Infections
Abstract
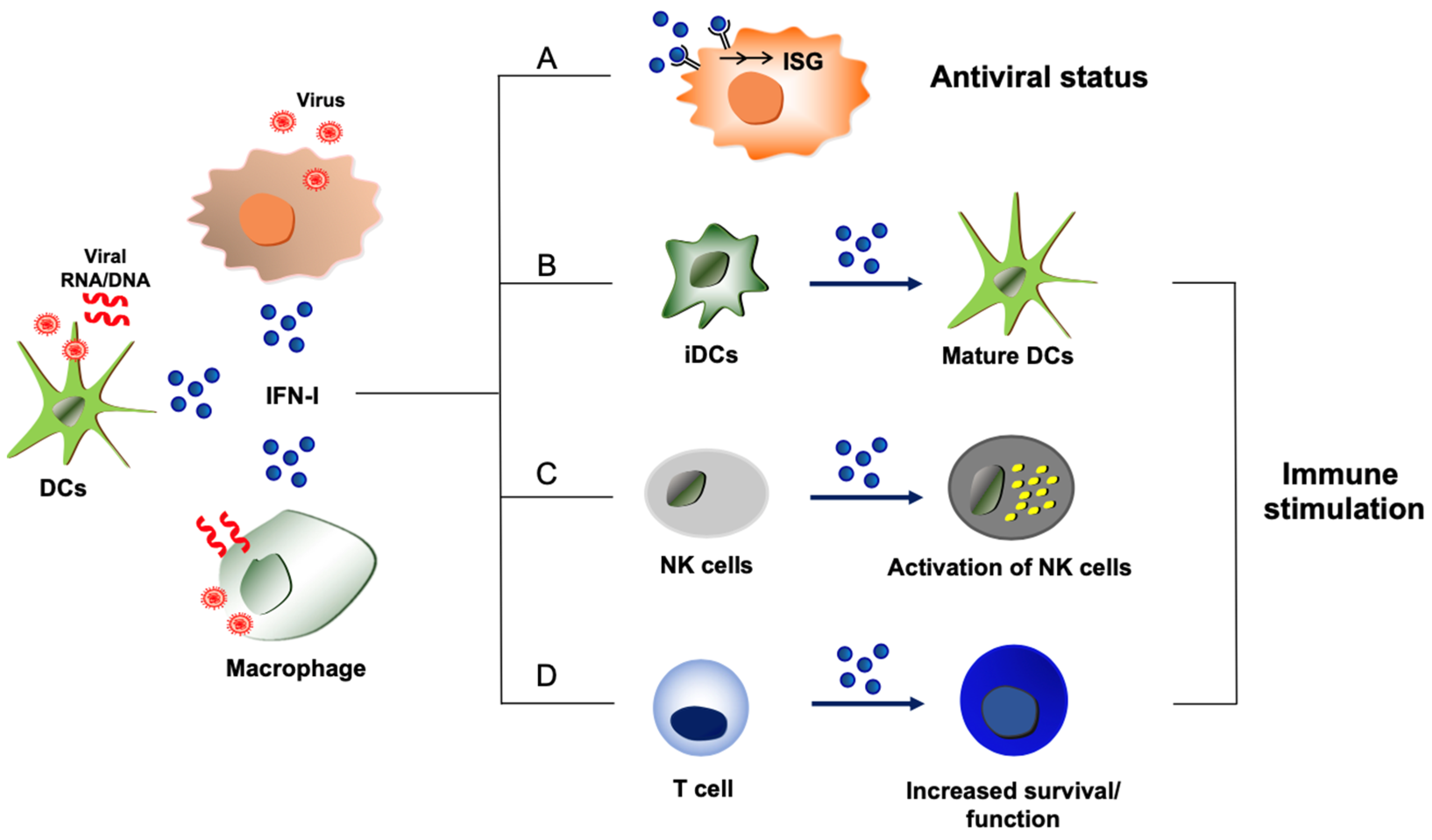
:1. Introduction
2. Protective Functions of Type I IFN
2.1. Antiviral Activity
2.1.1. Induction of Type I IFN Response
2.1.2. Viral Strategies to Subvert Type I IFN Response
2.1.3. Type I IFN in COVID-19
2.1.4. Type I IFN from Microbiome
2.2. Immune Stimulatory Action of Type I IFNs
2.2.1. Type I IFN Production from Immune Cells
2.2.2. Immune Stimulation by Type I IFN
3. Pathogenic Functions of Type I IFN
3.1. Immune Suppression by Type I IFN
3.1.1. Inhibition of Cellular Development Process by Type I IFN
3.1.2. Suppression of Host Immunity during Virus Persistence
3.2. Inflammation and Tissue Injury Caused by Type I IFN
3.2.1. Inflammation
3.2.2. Impairment of Tissue Repair by Type I IFN
4. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Isaacs, A.; Lindenmann, J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957, 147, 258–267. [Google Scholar]
- González-Navajas, J.M.; Lee, J.; David, M.; Raz, E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012, 12, 125–135. [Google Scholar] [CrossRef] [PubMed]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Hahm, B. Type I interferon modulates the battle of host immune system against viruses. Adv. Appl. Microbiol. 2010, 73, 83–101. [Google Scholar]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Brown, H.M.; Hwang, S. Direct Antiviral Mechanisms of Interferon-Gamma. Immune. Netw. 2018, 18, e33. [Google Scholar] [CrossRef]
- Boulant, S.; Forero, A.; Santer, D.M.; Broggi, A. Editorial: Type III interferons: Emerging roles beyond antiviral barrier defense. Front. Immunol. 2022, 13, 1030812. [Google Scholar] [CrossRef]
- Ye, L.; Schnepf, D.; Staeheli, P. Interferon-λ orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 2019, 19, 614–625. [Google Scholar] [CrossRef]
- García-Sastre, A.; Biron, C.A. Type 1 interferons and the virus-host relationship: A lesson in détente. Science 2006, 312, 879–882. [Google Scholar] [CrossRef]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89 Pt 1, 1–47. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Ashkar, A.A. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 2018, 9, 2061. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal. Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Duan, T.; Du, Y.; Xing, C.; Wang, H.Y.; Wang, R.F. Toll-Like Receptor Signaling and Its Role in Cell-Mediated Immunity. Front. Immunol. 2022, 13, 812774. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-stimulated genes: Roles in viral pathogenesis. Curr. Opin. Virol. 2014, 6, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Samuel, C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.; Qiu, X.G. Type I interferon receptor knockout mice as models for infection of highly pathogenic viruses with outbreak potential. Zool. Res. 2018, 39, 3–14. [Google Scholar] [PubMed]
- Yoneyama, M.; Fujita, T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010, 20, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug. Discov. 2007, 6, 975–990. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Fujita, T. RIG-I-like receptors and autoimmune diseases. Curr. Opin. Immunol. 2015, 37, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Sadler, A.J.; Williams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef]
- Zhu, J.; Chiang, C.; Gack, M.U. Viral evasion of the interferon response at a glance. J. Cell Sci. 2023, 136. [Google Scholar] [CrossRef]
- Rosário-Ferreira, N.; Preto, A.J.; Melo, R.; Moreira, I.S.; Brito, R.M.M. The Central Role of Non-Structural Protein 1 (NS1) in Influenza Biology and Infection. Int. J. Mol. Sci. 2020, 21, 1511. [Google Scholar] [CrossRef]
- Xia, C.; Wolf, J.J.; Vijayan, M.; Studstill, C.J.; Ma, W.; Hahm, B. Casein Kinase 1α Mediates the Degradation of Receptors for Type I and Type II Interferons Caused by Hemagglutinin of Influenza A Virus. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Park, E.S.; Byun, Y.H.; Park, S.; Jang, Y.H.; Han, W.R.; Won, J.; Cho, K.C.; Kim, D.H.; Lee, A.R.; Shin, G.C.; et al. Co-degradation of interferon signaling factor DDX3 by PB1-F2 as a basis for high virulence of 1918 pandemic influenza. Embo J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef]
- Varga, Z.T.; Ramos, I.; Hai, R.; Schmolke, M.; García-Sastre, A.; Fernandez-Sesma, A.; Palese, P. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS. Pathog. 2011, 7, e1002067. [Google Scholar] [CrossRef]
- Graef, K.M.; Vreede, F.T.; Lau, Y.F.; McCall, A.W.; Carr, S.M.; Subbarao, K.; Fodor, E. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J. Virol. 2010, 84, 8433–8445. [Google Scholar] [CrossRef] [PubMed]
- Kochs, G.; García-Sastre, A.; Martínez-Sobrido, L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 2007, 81, 7011–7021. [Google Scholar] [CrossRef] [PubMed]
- Mibayashi, M.; Martínez-Sobrido, L.; Loo, Y.M.; Cárdenas, W.B.; Gale, M., Jr.; García-Sastre, A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007, 81, 514–524. [Google Scholar] [CrossRef]
- Trouillet-Assant, S.; Viel, S.; Gaymard, A.; Pons, S.; Richard, J.C.; Perret, M.; Villard, M.; Brengel-Pesce, K.; Lina, B.; Mezidi, M.; et al. Type I IFN immunoprofiling in COVID-19 patients. J. Allergy. Clin. Immunol. 2020, 146, 206–208.e2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Matuozzo, D.; Le Pen, J.; Lee, D.; Moens, L.; Asano, T.; Bohlen, J.; Liu, Z.; Moncada-Velez, M.; Kendir-Demirkol, Y.; et al. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J. Exp. Med. 2022, 219. [Google Scholar] [CrossRef]
- Kristiansen, H.; Gad, H.H.; Eskildsen-Larsen, S.; Despres, P.; Hartmann, R. The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J. Interferon. Cytokine. Res. 2011, 31, 41–47. [Google Scholar] [CrossRef]
- Banday, A.R.; Stanifer, M.L.; Florez-Vargas, O.; Onabajo, O.O.; Papenberg, B.W.; Zahoor, M.A.; Mirabello, L.; Ring, T.J.; Lee, C.H.; Albert, P.S.; et al. Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries. Nat. Genet. 2022, 54, 1103–1116. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, V.; Shannon, C.P.; Wei, X.S.; Xiang, X.; Wang, X.; Wang, Z.H.; Tebbutt, S.J.; Kollmann, T.R.; Fish, E.N. Interferon-α2b Treatment for COVID-19. Front. Immunol. 2020, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Vinh, D.C.; Abel, L.; Bastard, P.; Cheng, M.P.; Condino-Neto, A.; Gregersen, P.K.; Haerynck, F.; Cicalese, M.P.; Hagin, D.; Soler-Palacín, P.; et al. Harnessing Type I IFN Immunity Against SARS-CoV-2 with Early Administration of IFN-β. J. Clin. Immunol. 2021, 41, 1425–1442. [Google Scholar] [CrossRef] [PubMed]
- Schaupp, L.; Muth, S.; Rogell, L.; Kofoed-Branzk, M.; Melchior, F.; Lienenklaus, S.; Ganal-Vonarburg, S.C.; Klein, M.; Guendel, F.; Hain, T.; et al. Microbiota-Induced Type I Interferons Instruct a Poised Basal State of Dendritic Cells. Cell 2020, 181, 1080–1096.e19. [Google Scholar] [CrossRef] [PubMed]
- Bradley, K.C.; Finsterbusch, K.; Schnepf, D.; Crotta, S.; Llorian, M.; Davidson, S.; Fuchs, S.Y.; Staeheli, P.; Wack, A. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect from Influenza Virus Infection. Cell Rep. 2019, 28, 245–256.e4. [Google Scholar] [CrossRef]
- Stefan, K.L.; Kim, M.V.; Iwasaki, A.; Kasper, D.L. Commensal Microbiota Modulation of Natural Resistance to Virus Infection. Cell 2020, 183, 1312–1324.e10. [Google Scholar] [CrossRef]
- Kawashima, T.; Kosaka, A.; Yan, H.; Guo, Z.; Uchiyama, R.; Fukui, R.; Kaneko, D.; Kumagai, Y.; You, D.J.; Carreras, J.; et al. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-β. Immunity 2013, 38, 1187–1197. [Google Scholar] [CrossRef]
- Antunes, K.H.; Fachi, J.L.; de Paula, R.; da Silva, E.F.; Pral, L.P.; Dos Santos, A.; Dias, G.B.M.; Vargas, J.E.; Puga, R.; Mayer, F.Q.; et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 2019, 10, 3273. [Google Scholar] [CrossRef]
- Winkler, E.S.; Shrihari, S.; Hykes, B.L., Jr.; Handley, S.A.; Andhey, P.S.; Huang, Y.S.; Swain, A.; Droit, L.; Chebrolu, K.K.; Mack, M.; et al. The Intestinal Microbiome Restricts Alphavirus Infection and Dissemination through a Bile Acid-Type I IFN Signaling Axis. Cell 2020, 182, 901–918.e18. [Google Scholar] [CrossRef]
- Wirusanti, N.I.; Baldridge, M.T.; Harris, V.C. Microbiota regulation of viral infections through interferon signaling. Trends. Microbiol. 2022, 30, 778–792. [Google Scholar] [CrossRef]
- Ericsson, A.C.; Davis, J.W.; Spollen, W.; Bivens, N.; Givan, S.; Hagan, C.E.; McIntosh, M.; Franklin, C.L. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS ONE 2015, 10, e0116704. [Google Scholar] [CrossRef] [PubMed]
- Wolff, N.S.; Jacobs, M.C.; Haak, B.W.; Roelofs, J.; de Vos, A.F.; Hugenholtz, F.; Wiersinga, W.J. Vendor effects on murine gut microbiota and its influence on lipopolysaccharide-induced lung inflammation and Gram-negative pneumonia. Intensive Care. Med. Exp. 2020, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Radford, K.J. The role of dendritic cells in cancer. Int. Rev. Cell Mol. Biol. 2019, 348, 123–178. [Google Scholar]
- Bencze, D.; Fekete, T.; Pázmándi, K. Type I Interferon Production of Plasmacytoid Dendritic Cells under Control. Int. J. Mol. Sci. 2021, 22, 4190. [Google Scholar] [CrossRef]
- Sin, W.X.; Yeong, J.P.; Lim, T.J.F.; Su, I.H.; Connolly, J.E.; Chin, K.C. IRF-7 Mediates Type I IFN Responses in Endotoxin-Challenged Mice. Front. Immunol. 2020, 11, 640. [Google Scholar] [CrossRef]
- Hofmeyer, K.A.; Ray, A.; Zang, X. The contrasting role of B7-H3. Proc. Natl. Acad. Sci. USA 2008, 105, 10277–10278. [Google Scholar] [CrossRef]
- Swiecki, M.; Colonna, M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015, 15, 471–485. [Google Scholar] [CrossRef]
- Montoya, M.; Schiavoni, G.; Mattei, F.; Gresser, I.; Belardelli, F.; Borrow, P.; Tough, D.F. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood 2002, 99, 3263–3271. [Google Scholar] [CrossRef]
- Huber, J.P.; Farrar, J.D. Regulation of effector and memory T-cell functions by type I interferon. Immunology 2011, 132, 466–474. [Google Scholar] [CrossRef]
- Aichele, P.; Unsoeld, H.; Koschella, M.; Schweier, O.; Kalinke, U.; Vucikuja, S. CD8 T cells specific for lymphocytic choriomeningitis virus require type I IFN receptor for clonal expansion. J. Immunol. 2006, 176, 4525–4529. [Google Scholar] [CrossRef] [PubMed]
- Kolumam, G.A.; Thomas, S.; Thompson, L.J.; Sprent, J.; Murali-Krishna, K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 2005, 202, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Gongora, R.; Stephan, R.P.; Schreiber, R.D.; Cooper, M.D. Stat-1 is not essential for inhibition of B lymphopoiesis by type I IFNs. J. Immunol. 2000, 165, 2362–2366. [Google Scholar] [CrossRef]
- Gongora, R.; Stephan, R.P.; Zhang, Z.; Cooper, M.D. An essential role for Daxx in the inhibition of B lymphopoiesis by type I interferons. Immunity 2001, 14, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Dong, C.; Cooper, M.D. Impairment of T and B cell development by treatment with a type I interferon. J. Exp. Med. 1998, 187, 79–87. [Google Scholar] [CrossRef]
- Elsaesser, H.J.; Mohtashami, M.; Osokine, I.; Snell, L.M.; Cunningham, C.R.; Boukhaled, G.M.; McGavern, D.B.; Zúñiga-Pflücker, J.C.; Brooks, D.G. Chronic virus infection drives CD8 T cell-mediated thymic destruction and impaired negative selection. Proc. Natl. Acad. Sci. USA 2020, 117, 5420–5429. [Google Scholar] [CrossRef]
- Hahm, B.; Trifilo, M.J.; Zuniga, E.I.; Oldstone, M.B. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity 2005, 22, 247–257. [Google Scholar] [CrossRef]
- Sevilla, N.; McGavern, D.B.; Teng, C.; Kunz, S.; Oldstone, M.B. Viral targeting of hematopoietic progenitors and inhibition of DC maturation as a dual strategy for immune subversion. J. Clin. Investig. 2004, 113, 737–745. [Google Scholar] [CrossRef]
- Studstill, C.J.; Hahm, B. Chronic LCMV Infection Is Fortified with Versatile Tactics to Suppress Host T Cell Immunity and Establish Viral Persistence. Viruses 2021, 13, 1951. [Google Scholar] [CrossRef]
- Kahan, S.M.; Zajac, A.J. Immune Exhaustion: Past Lessons and New Insights from Lymphocytic Choriomeningitis Virus. Viruses 2019, 11, 156. [Google Scholar] [CrossRef]
- Oldstone, M.B. Viral persistence: Parameters, mechanisms and future predictions. Virology 2006, 344, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Sandu, I.; Cerletti, D.; Oetiker, N.; Borsa, M.; Wagen, F.; Spadafora, I.; Welten, S.P.M.; Stolz, U.; Oxenius, A.; Claassen, M. Landscape of Exhausted Virus-Specific CD8 T Cells in Chronic LCMV Infection. Cell Rep. 2020, 32, 108078. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.L.; Wherry, E.J.; Masopust, D.; Zhu, B.; Allison, J.P.; Sharpe, A.H.; Freeman, G.J.; Ahmed, R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006, 439, 682–687. [Google Scholar] [CrossRef]
- Studstill, C.J.; Pritzl, C.J.; Seo, Y.J.; Kim, D.Y.; Xia, C.; Wolf, J.J.; Nistala, R.; Vijayan, M.; Cho, Y.B.; Kang, K.W.; et al. Sphingosine kinase 2 restricts T cell immunopathology but permits viral persistence. J. Clin. Investig. 2020, 130, 6523–6538. [Google Scholar] [CrossRef]
- Jin, H.T.; Anderson, A.C.; Tan, W.G.; West, E.E.; Ha, S.J.; Araki, K.; Freeman, G.J.; Kuchroo, V.K.; Ahmed, R. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA 2010, 107, 14733–14738. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J.; Ha, S.J.; Kaech, S.M.; Haining, W.N.; Sarkar, S.; Kalia, V.; Subramaniam, S.; Blattman, J.N.; Barber, D.L.; Ahmed, R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 2007, 27, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Teijaro, J.R.; Ng, C.; Lee, A.M.; Sullivan, B.M.; Sheehan, K.C.; Welch, M.; Schreiber, R.D.; de la Torre, J.C.; Oldstone, M.B. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 2013, 340, 207–211. [Google Scholar] [CrossRef]
- Wilson, E.B.; Yamada, D.H.; Elsaesser, H.; Herskovitz, J.; Deng, J.; Cheng, G.; Aronow, B.J.; Karp, C.L.; Brooks, D.G. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 2013, 340, 202–207. [Google Scholar] [CrossRef]
- Huang, Z.; Kang, S.G.; Li, Y.; Zak, J.; Shaabani, N.; Deng, K.; Shepherd, J.; Bhargava, R.; Teijaro, J.R.; Xiao, C. IFNAR1 signaling in NK cells promotes persistent virus infection. Sci. Adv. 2021, 7, eabb8087. [Google Scholar] [CrossRef]
- Zhen, A.; Rezek, V.; Youn, C.; Lam, B.; Chang, N.; Rick, J.; Carrillo, M.; Martin, H.; Kasparian, S.; Syed, P.; et al. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J. Clin. Investig. 2017, 127, 260–268. [Google Scholar] [CrossRef]
- Cheng, L.; Yu, H.; Li, G.; Li, F.; Ma, J.; Li, J.; Chi, L.; Zhang, L.; Su, L. Type I interferons suppress viral replication but contribute to T cell depletion and dysfunction during chronic HIV-1 infection. JCI Insight 2017, 2, e94366. [Google Scholar] [CrossRef]
- Ho, N.I.; Huis In’t Veld, L.G.M.; Raaijmakers, T.K.; Adema, G.J. Adjuvants Enhancing Cross-Presentation by Dendritic Cells: The Key to More Effective Vaccines? Front. Immunol. 2018, 9, 2874. [Google Scholar] [CrossRef]
- Green, N.M.; Laws, A.; Kiefer, K.; Busconi, L.; Kim, Y.M.; Brinkmann, M.M.; Trail, E.H.; Yasuda, K.; Christensen, S.R.; Shlomchik, M.J.; et al. Murine B cell response to TLR7 ligands depends on an IFN-beta feedback loop. J. Immunol. 2009, 183, 1569–1576. [Google Scholar] [CrossRef]
- Kash, J.C.; Tumpey, T.M.; Proll, S.C.; Carter, V.; Perwitasari, O.; Thomas, M.J.; Basler, C.F.; Palese, P.; Taubenberger, J.K.; García-Sastre, A.; et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 2006, 443, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Kobasa, D.; Jones, S.M.; Shinya, K.; Kash, J.C.; Copps, J.; Ebihara, H.; Hatta, Y.; Kim, J.H.; Halfmann, P.; Hatta, M.; et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007, 445, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Perrone, L.A.; Plowden, J.K.; García-Sastre, A.; Katz, J.M.; Tumpey, T.M. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008, 4, e1000115. [Google Scholar] [CrossRef] [PubMed]
- Elshina, E.; Te Velthuis, A.J.W. The influenza virus RNA polymerase as an innate immune agonist and antagonist. Cell. Mol. Life Sci. 2021, 78, 7237–7256. [Google Scholar] [CrossRef]
- Billharz, R.; Zeng, H.; Proll, S.C.; Korth, M.J.; Lederer, S.; Albrecht, R.; Goodman, A.G.; Rosenzweig, E.; Tumpey, T.M.; García-Sastre, A.; et al. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J. Virol. 2009, 83, 10557–10570. [Google Scholar] [CrossRef]
- Dubrow, A.; Lin, S.; Savage, N.; Shen, Q.; Cho, J.H. Molecular Basis of the Ternary Interaction between NS1 of the 1918 Influenza A Virus, PI3K, and CRK. Viruses 2020, 12, 338. [Google Scholar] [CrossRef]
- Thangavel, R.R.; Bouvier, N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. J. Immunol. Methods 2014, 410, 60–79. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Israelow, B.; Song, E.; Mao, T.; Lu, P.; Meir, A.; Liu, F.; Alfajaro, M.M.; Wei, J.; Dong, H.; Homer, R.J.; et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020, 217, e20201241. [Google Scholar] [CrossRef]
- Xu, H.; Akinyemi, I.A.; Chitre, S.A.; Loeb, J.C.; Lednicky, J.A.; McIntosh, M.T.; Bhaduri-McIntosh, S. SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway. Virology 2022, 568, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.W.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. eLife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, A.M.; Suptela, A.J.; Marriott, I. Z-DNA binding protein 1 mediates necroptotic and apoptotic cell death pathways in murine astrocytes following herpes simplex virus-1 infection. J. Neuroinflamm. 2022, 19, 109. [Google Scholar] [CrossRef]
- Zhang, T.; Yin, C.; Boyd, D.F.; Quarato, G.; Ingram, J.P.; Shubina, M.; Ragan, K.B.; Ishizuka, T.; Crawford, J.C.; Tummers, B.; et al. Influenza Virus Z-RNAs Induce ZBP1-Mediated Necroptosis. Cell 2020, 180, 1115–1129.e13. [Google Scholar] [CrossRef]
- Karki, R.; Lee, S.; Mall, R.; Pandian, N.; Wang, Y.; Sharma, B.R.; Malireddi, R.S.; Yang, D.; Trifkovic, S.; Steele, J.A.; et al. ZBP1-dependent inflammatory cell death, PANoptosis, and cytokine storm disrupt IFN therapeutic efficacy during coronavirus infection. Sci. Immunol. 2022, 7, eabo6294. [Google Scholar] [CrossRef]
- Major, J.; Crotta, S.; Llorian, M.; McCabe, T.M.; Gad, H.H.; Priestnall, S.L.; Hartmann, R.; Wack, A. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 2020, 369, 712–717. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, K.I.; McKenna, S.; Vijayamahantesh, V.; He, Y.; Hahm, B. Protective versus Pathogenic Type I Interferon Responses during Virus Infections. Viruses 2023, 15, 1916. https://doi.org/10.3390/v15091916
Jung KI, McKenna S, Vijayamahantesh V, He Y, Hahm B. Protective versus Pathogenic Type I Interferon Responses during Virus Infections. Viruses. 2023; 15(9):1916. https://doi.org/10.3390/v15091916
Chicago/Turabian StyleJung, Kwang Il, Savannah McKenna, Vijayamahantesh Vijayamahantesh, Ying He, and Bumsuk Hahm. 2023. "Protective versus Pathogenic Type I Interferon Responses during Virus Infections" Viruses 15, no. 9: 1916. https://doi.org/10.3390/v15091916
APA StyleJung, K. I., McKenna, S., Vijayamahantesh, V., He, Y., & Hahm, B. (2023). Protective versus Pathogenic Type I Interferon Responses during Virus Infections. Viruses, 15(9), 1916. https://doi.org/10.3390/v15091916






