Fusarium sacchari hypovirus 1, a Member of Hypoviridae with Virulence Attenuation Capacity in Phytopathogenic Fusarium Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Sugarcane Plants
2.2. Isolation and Identification of Fusarium Species
2.3. Extraction of DNA and RNA
2.4. Purification of dsRNA, cDNA Synthesis, and RT-PCR
2.5. Sequence Assembly, Alignment, and Phylogenetic Analysis
2.6. Horizontal Transmission of the Viruses
2.7. Measurement of Phenotypic Traits
3. Results
3.1. Identification of Fusarium Species Containing FsHV1
3.2. Comparative Analysis of Full-Length Sequences of 12 FsHV1 Variants
3.3. Comparative Analysis of the Sequences of Six FsHV1-Defective RNAs
3.4. FsHV1 Virus Reduces Sporulation, Pigmentation, and Virulence of the Host Fungus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- SK, V. Pokkah Boeng: An Emerging Disease of Sugarcane. J. Plant Pathol. Microbiol. 2013, 4, 3. [Google Scholar] [CrossRef]
- Bao, Y.; Xu, Y.; Wang, S.; Yao, Z.; Rao, G.P.; Zhang, M. First Report of Fusarium sacchari that Causes Sugarcane Wilt Disease in China. Plant Dis. 2020, 8, 104. [Google Scholar] [CrossRef]
- Meng, J.; Haung, H.; Li, Y.; Li, Y.; Chen, B. First Report of Fusarium sacchari Causing Sugarcane Pokkah Boeng in China. Plant Dis. 2019, 5, 104. [Google Scholar] [CrossRef]
- Bao, Y.; Huang, Z.; Li, T.; Yao, Z.; Powell, C.A.; Zhang, M. First Report of Fusarium andiyazi Causing Sugarcane Pokkah Boeng Disease in China. Plant Dis. 2019, 1, 104. [Google Scholar] [CrossRef]
- Bao, Y.; Sun, H.; Li, Y.; Duan, Z.; Zhang, M. First Report of Fusarium oxysporum isolate gx3 Causing Sugarcane Pokkah Boeng in Guangxi of China. Plant Dis. 2016, 8, 100. [Google Scholar] [CrossRef]
- Lin, Z.; Xu, S.; Que, Y.; Wang, J.; Comstock, J.C.; Wei, J.; McCord, P.H.; Chen, B.; Chen, R.; Zhang, M. Species-Specific Detection and Identification of Fusarium Species Complex, the Causal Agent of Sugarcane Pokkah Boeng in China. PLoS ONE 2014, 9, e104195. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Abd, M.N.; Mohamed, N.N.; Shohaimi, S.; Mohd, Z.N. Genetic Diversity and Pathogenicity of Fusarium Species Associated with Fruit Rot Disease in Banana across Peninsular Malaysia. J. Appl. Microbiol. 2017, 123, 1533–1546. [Google Scholar]
- Xu, S.; Wang, J.; Wang, H.; Bao, Y.; Li, Y.; Govindaraju, M.; Yao, W.; Chen, B.; Zhang, M. Molecular Characterization of Carbendazim Resistance of Fusarium Species Complex that Causes Sugarcane Pokkah Boeng Disease. BMC Genom. 2019, 20, 115. [Google Scholar] [CrossRef]
- Xie, J.; Jiang, D. New Insights into Mycoviruses and Exploration for the Biological Control of Crop Fungal Diseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Suzuki, N. Viruses of Plant Pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, B.; Fu, Y.; Jiang, D.; Ghabrial, S.A.; Li, G.; Peng, Y.; Xie, J.; Cheng, J.; Huang, J.; et al. A Geminivirus-Related DNA Mycovirus that Confers Hypovirulence to a Plant Pathogenic Fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef] [PubMed]
- Torres-Trenas, A.; Canizares, M.C.; Garcia-Pedrajas, M.D.; Perez-Artes, E. Molecular and Biological Characterization of the First Hypovirus Identified in Fusarium Oxysporum. Front. Microbiol. 2019, 10, 3131. [Google Scholar] [CrossRef]
- Wang, S.; Kondo, H.; Liu, L.; Guo, L.; Qiu, D. A Novel Virus in the Family Hypoviridae from the Plant Pathogenic Fungus Fusarium graminearum. Virus Res. 2013, 174, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Yaegashi, H.; Kanematsu, S.; Ito, T. Molecular Characterization of a New Hypovirus Infecting a Phytopathogenic Fungus, Valsa ceratosperma. Virus Res. 2012, 165, 150. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Biological Control of Chestnut Blight: Biological control of Chestnut Blight: An Example of Virus-Mediated Attenuation of Fungal Pathogenesis. Microbiol. Rev. 1992, 56, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Kanematsu, S.; Arakawa, M.; Suzaki, K.; Yoshida, K.; Matsumoto, N.; Oikawa, Y.; Onoue, M.; Osaki, H.; Nakamura, H.; Ikeda, K.; et al. A Reovirus Causes Hypovirulence of Rosellinia necatrix. Phytopathology 2004, 94, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, B.; Fu, Y.; Xie, J.; Cheng, J.; Ghabrial, S.A.; Li, G.; Yi, X.; Jiang, D. Extracellular Transmission of a DNA Mycovirus and its use as a Natural Fungicide. Proc. Natl. Acad. Sci. USA 2013, 110, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, M.E.P.M. Characterisation of a Novel Hypovirus from Sclerotinia sclerotiorum Potentially Representing a New Genus within the Hypoviridae. Virology 2014, 464–465, 441–449. [Google Scholar] [CrossRef]
- Shamsi, W.; Mittelstrass, J.; Ulrich, S.; Kondo, H.; Rigling, D.; Prospero, S. Possible Biological Control of Ash Dieback using the Mycoparasite Hymenoscyphus fraxineus mitovirus 2. Phytopathology 2023. [Google Scholar] [CrossRef]
- Li, P.; Lin, Y.; Zhang, H.; Wang, S.; Qiu, D.; Guo, L. Molecular Characterization of a Novel Mycovirus of the Family Tymoviridae Isolated from the plant Pathogenic Fungus Fusarium graminearum. Virology 2016, 489, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.M.; Jeon, J.J.; Yea, S.J.; Kim, Y.H.; Yun, S.H.; Lee, Y.W.; Kim, K.H. Double-Stranded RNA Mycovirus from Fusarium graminearum. Appl. Environ. Microbiol. 2002, 5, 2529–2534. [Google Scholar] [CrossRef] [PubMed]
- Lemus-Minor, C.G.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Fusarium oxysporum f. sp. dianthi virus 1 Accumulation Is Correlated with Changes in Virulence and Other Phenotypic Traits of Its Fungal Host. Phytopathology 2018, 108, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Velasco, L.; Ayllon, M.A.; Suzuki, N.; Lee-Marzano, S.Y.; Sun, L.; Sabanadzovic, S.; Turina, M. ICTV Virus Taxonomy Profile: Hypoviridae 2023. J. Gen. Virol. 2023, 104. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Zou, C.; Peng, N.; Zhu, Y.; Bao, Y.; Zhou, Q.; Wu, Q.; Chen, B.; Zhang, M. Virome Identification and Characterization of Fusarium sacchari and F. andiyazi: Causative Agents of Pokkah Boeng Disease in Sugarcane. Front. Microbiol. 2020, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.; Wei, W. Comparing Different Methods for lsolating Fusarium from Soybean Rhizosphere Soil. Soybean Sci. 2008, 27, 106–112. [Google Scholar]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple Evolutionary Origins of the Fungus Causing Panama Disease of Banana: Concordant Evidence from Nuclear and Mitochondrial Gene Genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Frandsen, R.J.N.; Lysøe, E.; Rehner, S.A.; Aoki, T.; et al. Phylogenetic Analyses of RPB1 and RPB2 Support a Middle Cretaceous Origin for a Clade Comprising all Agriculturally and Medically Important Fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic Relationships among Ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Suzuki, N.; Nuss, D.L. Contribution of Protein p40 to Hypovirus-Mediated Modulation of Fungal Host Phenotype and Viral RNA accumulation. J. Virol. 2002, 76, 7747–7759. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, H.; Guo, Q.; Xu, S.; Wang, J.; Lin, S.; Zhang, M. Artificial Inoculation Method of Pokkah Boeng Disease of Sugarcane and Screening of Resistant Germplasm Resources in Subtropical China. Sugar Tech 2017, 19, 283–292. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. F. sacchari. In The Fusarium Laboratory Manual; Blackwell Publishing: Ames, IA, USA, 2006; pp. 240–241. [Google Scholar]
- Leslie, J.F.; Summerell, B.A. F. andiyazi. In The Fusarium Laboratory Manual; Blackwell Publishing: Ames, IA, USA, 2006; pp. 126–127. [Google Scholar]
- Leslie, J.F.; Summerell, B.A. F. solani. In The Fusarium Laboratory Manual; Blackwell Publishing: Ames, IA, USA, 2006; pp. 250–254. [Google Scholar]
- Sun, A.; Zhao, L.; Sun, Y.; Chen, Y.; Li, C.; Dong, W.; Yang, G. Horizontal and Vertical Transmission of a Mycovirus Closely Related to the Partitivirus RhsV717 That Confers Hypovirulence in Rhizoctonia solani. Viruses 2023, 15, 2088. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Hypovirulence: Mycoviruses at the Fungal–Plant interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, C.; Hua, H.; Cai, Q.; Wu, X. Characterization of the First Alternavirus Identified in Fusarium Avenaceum, the Causal Agent of Potato Dry Rot. Viruses 2023, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Vignuzzi, M.; López, C.B. Defective Viral Genomes are Key Drivers of the Virus–Host Interaction. Nat. Microbiol. 2019, 4, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Lin, Y.; Kondo, H.; Kanematsu, S.; Suzuki, N. Effects of Defective Interfering RNA on Symptom Induction by, and Replication of, a Novel Partitivirus from a Phytopathogenic fungus, Rosellinia necatrix. J. Virol. 2013, 87, 2330–2341. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lott, W.B.; Lowry, K.; Jones, A.; Thu, H.M.; Aaskov, J.; Coffey, L.L. Defective Interfering Viral Particles in Acute Dengue Infections. PLoS ONE 2011, 6, e19447. [Google Scholar] [CrossRef] [PubMed]
- Tapia, K.; Kim, W.K.; Sun, Y.; Mercado-Lopez, X.; Dunay, E.; Wise, M.; Adu, M.; Lopez, C.B. Defective Viral Genomes Arising in Vivo Provide Critical Danger Signals for the Triggering of Lung Antiviral Immunity. PLoS Pathog. 2013, 9, e1003703. [Google Scholar] [CrossRef] [PubMed]
- Shapira, R.; Choi, G.H.; Hillman, B.I.; Nuss, D.L. The Contribution of Defective RNAs to the Complexity of Viral-Encoded Double-Stranded RNA Populations Present in Hypovirulent Strains of the Chestnut Blight Fungus Cryphonectria parasitica. EMBO J. 1991, 10, 741–746. [Google Scholar] [CrossRef]
- Hillman, B.I.; Foglia, R.; Yuan, W. Satellite and Defective RNAs of Cryphonectria Hypovirus 3-Grand Haven 2, a Virus Species in the Family Hypoviridae with a Single Open Reading Frame. Virology 2000, 276, 181–189. [Google Scholar] [CrossRef]
- Li, H.; Bian, R.; Liu, Q.; Yang, L.; Pang, T.; Salaipeth, L.; Andika, I.B.; Kondo, H.; Sun, L. Identification of a Novel Hypovirulence-Inducing Hypovirus from Alternaria alternata. Front. Microbiol. 2019, 10, 1076. [Google Scholar] [CrossRef] [PubMed]
- Mahillon, M.; Decroës, A.; Caulier, S.; Tiendrebeogo, A.; Legrève, A.; Bragard, C. Genomic and Biological Characterization of a Novel Partitivirus Infecting Fusarium equiseti. Virus Res. 2021, 297, 198386. [Google Scholar] [CrossRef] [PubMed]
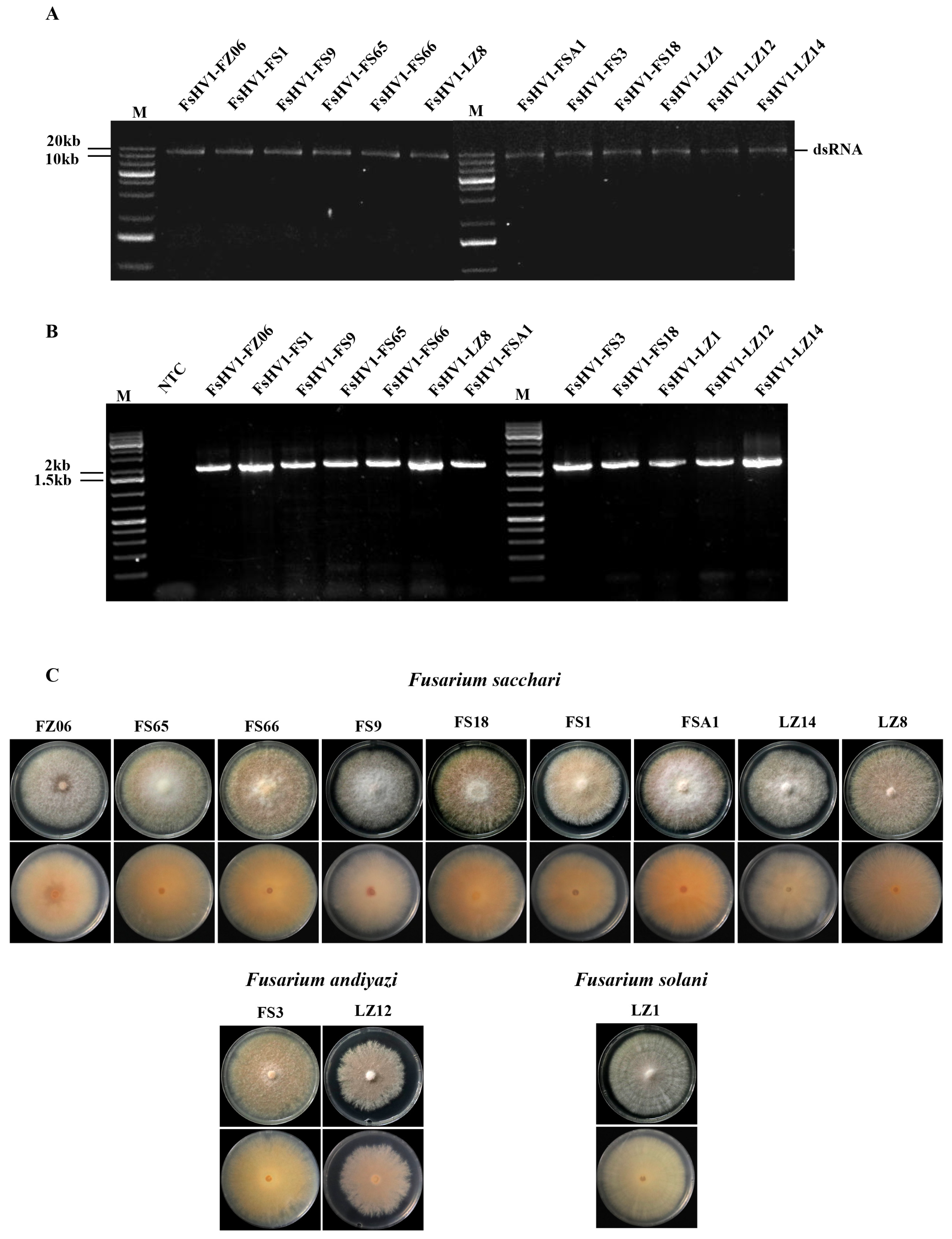
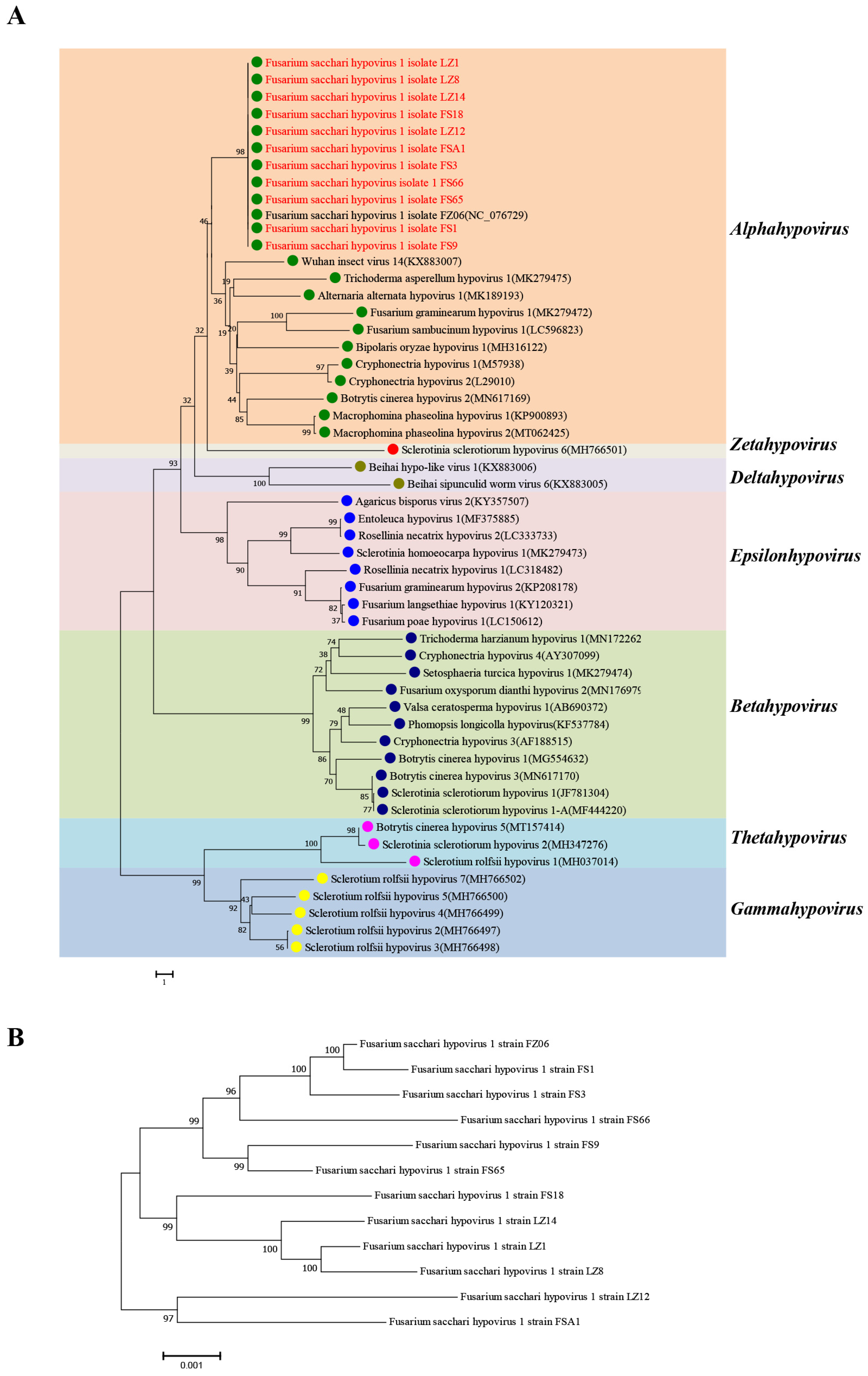

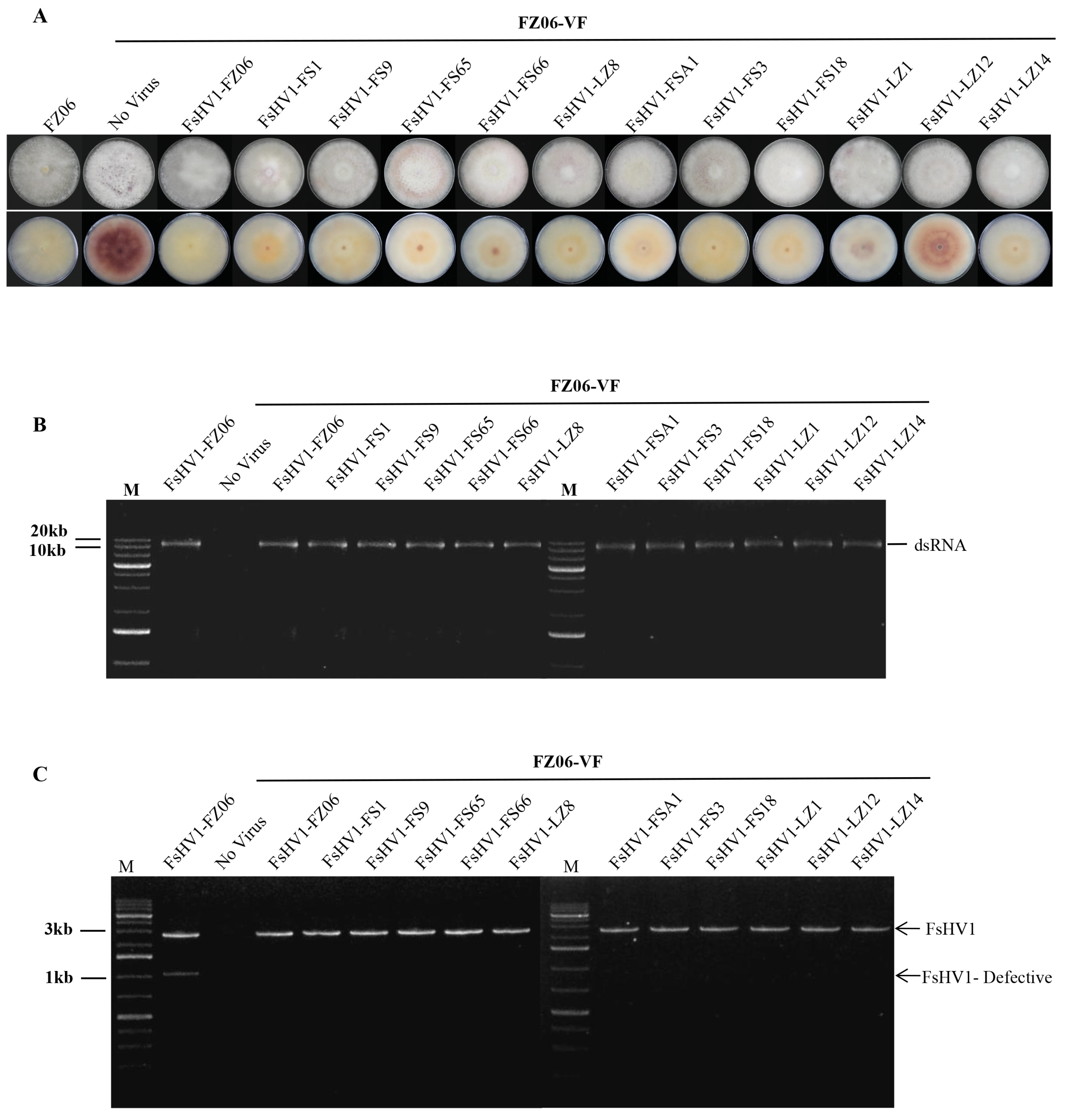
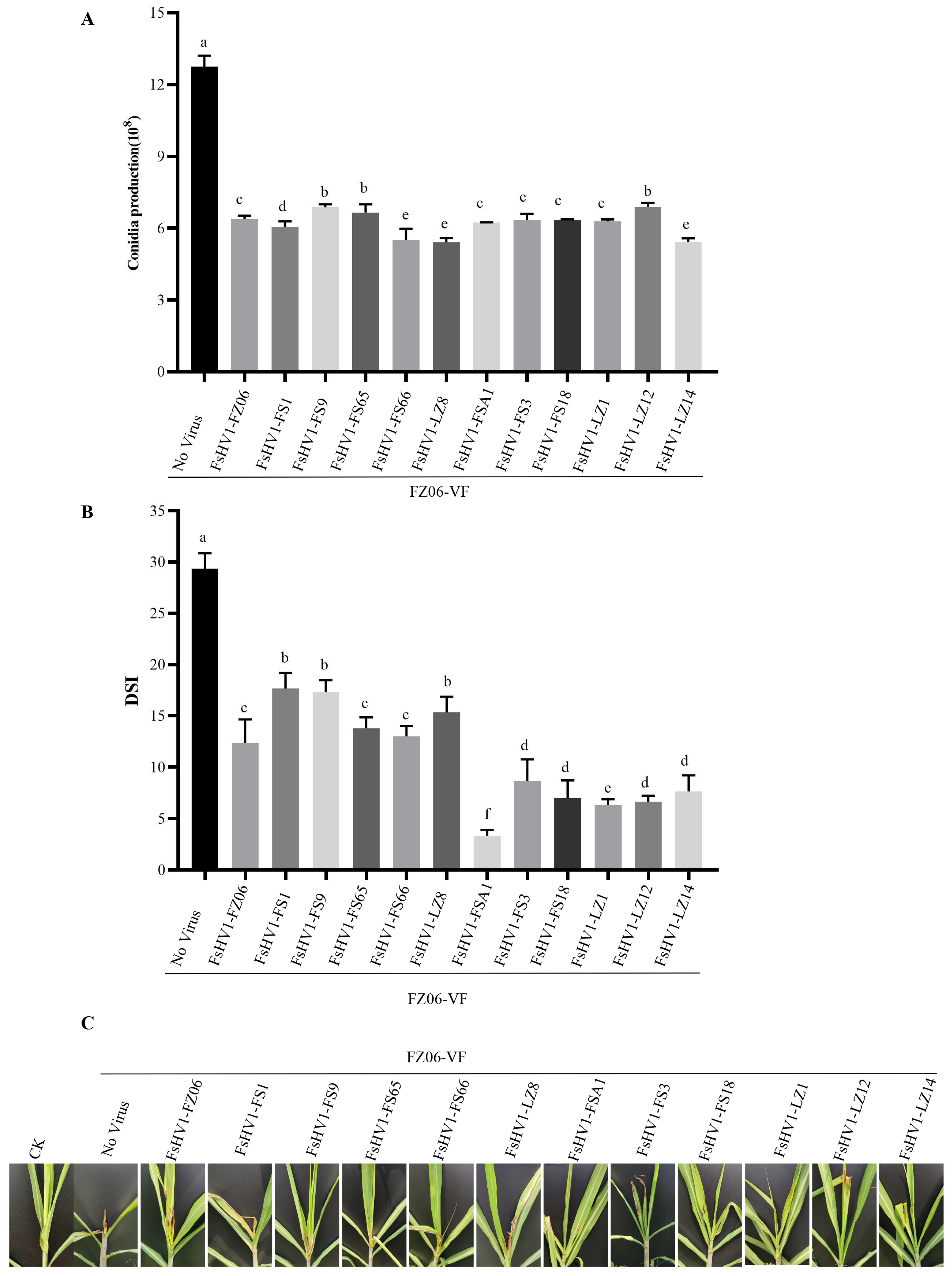
| No. | Name of FsHV1 Isolate | Abbreviation | Genome Size | 5′ UTR | 3′ UTR |
|---|---|---|---|---|---|
| 1 | Fusarium sacchari hypovirus 1 isolate FZ06 | FsHV1-FZ06 | 13,975 bp | 735 | 466 |
| 2 | Fusarium sacchari hypovirus 1 isolate FS65 | FsHV1-FS65 | 13,983 bp | 735 | 474 |
| 3 | Fusarium sacchari hypovirus 1 isolate FS66 | FsHV1-FS66 | 13,980 bp | 735 | 471 |
| 4 | Fusarium sacchari hypovirus 1 isolate FS9 | FsHV1-FS9 | 13,979 bp | 735 | 470 |
| 5 | Fusarium sacchari hypovirus 1 isolate FS18 | FsHV1-FS18 | 13,981 bp | 735 | 472 |
| 6 | Fusarium sacchari hypovirus 1 isolate FS1 | FsHV1-FS1 | 13,975 bp | 735 | 466 |
| 7 | Fusarium sacchari hypovirus 1 isolate FSA1 | FsHV1-FSA1 | 13,966 bp | 735 | 458 |
| 8 | Fusarium sacchari hypovirus 1 isolate LZ14 | FsHV1-LZ14 | 13,980 bp | 735 | 471 |
| 9 | Fusarium sacchari hypovirus 1 isolate LZ8 | FsHV1-LZ8 | 13,982 bp | 735 | 473 |
| 10 | Fusarium sacchari hypovirus 1 isolate FS3 | FsHV1-FS3 | 13,982 bp | 734 | 474 |
| 11 | Fusarium sacchari hypovirus 1 isolate LZ12 | FsHV1-LZ12 | 13,967 bp | 735 | 458 |
| 12 | Fusarium sacchari hypovirus 1 isolate LZ1 | FsHV1-LZ1 | 13,971 bp | 735 | 462 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Yao, Z.; Cao, X.; Chen, Y.; Zou, C.; Chen, B. Fusarium sacchari hypovirus 1, a Member of Hypoviridae with Virulence Attenuation Capacity in Phytopathogenic Fusarium Species. Viruses 2024, 16, 608. https://doi.org/10.3390/v16040608
Zhou Q, Yao Z, Cao X, Chen Y, Zou C, Chen B. Fusarium sacchari hypovirus 1, a Member of Hypoviridae with Virulence Attenuation Capacity in Phytopathogenic Fusarium Species. Viruses. 2024; 16(4):608. https://doi.org/10.3390/v16040608
Chicago/Turabian StyleZhou, Qiujuan, Ziting Yao, Xueying Cao, Yuejia Chen, Chengwu Zou, and Baoshan Chen. 2024. "Fusarium sacchari hypovirus 1, a Member of Hypoviridae with Virulence Attenuation Capacity in Phytopathogenic Fusarium Species" Viruses 16, no. 4: 608. https://doi.org/10.3390/v16040608
APA StyleZhou, Q., Yao, Z., Cao, X., Chen, Y., Zou, C., & Chen, B. (2024). Fusarium sacchari hypovirus 1, a Member of Hypoviridae with Virulence Attenuation Capacity in Phytopathogenic Fusarium Species. Viruses, 16(4), 608. https://doi.org/10.3390/v16040608







