Mechanism and Kinetics of HIV-1 Protease Activation
Abstract
1. Introduction

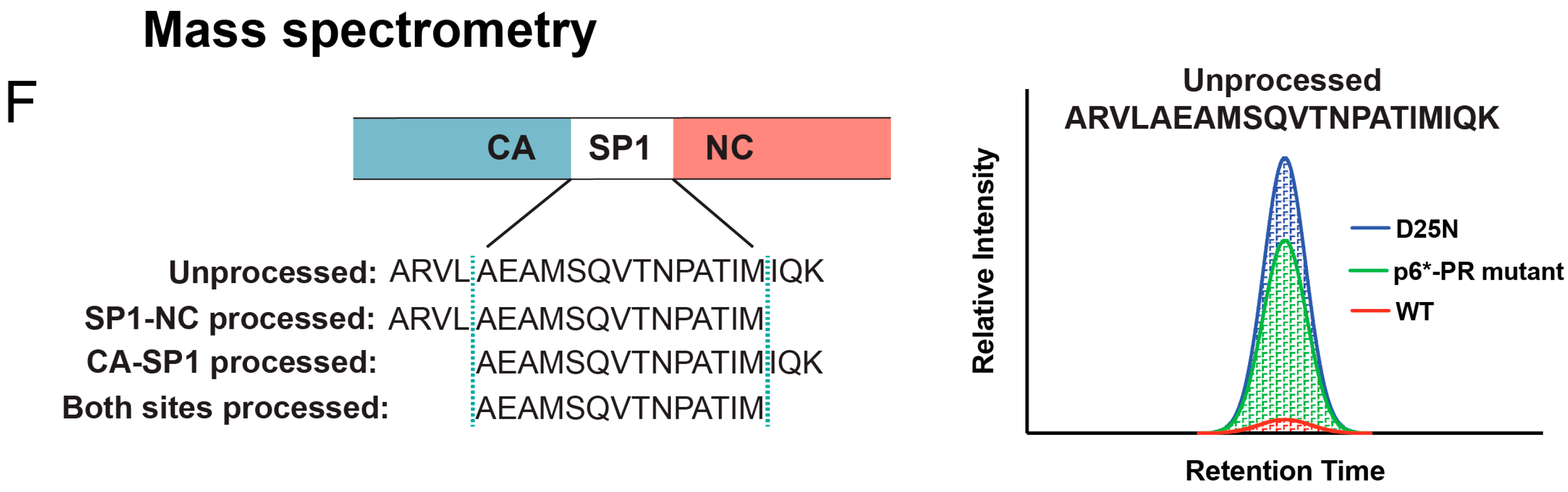
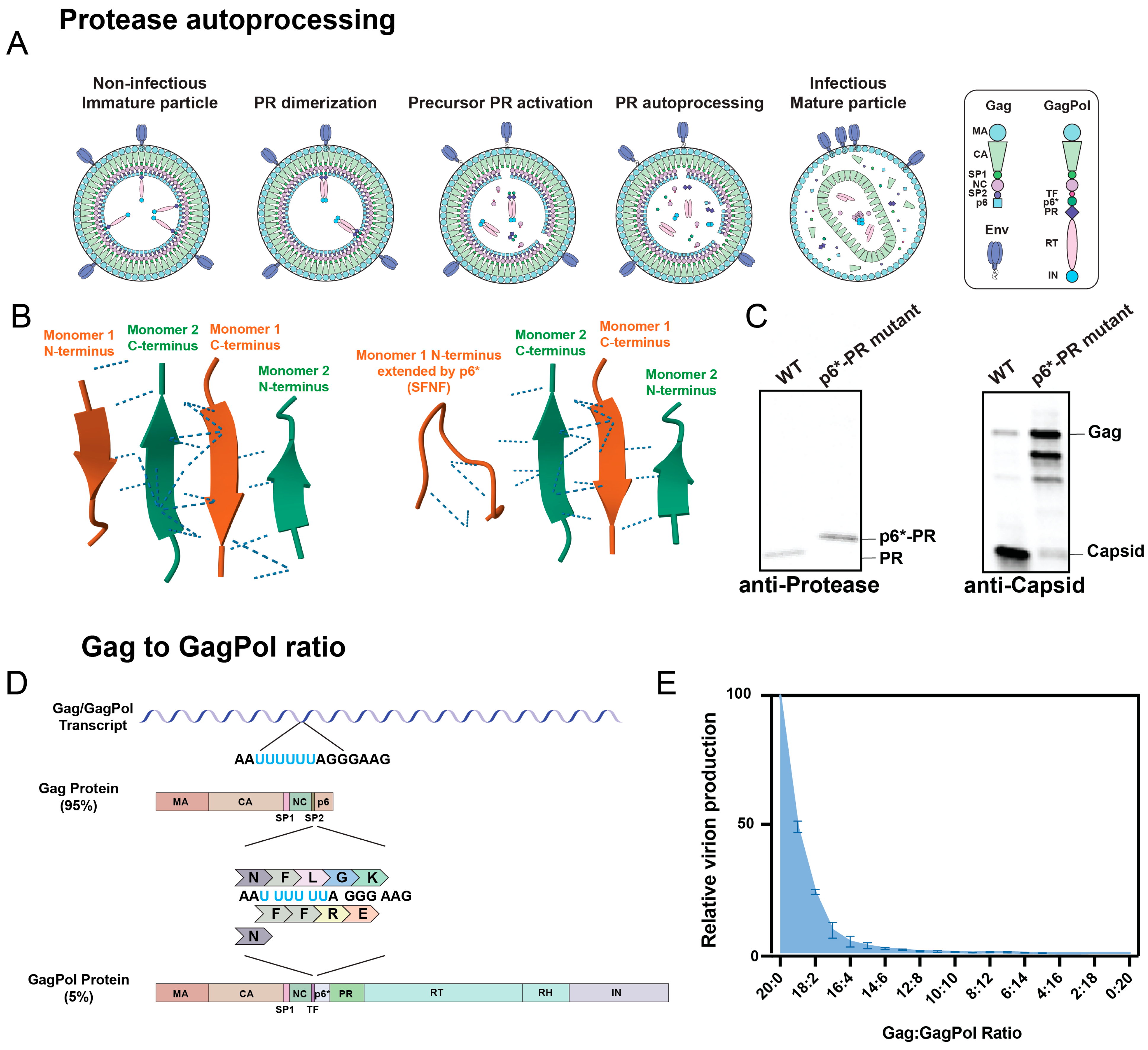
2. Mechanism of Protease Activation
3. Methods to Analyze Protease Activation
3.1. Traditional Methods
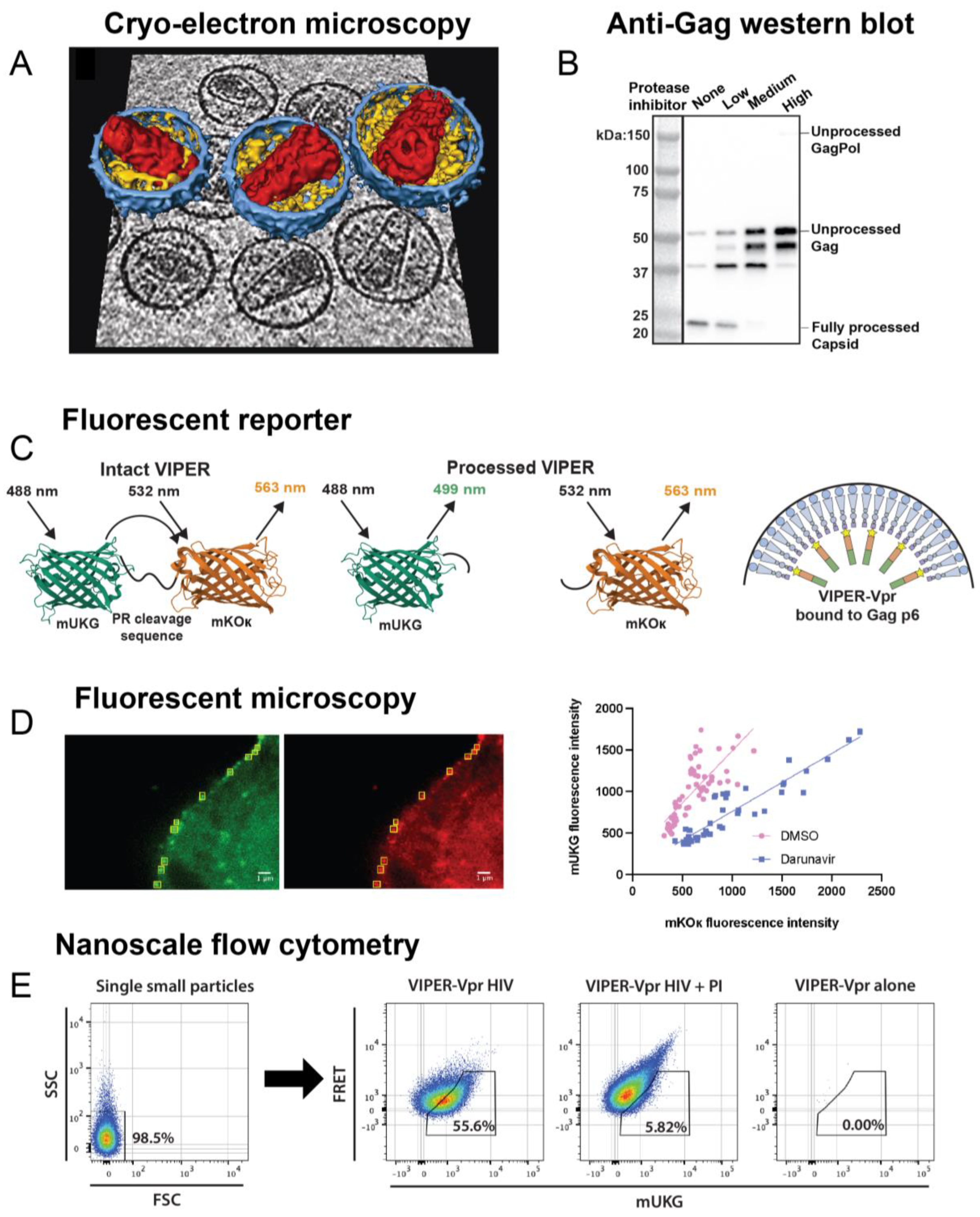
3.2. Nanoscale Flow Cytometry
3.3. Mass Spectrometry (MS)
4. Discoveries Concerning Protease Activation Kinetics
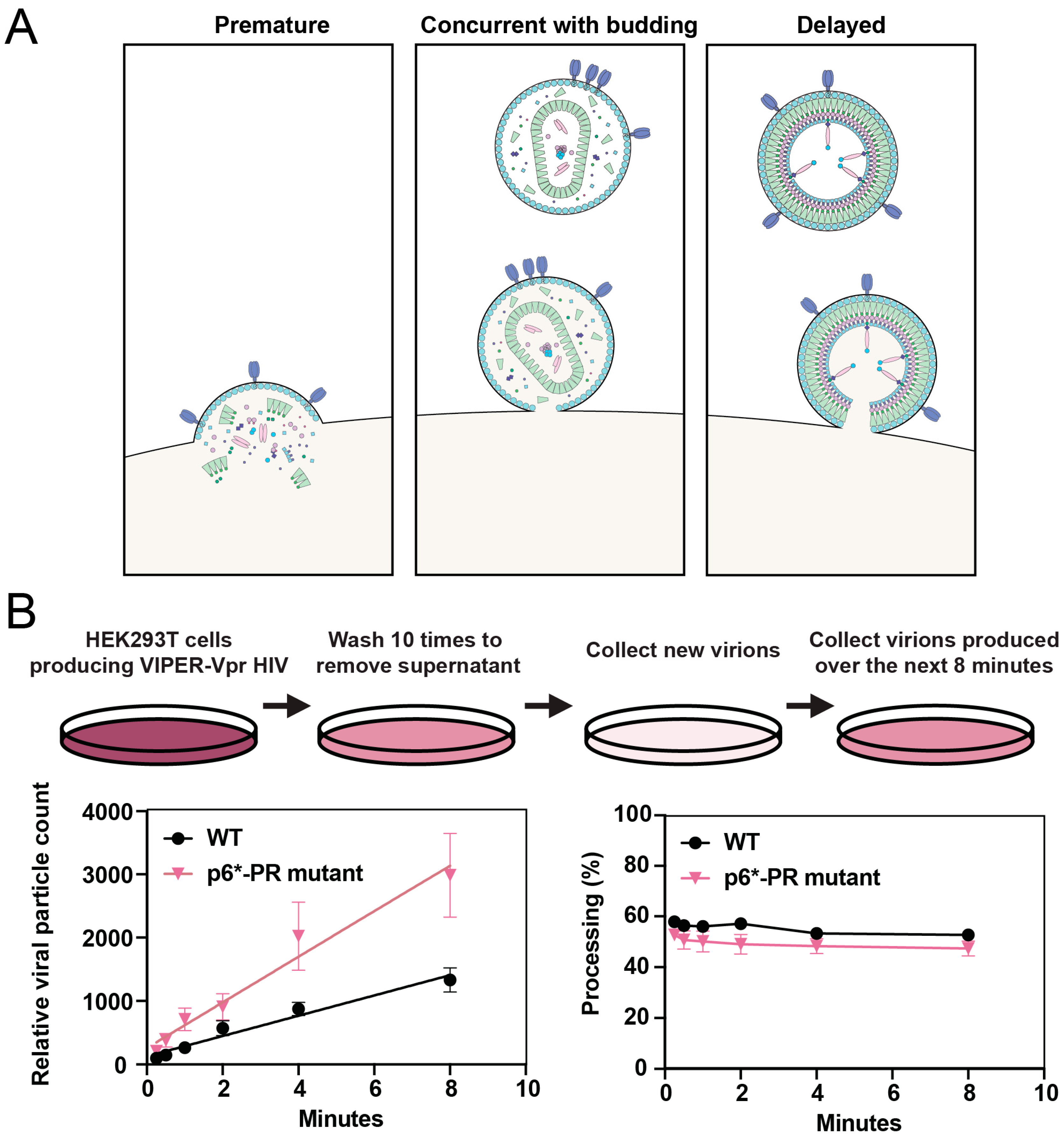
4.1. Timing of Protease Activation
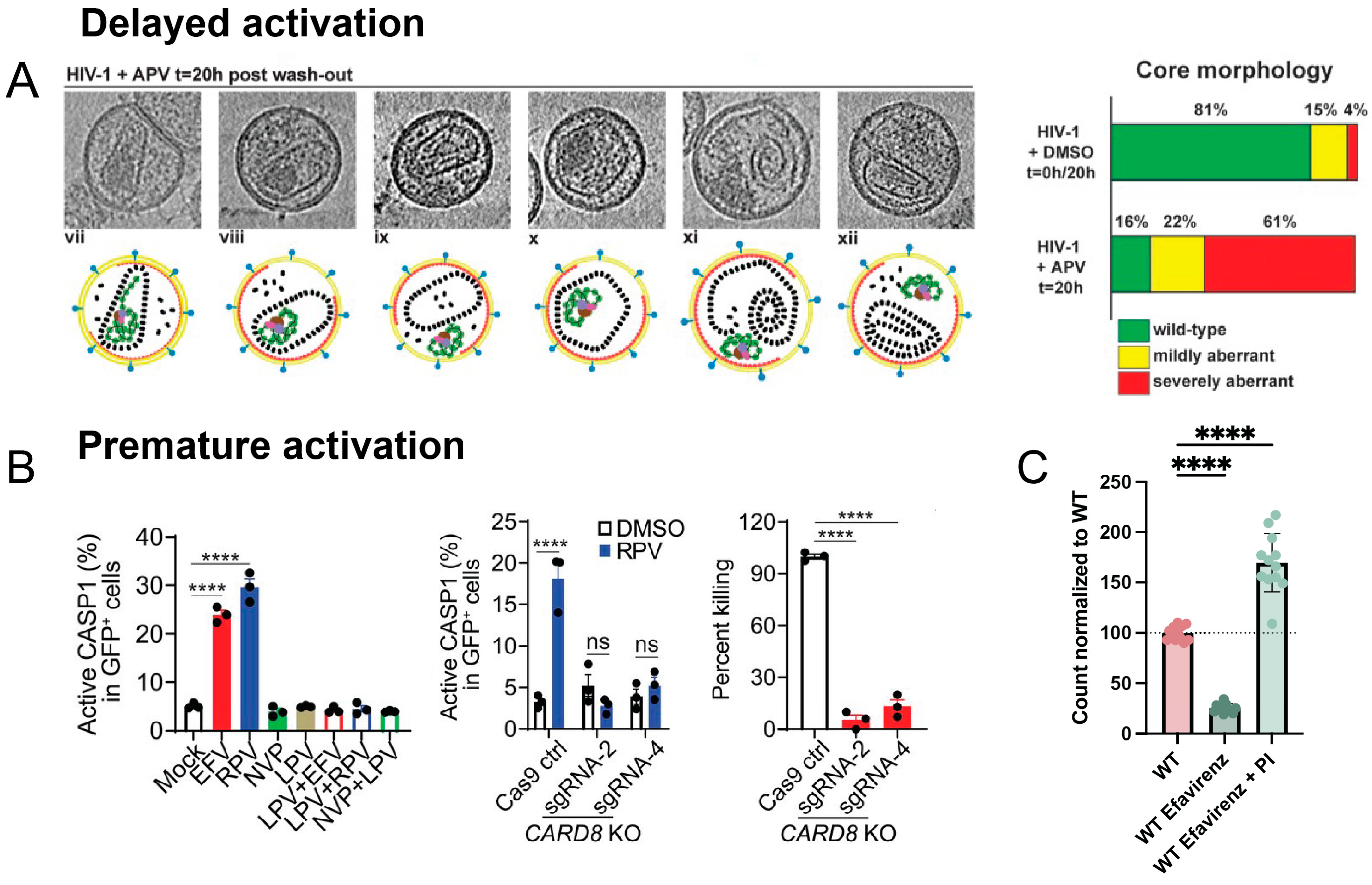
4.2. Delaying Protease Activation
4.3. Promoting Protease Activation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kohl, N.E.; Emini, E.A.; Schleif, W.A.; Davis, L.J.; Heimbach, J.C.; Dixon, R.A.; Scolnick, E.M.; Sigal, I.S. Active Human Immunodeficiency Virus Protease Is Required for Viral Infectivity. Proc. Natl. Acad. Sci. USA 1988, 85, 4686–4690. [Google Scholar] [CrossRef] [PubMed]
- Wlodawer, A.; Miller, M.; Jaskólski, M.; Sathyanarayana, B.K.; Baldwin, E.; Weber, I.T.; Selk, L.M.; Clawson, L.; Schneider, J.; Kent, S.B. Conserved Folding in Retroviral Proteases: Crystal Structure of a Synthetic HIV-1 Protease. Science 1989, 245, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Samal, A.B.; Vlach, J.; Saad, J.S. Solution Structure and Membrane Interaction of the Cytoplasmic Tail of HIV-1 Gp41 Protein. Structure 2017, 25, 1708–1718.e5. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, T.; Mammano, F.; Haseltine, W.A.; Göttlinger, H.G. Role of the Matrix Protein in the Virion Association of the Human Immunodeficiency Virus Type 1 Envelope Glycoprotein. J. Virol. 1994, 68, 1689–1696. [Google Scholar] [CrossRef]
- Freed, E.O.; Martin, M.A. Domains of the Human Immunodeficiency Virus Type 1 Matrix and Gp41 Cytoplasmic Tail Required for Envelope Incorporation into Virions. J. Virol. 1996, 70, 341–351. [Google Scholar] [CrossRef]
- Wyma, D.J.; Kotov, A.; Aiken, C. Evidence for a Stable Interaction of Gp41 with Pr55(Gag) in Immature Human Immunodeficiency Virus Type 1 Particles. J. Virol. 2000, 74, 9381–9387. [Google Scholar] [CrossRef]
- Kol, N.; Shi, Y.; Tsvitov, M.; Barlam, D.; Shneck, R.Z.; Kay, M.S.; Rousso, I. A Stiffness Switch in Human Immunodeficiency Virus. Biophys. J. 2007, 92, 1777–1783. [Google Scholar] [CrossRef]
- Chojnacki, J.; Staudt, T.; Glass, B.; Bingen, P.; Engelhardt, J.; Anders, M.; Schneider, J.; Müller, B.; Hell, S.W.; Kräusslich, H.-G. Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 2012, 338, 524–528. [Google Scholar] [CrossRef]
- Roy, N.H.; Chan, J.; Lambelé, M.; Thali, M. Clustering and Mobility of HIV-1 Env at Viral Assembly Sites Predict Its Propensity to Induce Cell-Cell Fusion. J. Virol. 2013, 87, 7516–7525. [Google Scholar] [CrossRef]
- Joyner, A.S.; Willis, J.R.; Crowe, J.E., Jr.; Aiken, C. Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles. PLoS Pathog. 2011, 7, e1002234. [Google Scholar] [CrossRef]
- Wyma, D.J.; Jiang, J.; Shi, J.; Zhou, J.; Lineberger, J.E.; Miller, M.D.; Aiken, C. Coupling of Human Immunodeficiency Virus Type 1 Fusion to Virion Maturation: A Novel Role of the Gp41 Cytoplasmic Tail. J. Virol. 2004, 78, 3429–3435. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Waithe, D.; Carravilla, P.; Huarte, N.; Galiani, S.; Enderlein, J.; Eggeling, C. Envelope Glycoprotein Mobility on HIV-1 Particles Depends on the Virus Maturation State. Nat. Commun. 2017, 8, 545. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Ablan, S.; Freed, E.O.; Tanaka, Y. Regulation of Human Immunodeficiency Virus Type 1 Env-Mediated Membrane Fusion by Viral Protease Activity. J. Virol. 2004, 78, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Aiken, C. Maturation-Dependent Human Immunodeficiency Virus Type 1 Particle Fusion Requires a Carboxyl-Terminal Region of the Gp41 Cytoplasmic Tail. J. Virol. 2007, 81, 9999–10008. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, X.; Satoh, T.; Gentili, M.; Cerboni, S.; Conrad, C.; Hurbain, I.; El Marjou, A.; Lacabaratz, C.; Lelièvre, J.-D.; Manel, N. The Capsids of HIV-1 and HIV-2 Determine Immune Detection of the Viral CDNA by the Innate Sensor CGAS in Dendritic Cells. Immunity 2013, 39, 1132–1142. [Google Scholar] [CrossRef]
- Rasaiyaah, J.; Tan, C.P.; Fletcher, A.J.; Price, A.J.; Blondeau, C.; Hilditch, L.; Jacques, D.A.; Selwood, D.L.; James, L.C.; Noursadeghi, M.; et al. HIV-1 Evades Innate Immune Recognition through Specific Cofactor Recruitment. Nature 2013, 503, 402–405. [Google Scholar] [CrossRef]
- Achuthan, V.; Perreira, J.M.; Sowd, G.A.; Puray-Chavez, M.; McDougall, W.M.; Paulucci-Holthauzen, A.; Wu, X.; Fadel, H.J.; Poeschla, E.M.; Multani, A.S.; et al. Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe 2018, 24, 392–404.e8. [Google Scholar] [CrossRef]
- Francis, A.C.; Melikyan, G.B. Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell Host Microbe 2018, 23, 536–548.e6. [Google Scholar] [CrossRef]
- Abram, M.E.; Parniak, M.A. Virion Instability of Human Immunodeficiency Virus Type 1 Reverse Transcriptase (RT) Mutated in the Protease Cleavage Site between RT P51 and the RT RNase H Domain. J. Virol. 2005, 79, 11952–11961. [Google Scholar] [CrossRef]
- Müller, T.G.; Zila, V.; Müller, B.; Kräusslich, H.-G. Nuclear Capsid Uncoating and Reverse Transcription of HIV-1. Annu. Rev. Virol. 2022, 9, 261–284. [Google Scholar] [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Kožíšek, M.; Lepšík, M.; Grantz Šašková, K.; Brynda, J.; Konvalinka, J.; Rezáčová, P. Thermodynamic and Structural Analysis of HIV Protease Resistance to Darunavir-Analysis of Heavily Mutated Patient-Derived HIV-1 Proteases. FEBS J. 2014, 281, 1834–1847. [Google Scholar] [CrossRef]
- Grantz Saskova, K.; Rezacova, P.; Brynda, J.; Kozisek, M.; Konvalinka, J. Structure of Wild-Type HIV Protease in Complex with Darunavir 2014. PDB. [CrossRef]
- Hornak, V.; Okur, A.; Rizzo, R.C.; Simmerling, C. HIV-1 Protease Flaps Spontaneously Open and Reclose in Molecular Dynamics Simulations. Proc. Natl. Acad. Sci. USA 2006, 103, 915–920. [Google Scholar] [CrossRef]
- Deshmukh, L.; Tugarinov, V.; Louis, J.M.; Clore, G.M. Binding Kinetics and Substrate Selectivity in HIV-1 Protease-Gag Interactions Probed at Atomic Resolution by Chemical Exchange NMR. Proc. Natl. Acad. Sci. USA 2017, 114, E9855–E9862. [Google Scholar] [CrossRef]
- Laco, G.S. HIV-1 Protease Substrate-Groove: Role in Substrate Recognition and Inhibitor Resistance. Biochimie 2015, 118, 90–103. [Google Scholar] [CrossRef]
- Lee, S.-K.; Potempa, M.; Kolli, M.; Özen, A.; Schiffer, C.A.; Swanstrom, R. Context Surrounding Processing Sites Is Crucial in Determining Cleavage Rate of a Subset of Processing Sites in HIV-1 Gag and Gag-Pro-Pol Polyprotein Precursors by Viral Protease. J. Biol. Chem. 2012, 287, 13279–13290. [Google Scholar] [CrossRef] [PubMed]
- Kontijevskis, A.; Wikberg, J.E.S.; Komorowski, J. Computational Proteomics Analysis of HIV-1 Protease Interactome. Proteins 2007, 68, 305–312. [Google Scholar] [CrossRef]
- Singh, O.; Su, E.C.-Y. Prediction of HIV-1 Protease Cleavage Site Using a Combination of Sequence, Structural, and Physicochemical Features. BMC Bioinform. 2016, 17, 478. [Google Scholar] [CrossRef]
- Shen, H.-B.; Chou, K.-C. HIVcleave: A Web-Server for Predicting Human Immunodeficiency Virus Protease Cleavage Sites in Proteins. Anal. Biochem. 2008, 375, 388–390. [Google Scholar] [CrossRef]
- Samant, N.; Nachum, G.; Tsepal, T.; Bolon, D.N.A. Sequence Dependencies and Biophysical Features Both Govern Cleavage of Diverse Cut-Sites by HIV Protease. Protein Sci. 2022, 31, e4366. [Google Scholar] [CrossRef]
- Prabu-Jeyabalan, M.; Nalivaika, E.; Schiffer, C.A. Substrate Shape Determines Specificity of Recognition for HIV-1 Protease: Analysis of Crystal Structures of Six Substrate Complexes. Structure 2002, 10, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Potempa, M.; Lee, S.-K.; Kurt Yilmaz, N.; Nalivaika, E.A.; Rogers, A.; Spielvogel, E.; Carter, C.W., Jr.; Schiffer, C.A.; Swanstrom, R. HIV-1 Protease Uses Bi-Specific S2/S2’ Subsites to Optimize Cleavage of Two Classes of Target Sites. J. Mol. Biol. 2018, 430, 5182–5195. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.C.; Lindquist, J.N.; Kaplan, A.H.; Swanstrom, R. Processing Sites in the Human Immunodeficiency Virus Type 1 (HIV-1) Gag-Pro-Pol Precursor Are Cleaved by the Viral Protease at Different Rates. Retrovirology 2005, 2, 66. [Google Scholar] [CrossRef]
- Pettit, S.C.; Everitt, L.E.; Choudhury, S.; Dunn, B.M.; Kaplan, A.H. Initial Cleavage of the Human Immunodeficiency Virus Type 1 GagPol Precursor by Its Activated Protease Occurs by an Intramolecular Mechanism. J. Virol. 2004, 78, 8477–8485. [Google Scholar] [CrossRef]
- Pettit, S.C.; Clemente, J.C.; Jeung, J.A.; Dunn, B.M.; Kaplan, A.H. Ordered Processing of the Human Immunodeficiency Virus Type 1 GagPol Precursor Is Influenced by the Context of the Embedded Viral Protease. J. Virol. 2005, 79, 10601–10607. [Google Scholar] [CrossRef]
- Centazzo, M.; Manganaro, L.; Alvisi, G. Cellular Targets of HIV-1 Protease: Just the Tip of the Iceberg? Viruses 2023, 15, 712. [Google Scholar] [CrossRef]
- Impens, F.; Timmerman, E.; Staes, A.; Moens, K.; Ariën, K.K.; Verhasselt, B.; Vandekerckhove, J.; Gevaert, K. A Catalogue of Putative HIV-1 Protease Host Cell Substrates. Biol. Chem. 2012, 393, 915–931. [Google Scholar] [CrossRef]
- Jurczyszak, D.; Zhang, W.; Terry, S.N.; Kehrer, T.; Bermúdez González, M.C.; McGregor, E.; Mulder, L.C.F.; Eckwahl, M.J.; Pan, T.; Simon, V. HIV Protease Cleaves the Antiviral M6A Reader Protein YTHDF3 in the Viral Particle. PLoS Pathog. 2020, 16, e1008305. [Google Scholar] [CrossRef]
- Yang, H.; Nkeze, J.; Zhao, R.Y. Effects of HIV-1 Protease on Cellular Functions and Their Potential Applications in Antiretroviral Therapy. Cell Biosci. 2012, 2, 32. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, H.; Clark, K.M.; Mugisha, C.S.; Davis, K.; Tang, J.P.; Harlan, G.H.; DeSelm, C.J.; Presti, R.M.; Kutluay, S.B.; et al. CARD8 Is an Inflammasome Sensor for HIV-1 Protease Activity. Science 2021, 371, eabe1707. [Google Scholar] [CrossRef]
- Strack, P.R.; Frey, M.W.; Rizzo, C.J.; Cordova, B.; George, H.J.; Meade, R.; Ho, S.P.; Corman, J.; Tritch, R.; Korant, B.D. Apoptosis Mediated by HIV Protease Is Preceded by Cleavage of Bcl-2. Proc. Natl. Acad. Sci. USA 1996, 93, 9571–9576. [Google Scholar] [CrossRef]
- Nie, Z.; Bren, G.D.; Vlahakis, S.R.; Schimnich, A.A.; Brenchley, J.M.; Trushin, S.A.; Warren, S.; Schnepple, D.J.; Kovacs, C.M.; Loutfy, M.R.; et al. Human Immunodeficiency Virus Type 1 Protease Cleaves Procaspase 8 in Vivo. J. Virol. 2007, 81, 6947–6956. [Google Scholar] [CrossRef]
- Koh, Y.; Matsumi, S.; Das, D.; Amano, M.; Davis, D.A.; Li, J.; Leschenko, S.; Baldridge, A.; Shioda, T.; Yarchoan, R.; et al. Potent Inhibition of HIV-1 Replication by Novel Non-Peptidyl Small Molecule Inhibitors of Protease Dimerization. J. Biol. Chem. 2007, 282, 28709–28720. [Google Scholar] [CrossRef]
- Rock, B.M.; Hengel, S.M.; Rock, D.A.; Wienkers, L.C.; Kunze, K.L. Characterization of Ritonavir-Mediated Inactivation of Cytochrome P450 3A4. Mol. Pharmacol. 2014, 86, 665–674. [Google Scholar] [CrossRef]
- De Meyer, S.; Hill, A.; Picchio, G.; DeMasi, R.; De Paepe, E.; de Béthune, M.-P. Influence of Baseline Protease Inhibitor Resistance on the Efficacy of Darunavir/Ritonavir or Lopinavir/Ritonavir in the TITAN Trial. J. Acquir. Immune Defic. Syndr. 2008, 49, 563–564. [Google Scholar] [CrossRef]
- Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIVS. Available online: https://www.ncbi.nlm.nih.gov/books/NBK586306/ (accessed on 12 September 2024).
- Todd, M.J.; Semo, N.; Freire, E. The Structural Stability of the HIV-1 Protease. J. Mol. Biol. 1998, 283, 475–488. [Google Scholar] [CrossRef]
- Tessmer, U.; Kräusslich, H.G. Cleavage of Human Immunodeficiency Virus Type 1 Proteinase from the N-Terminally Adjacent P6* Protein Is Essential for Efficient Gag Polyprotein Processing and Viral Infectivity. J. Virol. 1998, 72, 3459–3463. [Google Scholar] [CrossRef]
- Tang, C.; Louis, J.M.; Aniana, A.; Suh, J.-Y.; Clore, G.M. Visualizing Transient Events in Amino-Terminal Autoprocessing of HIV-1 Protease. Nature 2008, 455, 693–696. [Google Scholar] [CrossRef]
- Louis, J.M.; Clore, G.M.; Gronenborn, A.M. Autoprocessing of HIV-1 Protease Is Tightly Coupled to Protein Folding. Nat. Struct. Biol. 1999, 6, 868–875. [Google Scholar] [CrossRef]
- Agniswamy, J.; Sayer, J.M.; Weber, I.T.; Louis, J.M. Terminal Interface Conformations Modulate Dimer Stability Prior to Amino Terminal Autoprocessing of HIV-1 Protease. Biochemistry 2012, 51, 1041–1050. [Google Scholar] [CrossRef]
- Louis, J.M.; Aniana, A.; Weber, I.T.; Sayer, J.M. Inhibition of Autoprocessing of Natural Variants and Multidrug Resistant Mutant Precursors of HIV-1 Protease by Clinical Inhibitors. Proc. Natl. Acad. Sci. USA 2011, 108, 9072–9077. [Google Scholar] [CrossRef]
- Humpolíčková, J.; Weber, J.; Starková, J.; Mašínová, E.; Günterová, J.; Flaisigová, I.; Konvalinka, J.; Majerová, T. Inhibition of the Precursor and Mature Forms of HIV-1 Protease as a Tool for Drug Evaluation. Sci. Rep. 2018, 8, 10438. [Google Scholar] [CrossRef]
- Davis, D.A.; Soule, E.E.; Davidoff, K.S.; Daniels, S.I.; Naiman, N.E.; Yarchoan, R. Activity of Human Immunodeficiency Virus Type 1 Protease Inhibitors against the Initial Autocleavage in Gag-Pol Polyprotein Processing. Antimicrob. Agents Chemother. 2012, 56, 3620–3628. [Google Scholar] [CrossRef]
- Ludwig, C.; Leiherer, A.; Wagner, R. Importance of Protease Cleavage Sites within and Flanking Human Immunodeficiency Virus Type 1 Transframe Protein P6* for Spatiotemporal Regulation of Protease Activation. J. Virol. 2008, 82, 4573–4584. [Google Scholar] [CrossRef]
- Tabler, C.O.; Wegman, S.J.; Chen, J.; Shroff, H.; Alhusaini, N.; Tilton, J.C. The HIV-1 Viral Protease Is Activated during Assembly and Budding Prior to Particle Release. J. Virol. 2022, 96, e0219821. [Google Scholar] [CrossRef]
- Agniswamy, J.; Sayer, J.; Weber, I.; Louis, J. Crystal Structure of HIV Protease Model Precursor/Darunavir Complex 2012. [CrossRef]
- Partin, K.; Zybarth, G.; Ehrlich, L.; DeCrombrugghe, M.; Wimmer, E.; Carter, C. Deletion of Sequences Upstream of the Proteinase Improves the Proteolytic Processing of Human Immunodeficiency Virus Type 1. Proc. Natl. Acad. Sci. USA 1991, 88, 4776–4780. [Google Scholar] [CrossRef]
- Yu, F.-H.; Chou, T.-A.; Liao, W.-H.; Huang, K.-J.; Wang, C.-T. Gag-Pol Transframe Domain P6* Is Essential for HIV-1 Protease-Mediated Virus Maturation. PLoS ONE 2015, 10, e0127974. [Google Scholar] [CrossRef]
- Jacks, T.; Power, M.D.; Masiarz, F.R.; Luciw, P.A.; Barr, P.J.; Varmus, H.E. Characterization of Ribosomal Frameshifting in HIV-1 Gag-Pol Expression. Nature 1988, 331, 280–283. [Google Scholar] [CrossRef]
- Lee, S.-K.; Potempa, M.; Swanstrom, R. The Choreography of HIV-1 Proteolytic Processing and Virion Assembly. J. Biol. Chem. 2012, 287, 40867–40874. [Google Scholar] [CrossRef]
- Takagi, S.; Momose, F.; Morikawa, Y. FRET Analysis of HIV-1 Gag and GagPol Interactions. FEBS Open Bio 2017, 7, 1815–1825. [Google Scholar] [CrossRef]
- Hsieh, S.-H.; Yu, F.-H.; Huang, K.-J.; Wang, C.-T. HIV-1 Reverse Transcriptase Stability Correlates with Gag Cleavage Efficiency: Reverse Transcriptase Interaction Implications for Modulating Protease Activation. J. Virol. 2023, 97, e0094823. [Google Scholar] [CrossRef] [PubMed]
- Göttlinger, H.G.; Sodroski, J.G.; Haseltine, W.A. Role of Capsid Precursor Processing and Myristoylation in Morphogenesis and Infectivity of Human Immunodeficiency Virus Type 1. Proc. Natl. Acad. Sci. USA 1989, 86, 5781–5785. [Google Scholar] [CrossRef]
- Bryant, M.; Ratner, L. Myristoylation-Dependent Replication and Assembly of Human Immunodeficiency Virus 1. Proc. Natl. Acad. Sci. USA 1990, 87, 523–527. [Google Scholar] [CrossRef]
- Lee, Y.M.; Tian, C.J.; Yu, X.F. A Bipartite Membrane-Binding Signal in the Human Immunodeficiency Virus Type 1 Matrix Protein Is Required for the Proteolytic Processing of Gag Precursors in a Cell Type-Dependent Manner. J. Virol. 1998, 72, 9061–9068. [Google Scholar] [CrossRef]
- Bendjennat, M.; Saffarian, S. The Race against Protease Activation Defines the Role of ESCRTs in HIV Budding. PLoS Pathog. 2016, 12, e1005657. [Google Scholar] [CrossRef] [PubMed]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Côté, M.; Rich, R.L.; et al. Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef]
- Strack, B.; Calistri, A.; Craig, S.; Popova, E.; Göttlinger, H.G. AIP1/ALIX Is a Binding Partner for HIV-1 P6 and EIAV P9 Functioning in Virus Budding. Cell 2003, 114, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Orenstein, J.M.; Martin, M.A.; Freed, E.O. P6Gag Is Required for Particle Production from Full-Length Human Immunodeficiency Virus Type 1 Molecular Clones Expressing Protease. J. Virol. 1995, 69, 6810–6818. [Google Scholar] [CrossRef]
- Kaplan, A.H.; Manchester, M.; Swanstrom, R. The Activity of the Protease of Human Immunodeficiency Virus Type 1 Is Initiated at the Membrane of Infected Cells before the Release of Viral Proteins and Is Required for Release to Occur with Maximum Efficiency. J. Virol. 1994, 68, 6782–6786. [Google Scholar] [CrossRef]
- Kaplan, A.H.; Swanstrom, R. Human Immunodeficiency Virus Type 1 Gag Proteins Are Processed in Two Cellular Compartments. Proc. Natl. Acad. Sci. USA 1991, 88, 4528–4532. [Google Scholar] [CrossRef]
- Neefjes, J.; Dantuma, N.P. Fluorescent Probes for Proteolysis: Tools for Drug Discovery. Nat. Rev. Drug Discov. 2004, 3, 58–69. [Google Scholar] [CrossRef]
- Meng, J.; Lai, M.-T.; Munshi, V.; Grobler, J.; McCauley, J.; Zuck, P.; Johnson, E.N.; Uebele, V.N.; Hermes, J.D.; Adam, G.C. Screening of HIV-1 Protease Using a Combination of an Ultra-High-Throughput Fluorescent-Based Assay and RapidFire Mass Spectrometry. J. Biomol. Screen. 2015, 20, 606–615. [Google Scholar] [CrossRef]
- Gaber, R.; Majerle, A.; Jerala, R.; Benčina, M. Noninvasive High-Throughput Single-Cell Analysis of HIV Protease Activity Using Ratiometric Flow Cytometry. Sensors 2013, 13, 16330–16346. [Google Scholar] [CrossRef]
- Jin, S.; Ellis, E.; Veetil, J.V.; Yao, H.; Ye, K. Visualization of Human Immunodeficiency Virus Protease Inhibition Using a Novel Förster Resonance Energy Transfer Molecular Probe. Biotechnol. Prog. 2011, 27, 1107–1114. [Google Scholar] [CrossRef]
- Sood, C.; Francis, A.C.; Desai, T.M.; Melikyan, G.B. An Improved Labeling Strategy Enables Automated Detection of Single-Virus Fusion and Assessment of HIV-1 Protease Activity in Single Virions. J. Biol. Chem. 2017, 292, 20196–20207. [Google Scholar] [CrossRef]
- Briggs, J.A.G.; Grünewald, K.; Glass, B.; Förster, F.; Kräusslich, H.-G.; Fuller, S.D. The Mechanism of HIV-1 Core Assembly: Insights from Three-Dimensional Reconstructions of Authentic Virions. Structure 2006, 14, 15–20. [Google Scholar] [CrossRef]
- Selig, L.; Pages, J.C.; Tanchou, V.; Prévéral, S.; Berlioz-Torrent, C.; Liu, L.X.; Erdtmann, L.; Darlix, J.; Benarous, R.; Benichou, S. Interaction with the P6 Domain of the Gag Precursor Mediates Incorporation into Virions of Vpr and Vpx Proteins from Primate Lentiviruses. J. Virol. 1999, 73, 592–600. [Google Scholar] [CrossRef]
- Shu, X.; Remington, S.J. GFP/S205V Mutant 2008. PDB. [CrossRef]
- Shu, X.; Leiderman, P.; Gepshtein, R.; Smith, N.R.; Kallio, K.; Huppert, D.; Remington, S.J. An Alternative Excited-State Proton Transfer Pathway in Green Fluorescent Protein Variant S205V. Protein Sci. 2007, 16, 2703–2710. [Google Scholar] [CrossRef]
- Tabler, C.O.; Wegman, S.J.; Alhusaini, N.; Lee, N.F.; Tilton, J.C. Premature Activation of the HIV-1 Protease Is Influenced by Polymorphisms in the Hinge Region. Viruses 2024, 16, 849. [Google Scholar] [CrossRef]
- Tabler, C.O.; Tilton, J.C. Analysis of Individual Viral Particles by Flow Virometry. Viruses 2024, 16, 802. [Google Scholar] [CrossRef]
- Vestad, B.; Llorente, A.; Neurauter, A.; Phuyal, S.; Kierulf, B.; Kierulf, P.; Skotland, T.; Sandvig, K.; Haug, K.B.F.; Øvstebø, R. Size and Concentration Analyses of Extracellular Vesicles by Nanoparticle Tracking Analysis: A Variation Study. J. Extracell. Vesicles 2017, 6, 1344087. [Google Scholar] [CrossRef] [PubMed]
- Bonar, M.M.; Tabler, C.O.; Haqqani, A.A.; Lapointe, L.E.; Galiatsos, J.A.; Joussef-Piña, S.; Quiñones-Mateu, M.E.; Tilton, J.C. Nanoscale Flow Cytometry Reveals Interpatient Variability in HIV Protease Activity That Correlates with Viral Infectivity and Identifies Drug-Resistant Viruses. Sci. Rep. 2020, 10, 18101. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, P.V.; Chelius, D.; Shaler, T.A. Identification and Relative Quantitation of Protein Mixtures by Enzymatic Digestion Followed by Capillary Reversed-Phase Liquid Chromatography-Tandem Mass Spectrometry. Anal. Chem. 2002, 74, 4741–4749. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Cantone, J.; Lu, H.; Nowicka-Sans, B.; Protack, T.; Yuan, T.; Yang, H.; Liu, Z.; Drexler, D.; Regueiro-Ren, A.; et al. Mechanistic Studies and Modeling Reveal the Origin of Differential Inhibition of Gag Polymorphic Viruses by HIV-1 Maturation Inhibitors. PLoS Pathog. 2016, 12, e1005990. [Google Scholar] [CrossRef]
- Schimer, J.; Pávová, M.; Anders, M.; Pachl, P.; Šácha, P.; Cígler, P.; Weber, J.; Majer, P.; Řezáčová, P.; Kräusslich, H.-G.; et al. Triggering HIV Polyprotein Processing by Light Using Rapid Photodegradation of a Tight-Binding Protease Inhibitor. Nat. Commun. 2015, 6, 6461. [Google Scholar] [CrossRef]
- Dale, B.M.; McNerney, G.P.; Thompson, D.L.; Hubner, W.; de Los Reyes, K.; Chuang, F.Y.S.; Huser, T.; Chen, B.K. Cell-to-Cell Transfer of HIV-1 via Virological Synapses Leads to Endosomal Virion Maturation That Activates Viral Membrane Fusion. Cell Host Microbe 2011, 10, 551–562. [Google Scholar] [CrossRef]
- Neil, S.J.D.; Zang, T.; Bieniasz, P.D. Tetherin Inhibits Retrovirus Release and Is Antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.H.; Zack, J.A.; Knigge, M.; Paul, D.A.; Kempf, D.J.; Norbeck, D.W.; Swanstrom, R. Partial Inhibition of the Human Immunodeficiency Virus Type 1 Protease Results in Aberrant Virus Assembly and the Formation of Noninfectious Particles. J. Virol. 1993, 67, 4050–4055. [Google Scholar] [CrossRef]
- Mattei, S.; Anders, M.; Konvalinka, J.; Kräusslich, H.-G.; Briggs, J.A.G.; Müller, B. Induced Maturation of Human Immunodeficiency Virus. J. Virol. 2014, 88, 13722–13731. [Google Scholar] [CrossRef]
- Arrigo, S.J.; Huffman, K. Potent Inhibition of Human Immunodeficiency Virus Type 1 (HIV-1) Replication by Inducible Expression of HIV-1 PR Multimers. J. Virol. 1995, 69, 5988–5994. [Google Scholar] [CrossRef]
- Kräusslich, H.G. Human Immunodeficiency Virus Proteinase Dimer as Component of the Viral Polyprotein Prevents Particle Assembly and Viral Infectivity. Proc. Natl. Acad. Sci. USA 1991, 88, 3213–3217. [Google Scholar] [CrossRef] [PubMed]
- Trinité, B.; Zhang, H.; Levy, D.N. NNRTI-Induced HIV-1 Protease-Mediated Cytotoxicity Induces Rapid Death of CD4 T Cells during Productive Infection and Latency Reversal. Retrovirology 2019, 16, 17. [Google Scholar] [CrossRef]
- Jochmans, D.; Anders, M.; Keuleers, I.; Smeulders, L.; Kräusslich, H.-G.; Kraus, G.; Müller, B. Selective Killing of Human Immunodeficiency Virus Infected Cells by Non-Nucleoside Reverse Transcriptase Inhibitor-Induced Activation of HIV Protease. Retrovirology 2010, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Karacostas, V.; Wolffe, E.J.; Nagashima, K.; Gonda, M.A.; Moss, B. Overexpression of the HIV-1 Gag-Pol Polyprotein Results in Intracellular Activation of HIV-1 Protease and Inhibition of Assembly and Budding of Virus-like Particles. Virology 1993, 193, 661–671. [Google Scholar] [CrossRef]
- Park, J.; Morrow, C.D. Overexpression of the Gag-Pol Precursor from Human Immunodeficiency Virus Type 1 Proviral Genomes Results in Efficient Proteolytic Processing in the Absence of Virion Production. J. Virol. 1991, 65, 5111–5117. [Google Scholar] [CrossRef] [PubMed]
- Anokhina, V.S.; McAnany, J.D.; Ciesla, J.H.; Hilimire, T.A.; Santoso, N.; Miao, H.; Miller, B.L. Enhancing the Ligand Efficiency of Anti-HIV Compounds Targeting Frameshift-Stimulating RNA. Bioorg. Med. Chem. 2019, 27, 2972–2977. [Google Scholar] [CrossRef]
- Brakier-Gingras, L.; Charbonneau, J.; Butcher, S.E. Targeting Frameshifting in the Human Immunodeficiency Virus. Expert Opin. Ther. Targets 2012, 16, 249–258. [Google Scholar] [CrossRef]
- Rheinemann, L.; Downhour, D.M.; Bredbenner, K.; Mercenne, G.; Davenport, K.A.; Schmitt, P.T.; Necessary, C.R.; McCullough, J.; Schmitt, A.P.; Simon, S.M.; et al. RetroCHMP3 Blocks Budding of Enveloped Viruses without Blocking Cytokinesis. Cell 2021, 184, 5419–5431.e16. [Google Scholar] [CrossRef]
- Figueiredo, A.; Moore, K.L.; Mak, J.; Sluis-Cremer, N.; de Bethune, M.-P.; Tachedjian, G. Potent Nonnucleoside Reverse Transcriptase Inhibitors Target HIV-1 Gag-Pol. PLoS Pathog. 2006, 2, e119. [Google Scholar] [CrossRef]
- Balibar, C.J.; Klein, D.J.; Zamlynny, B.; Diamond, T.L.; Fang, Z.; Cheney, C.A.; Kristoff, J.; Lu, M.; Bukhtiyarova, M.; Ou, Y.; et al. Potent Targeted Activator of Cell Kill Molecules Eliminate Cells Expressing HIV-1. Sci. Transl. Med. 2023, 15, eabn2038. [Google Scholar] [CrossRef]
- Clark, K.M.; Kim, J.G.; Wang, Q.; Gao, H.; Presti, R.M.; Shan, L. Chemical Inhibition of DPP9 Sensitizes the CARD8 Inflammasome in HIV-1-Infected Cells. Nat. Chem. Biol. 2023, 19, 431–439. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabler, C.O.; Tilton, J.C. Mechanism and Kinetics of HIV-1 Protease Activation. Viruses 2024, 16, 1826. https://doi.org/10.3390/v16121826
Tabler CO, Tilton JC. Mechanism and Kinetics of HIV-1 Protease Activation. Viruses. 2024; 16(12):1826. https://doi.org/10.3390/v16121826
Chicago/Turabian StyleTabler, Caroline O., and John C. Tilton. 2024. "Mechanism and Kinetics of HIV-1 Protease Activation" Viruses 16, no. 12: 1826. https://doi.org/10.3390/v16121826
APA StyleTabler, C. O., & Tilton, J. C. (2024). Mechanism and Kinetics of HIV-1 Protease Activation. Viruses, 16(12), 1826. https://doi.org/10.3390/v16121826




