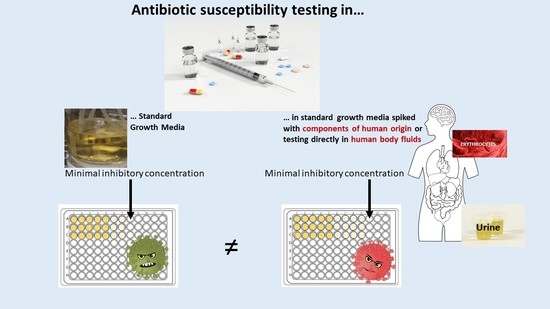
Use of Supplemented or Human Material to Simulate PD Behavior of Antibiotics at the Target Site In Vitro
Abstract
:1. Introduction
2. Complexity of Human Compartments and Their Physiologic Composition
3. Published In Vitro Studies
3.1. Adapted Growth Media
3.2. Body Fluids as Growth Media
4. Standard Practice Methods: Pros and Con for Adapted Media
4.1. MIC and MBC
4.2. TKC
5. Outlook
6. Summary
7. Methods
Literature Search
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Freires, I.A.; Sardi, J.D.C.O.; de Castro, R.D.; Rosalen, P.L. Alternative Animal and Non-Animal Models for Drug Discovery and Development: Bonus or Burden? Pharm. Res. 2017, 34, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lepak, A.J.; Andes, D.R. Animal models in the pharmacokinetic/pharmacodynamic evaluation of antimicrobial agents. Bioorganic Med. Chem. 2016, 24, 6390–6400. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Müller, M.; Derendorf, H. Rational dosing of antibiotics: The use of plasma concentrations versus tissue concentrations. Int. J. Antimicrob. Agents 2002, 19, 285–290. [Google Scholar] [CrossRef]
- Kumaraswamy, M.; Lin, L.; Olson, J.; Sun, C.F.; Nonejuie, P.; Corriden, R.; Döhrmann, S.; Ali, S.R.; Amaro, D.; Rohde, M.; et al. Standard susceptibility testing overlooks potent azithromycin activity and cationic peptide synergy against MDR Stenotrophomonas maltophilia. J. Antimicrob Chemother. 2016, 71, 1264–1269. [Google Scholar] [CrossRef] [Green Version]
- Faller, A.; Schünke, M. Der Körper des Menschen, 15 Auflage; Thieme Verlag: Stuttgart, Germany, 2009. [Google Scholar]
- Peterson, L.R.; Shanholtzer, C.J. Tests for bactericidal effects of antimicrobial agents: Technical performance and clinical relevance. Clin. Microbiol. Rev. 1992, 5, 420–432. [Google Scholar] [CrossRef]
- Horn, F. Bicohemie des Menschen, 6 Auflage; Thieme Verlag: Stuttgart, Germany, 2015. [Google Scholar]
- Neumeister, B.; Geiss, H.K.; Braun, R.W.; Kimming, P. Mikrobiologische Diagnostik; Bakteriologie, Mykologie, Virologie, Parasitologie, 2 Auflage; Thieme Verlag: Stuttgart, Germany, 2009. [Google Scholar]
- General Hospital Vienna, Klin. Abteilung für Medizinische und Chemische Labordiagnostik. 2018. Referenzliste. Available online: https://www.akhwien.at/default.aspx?pid=3986 (accessed on 13 June 2018).
- Zhanel, G.G.; Karlowsky, J.A.; Davidson, R.J.; Hoban, D.J. Effect of pooled human cerebrospinal fluid on the postantibiotic effects of cefotaxime, ciprofloxacin, and gentamicin against Escherichia coli. Antimicrob. Agents Chemother. 1992, 36, 1136–1139. [Google Scholar] [CrossRef] [Green Version]
- Matzneller, P.; Burian, A.; Zeitlinger, M.; Sauermann, R. Understanding the Activity of Antibiotics in Cerebrospinal Fluid in vitro. Pharmacology 2016, 97, 233–244. [Google Scholar] [CrossRef]
- Bicer, E.M. Compositional Characterisation of Human Respiratory Tract Lining Fluids for the Design of Disease Specific Simulants; King’s College London: London, UK, 2014. [Google Scholar]
- CIBA-Geigy AG. Pharmazeutische industrie, chemische industrie and NANWTG (1977). In Wissenschaftliche Tabellen Geigy; CIBA-Geigy AG: Basel, Switzerland, 1979. [Google Scholar]
- Zeitlinger, M.A.; Derendorf, H.; Mouton, J.W.; Cars, O.; Craig, W.A.; Andes, D.; Theuretzbacher, U. Protein binding: Do we ever learn? Antimicrob. Agents Chemother. 2011, 55, 3067–3074. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.; Gonzalez, D.; Derendorf, H. Significance of Protein Binding in Pharmacokinetics and Pharmacodynamics. J. Pharm. Sci. 2010, 99, 1107–1121. [Google Scholar] [CrossRef]
- Burian, A.; Wagner, C.; Stanek, J.; Manafi, M.; Böhmdorfer, M.; Jäger, W.; Zeitlinger, M. Plasma protein binding may reduce antimicrobial activity by preventing intra-bacterial uptake of antibiotics, for example clindamycin. J. Antimicrob. Chemother. 2011, 66, 134–137. [Google Scholar] [CrossRef] [Green Version]
- Zeitlinger, M.; Sauermann, R.; Fille, M.; Hausdorfer, J.; Leitner, I.; Müller, M. Plasma protein binding of fluoroquinolones affects antimicrobial activity. J. Antimicrob. Chemother. 2008, 61, 561–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevillano, D.; Giménez, M.J.; Alou, L.; Aguilar, L.; Cafini, F.; Torrico, M.; González, N.; Echeverría, O.; Coronel, P.; Prieto, J. Effects of human albumin and serum on the in vitro bactericidal activity of cefditoren against penicillin-resistant Streptococcus pneumoniae. J. Antimicrob. Chemother. 2007, 60, 156–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeitlinger, M.A.; Sauermann, R.; Traunmüller, F.; Georgopoulos, A.; Müller, M.; Joukhadar, C. Impact of plasma protein binding on antimicrobial activity using time-killing curves. J. Antimicrob. Chemother. 2004, 54, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Nix, D.E.; Matthias, K.R.; Ferguson, E.C. Effect of ertapenem protein binding on killing of bacteria. Antimicrob. Agents Chemother. 2004, 48, 3419–3424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balcabao, I.P. Influence of the decrease in ciprofloxacin susceptibility and the presence of human serum on the in vitro susceptibility of Streptococcus pneumoniae to five new quinolones. J. Antimicrob. Chemother. 2001, 48, 907–909. [Google Scholar] [CrossRef] [Green Version]
- Bedenić, B. Selection of Klebsiella pneumoniae Mutants with High-Level Cefotaxime Resistance during Growth in Serum Containing Therapeutic Concentrations of Cefotaxime. Chemotherapy 2002, 48, 10–14. [Google Scholar] [CrossRef]
- Edwards, J.R. Cefotetan: Antibacterial activity against Staphylococcus aureus in the presence of human serum. Chemioterapia 1988, 7, 271–273. [Google Scholar]
- Leuthner, K.D.; Cheung, C.M.; Rybak, M.J. Comparative activity of the new lipoglycopeptide telavancin in the presence and absence of serum against 50 glycopeptide non-susceptible staphylococci and three vancomycin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2006, 58, 338–343. [Google Scholar] [CrossRef]
- Odenholt, I.; Löwdin, E.; Cars, O. Pharmacodynamic effects of telavancin against methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains in the presence of human albumin or serum and in an in vitro kinetic model. Antimicrob. Agents Chemother. 2007, 51, 3311–3316. [Google Scholar] [CrossRef] [Green Version]
- Perl, T.M.; Pfaller, M.A.; Houston, A.; Wenzel, R.P. Effect of serum on the in vitro activities of 11 broad-spectrum antibiotics. Antimicrob. Agents Chemother. 1990, 34, 2234–2239. [Google Scholar] [CrossRef] [Green Version]
- Tsuji, B.T.; Leonard, S.N.; Rhomberg, P.R.; Jones, R.N.; Rybak, M.J. Evaluation of daptomycin, telavancin, teicoplanin, and vancomycin activity in the presence of albumin or serum. Diagn. Microbiol. Infect. Dis. 2008, 60, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, J.M.; Andrews, J.M.; Brenwald, N.P.; Ashby, J.P.; Wise, R. The in-vitro activity of faropenem, a novel oral penem. J. Antimicrob. Chemother. 1997, 39, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castanheira, M.; Duncan, L.R.; Rhomberg, P.R.; Sader, H.S. Enhanced activity of cefepime–tazobactam (WCK 4282) against KPC-producing Enterobacteriaceae when tested in media supplemented with human serum or sodium chloride. Diagn. Microbiol. Infect. Dis. 2017, 89, 305–309. [Google Scholar] [CrossRef]
- Cha, R.; Rybak, M.J. Influence of protein binding under controlled conditions on the bactericidal activity of daptomycin in an in vitro pharmacodynamic model. J. Antimicrob. Chemother. 2004, 54, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Laue, H.; Valensise, T.; Seguin, A.; Hawser, S.; Lociuro, S.; Islam, K. Effect of human plasma on the antimicrobial activity of iclaprim in vitro. J. Antimicrob. Chemother. 2007, 60, 1388–1390. [Google Scholar] [CrossRef]
- Traub, W.H.; Spohr, M.; Bauer, D. Susceptibility of Acinetobacter calcoaceticus to antimicrobial drugs, alone and combined, with and without defibrinated human blood. Chemotherapy 1989, 35, 95–104. [Google Scholar] [CrossRef]
- Traub, W.H.; Spohr, M.; Bauer, D. Teicoplanin combined with various antibiotics and human blood against a multiple-drug-resistant strain of staphylococcus aureus. Chemotherapy 1991, 37, 186–195. [Google Scholar] [CrossRef]
- Walter, H.; Leonhard, B.; Bauer, D. Enterococcus faecium: In vitro Activity of Antimicrobial Drugs, Singly and Combined, with and without Defibrinated Human Blood, against Multiple-Antibiotic-Resistant Strains. Chemotherapy 1993, 39, 254–264. [Google Scholar]
- Traub, W.; Leonhard, B.; Bauer, D. Stenotrophomonas (Xanthomonas) maltophilia: In vitro susceptibility to selected antimicrobial drugs, single and combined, with and without defibrinated human blood. Chemotherapy 1998, 44, 293–304. [Google Scholar] [CrossRef]
- Traub, W.H.; Sphor, M.; Bauer, D. Streptococcus faecalis: In vitro Susceptibility to Antimicrobial Drugs, Single and Combined, with and without Defibrinated Human Blood. Chemotherapy 1986, 32, 270–285. [Google Scholar] [CrossRef]
- Traub, W.H.; Spohr, M.; Bauer, D. Pseudomonas aeruginosa: In vitro susceptibility to antimicrobial drugs, single and combined, with and without defibrinated human blood. Chemotherapy 1988, 34, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Nussbaumer-Pröll, A.K.; Knotzer, S.; Eberl, S.; Reiter, B.; Stimpfl, T.; Jäger, W.; Poschner, S.; Zeitlinger, M. Impact of erythrocytes on bacterial growth and antimicrobial activity of selected antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nussbaumer-Pröll, A.K.; Eberl, S.; Reiter, B.; Stimpfl, T.; Jäger, W.; Poschner, S.; Zeitlinger, M. Impact of thrombocytes, on bacterial growth and antimicrobial activity of selected antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 593–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ausman, L.M.; Oliver, L.M.; Goldin, B.R.; Woods, M.N.; Gorbach, S.L.; Dwyer, J.T. Estimated Net Acid Excretion Inversely Correlates With Urine pH in Vegans, Lacto-Ovo Vegetarians, and Omnivores. J. Ren. Nutr. 2008, 18, 456–465. [Google Scholar] [CrossRef]
- So, W.; Crandon, J.L.; Nicolau, D.P. Effects of Urine Matrix and pH on the Potency of Delafloxacin and Ciprofloxacin against Urogenic Escherichia coli and Klebsiella pneumoniae. J. Urol. 2015, 194, 563–570. [Google Scholar] [CrossRef]
- Matzneller, P.; Strommer, S.; Österreicher, Z.; Mitteregger, D.; Zeitlinger, M. Target site antimicrobial activity of colistin might be misestimated if tested in conventional growth media. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1989–1994. [Google Scholar] [CrossRef]
- Jung, D.; Rozek, A.; Okon, M.; Hancock, R.E.W. Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin. Chem. Biol. 2004, 11, 949–957. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, R.; Ito-Horiyama, T.; Takemura, M.; Toba, S.; Matsumoto, S.; Ikehara, T.; Tsuji, M.; Sato, T.; Yamano, Y. In Vivo Pharmacodynamic Study of Cefiderocol, a Novel Parenteral Siderophore Cephalosporin, in Murine Thigh and Lung Infection Models. Antimicrob. Agents Chemother. 2019, 63, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Asempa, T.E.; Abdelraouf, K.; Nicolau, D.P. Metallo-β-lactamase resistance in Enterobacteriaceae is an artefact of currently utilized antimicrobial susceptibility testing methods. J. Antimicrob. Chemother. 2020, 75, 997–1005. [Google Scholar] [CrossRef]
- Van’t Veen, A.; Mouton, J.W.; Gommers, D.; Kluytmans, J.A.N.A.J.W.; Dekkers, P. Influence of Pulmonary Surfactant on In Vitro Bactericidal Activities of Amoxicillin, Ceftazidime, and Tobramycin. Antimicrob. Agents Chemother. 1995, 39, 329–333. [Google Scholar] [CrossRef] [Green Version]
- Silverman, J.A.; Mortin, L.I.; Vanpraagh, A.D.G.; Li, T.; Alder, J. Inhibition of Daptomycin by Pulmonary Surfactant: In Vitro Modeling and Clinical Impact. J. Infect. Dis. 2005, 191, 2149–2152. [Google Scholar] [CrossRef] [PubMed]
- Schwameis, R.; Erdogan-yildirim, Z.; Manafi, M.; Zeitlinger, M.A.; Strommer, S.; Sauermann, R. Effect of Pulmonary Surfactant on Antimicrobial Activity In Vitro. Antimicrob. Agents Chemother. 2013, 57, 5151–5154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotfried, M.H.; Shaw, J.; Benton, B.M.; Krause, K.M.; Goldberg, M.R.; Kitt, M.M.; Barriere, S.L. Intrapulmonary Distribution of Intravenous Telavancin in Healthy Subjects and Effect of Pulmonary Surfactant on In Vitro Activities of Telavancin and Other Antibiotics. Antimicrob. Agents Chemother. 2008, 52, 92–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dugourd, D.; Yang, H.; Elliott, M.; Siu, R.; Clement, J.J.; Straus, S.K.; Hancock, R.E.W.; Rubinchik, E. Antimicrobial Properties of MX-2401, an Expanded-Spectrum Lipopeptide Active in the Presence of Lung Surfactant. Antimicrob. Agents Chemother. 2011, 55, 3720–3728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oesterreicher, Z.; Eberl, S.; Peilensteiner, T.; Zeitlinger, M. Impact of different pathophysiological conditions on antimicrobial activity of glycopeptides in vitro. Clin. Microbiol. Infect. 2018, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, H.-J. Influence of pH and human urine on the antibacterial activity of ciprofloxacin, norfloxacin and ofloxacin. Drugs Exp. Clin. Res. 1985, 11, 335–338. [Google Scholar]
- Erdogan-Yildirim, Z.; Burian, A.; Manafi, M.; Zeitlinger, M. Impact of pH on bacterial growth and activity of recent fluoroquinolones in pooled urine. Res. Microbiol. 2011, 162, 249–252. [Google Scholar] [CrossRef]
- Burian, A.; Erdogan, Z.; Jandrisits, C.; Zeitlinger, M. Impact of pH on activity of trimethoprim, fosfomycin, amikacin, colistin and ertapenem in human urine. Pharmacology 2012, 90, 281–287. [Google Scholar] [CrossRef]
- Yang, L.; Wang, K.; Li, H.; Densted, J.D.; Cadieux, P.A. Re: The influence of urinary pH on antibiotic efficacy against bacterial uropathogens. J. Urol. 2015, 193, 151. [Google Scholar]
- Sauermann, R.; Schwameis, R.; Fille, M.; Camuz Ligios, M.L.; Zeitlinger, M. Antimicrobial activity of cefepime and rifampicin in cerebrospinal fluid in vitro. J. Antimicrob. Chemother. 2008, 62, 1057–1060. [Google Scholar] [CrossRef] [Green Version]
- Schwameis, R.; Fille, M.; Manafi, M.; Zeitlinger, M.; Sauermann, R. Enhanced activity of linezolid against Staphylococcus aureus in cerebrospinal fluid. Res. Microbiol. 2012, 163, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Sauermann, R.; Schwameis, R.; Fille, M.; Camuz ligios, M.L.; Zeitlinger, M. Cerebrospinal fluid impairs antimicrobial activity of fosfomycin in vitro. J. Antimicrob. Chemother. 2009, 64, 821–823. [Google Scholar] [CrossRef]
- Wulkersdorfer, B.; Jaros, D.; Poschner, S.; Jäger, W.; Cosentini, E.; Zeitlinger, M.; Schwameis, R. Human Bile Reduces Antimicrobial Activity of Selected Antibiotics against Enterococcus faecalis and Escherichia coli In Vitro. Antimicrob. Agents Chemother. 2017, 61, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Nussbaumer-Pröll, A.K.; Eberl, S.; Reiter, B.; Stimpfl, T.; Dorn, C.; Zeitlinger, M. Low pH reduces the activity of ceftolozane/tazobactam in human urine, but confirms current breakpoints for urinary tract infections. J. Antimicrob. Chemother. 2019, 488, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Thulin, E.; Thulin, M.; Andersson, D.I. Reversion of High-level Mecillinam Resistance to Susceptibility in Escherichia coli During Growth in Urine. EBioMedicine 2017, 23, 111–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouton, J.W.; Vinks, A.A. Relationship between minimum inhibitory concentration and stationary concentration revisited: Growth rates and minimum bactericidal concentrations. Clin. Pharmacokinet. 2005, 44, 767–768. [Google Scholar] [CrossRef]
- Mouton, J.W.; Vinks, A.A. Pharmacokinetic/pharmacodynamic modelling of antibacterials in vitro and in vivo using bacterial growth and kill kinetics: The minimum inhibitory concentration versus stationary concentration. Clin. Pharmacokinet. 2005, 44, 201–210. [Google Scholar] [CrossRef]
- Schuck, E.L.; Derendorf, H. Pharmacokinetic/pharmacodynamic evaluation of anti-infective agents. Expert Rev. Anti Infect. Ther. 2005, 3, 361–373. [Google Scholar] [CrossRef]
- Mueller, M.; Pena, A.; Derendorf, H. MINIREVIEW-Issues in Pharmacokinetics and Pharmacodynamics of Anti-Infective Agents: Distribution in Tissue. Antimicrob. Agents Chemother. 2004, 48, 1441–1453. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, E.I.; Viberg, A.; Löwdin, E.; Cars, O.; Karlsson, M.O.; Sandström, M. Semimechanistic pharmacokinetic/pharmacodynamic model for assessment of activity of antibacterial agents from time-kill curve experiments. Antimicrob. Agents Chemother. 2007, 51, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Lallemand, E.A.; Lacroix, M.Z.; Toutain, P.L.; Boullier, S.; Ferran, A.A.; Bousquet-Melou, A. In vitro degradation of antimicrobials during use of broth microdilution method can increase the measured minimal inhibitory and minimal bactericidal concentrations. Front. Microbiol. 2016, 7, 2–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, M.; de la Peña, A.; Derendorf, H. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: Kill curves versus MIC. Antimicrob. Agents Chemother. 2004, 48, 369–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, P.A.; Petersen, P.J.; Young, M.; Jones, C.H.; Tischler, M.; O’Connell, J. Tigecycline MIC testing by broth dilution requires use of fresh medium or addition of the biocatalytic oxygen-reducing reagent oxyrase to standardize the test method. Antimicrob. Agents Chemother. 2005, 49, 3903–3909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gloede, J.; Scheerans, C.; Derendorf, H.; Kloft, C. In vitro pharmacodynamic models to determine the effect of antibacterial drugs. J. Antimicrob. Chemother. 2009, 65, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Ledesma, K.R.; Chang, K.; Hou, J.; Prince, R.A.; Tam, V.H. Pharmacodynamics of moxifloxacin against a high inoculum of Escherichia coli in an in vitro infection model. J. Antimicrob. Chemother. 2009, 64, 556–562. [Google Scholar] [CrossRef] [Green Version]
- Marques, M.R.C.; Loebenger, R.; Almukainzi, M. Simulated Biological Fluids with Possible Application in Dissolution Testing. J. Dissolution Technol. 2011, 18, 15–28. [Google Scholar] [CrossRef]
- Budha, N.R.; Lee, R.B.; Hurdle, J.G.; Lee, R.E.; Meibohm, B. A simple in vitro PK/PD model system to determine time-kill curves of drugs against Mycobacteria. Tuberculosis 2009, 89, 378–385. [Google Scholar] [CrossRef] [Green Version]
- Breimer, D.D. PK/PD modelling and beyond: Impact on drug development. Pharm. Res. 2008, 25, 2720–2722. [Google Scholar] [CrossRef] [Green Version]
| Compartment | pH | Cell Type | Proteins | Minerals, Vitamins, Fats, and Additional Information | CO2 | O2 | Glucose | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| Blood | Hematocrit 45% | 7.4 | erythrocytes (4.5–5.5 million/µL) | / | / | arterial blood (32–48 mm Hg) venous blood (37–50 mm Hg) | arterial blood (83–108 mm Hg) venous blood (36–43 mm Hg) | <110 mg/dL sober, 130–140 mg/dL after carbohydrate-rich diet | [5,7,8] |
| leukocytes (4000–8000/µL) → 60–70% neutrophil granulocytes, 2–3% eosinophil granulocytes, 20–30% lymphocytes, 4–5% monocytes | / | / | |||||||
| thrombocytes (150,000–350,000/µL) | |||||||||
| Blood plasma 55% | 90% water + 10% dissolved substances, which are composed of 70% plasma proteins/other proteins → albumin (35–40 g/L plasma), α1 globulin (3–6 g/L plasma), α2 globulin (4–9 g/L plasma), β globulin (6–11 g/L plasma), ϒ globulin (13–17 g/L plasma) | 20% vitamins/minerals: urea, uric acid, creatinine, hormones, enzymes, fats (cholesterol, phospholipids, triglycerides, free fatty acids, 10% electrolytes → Na+, Cl−, Ca2+, K+, Mg2+, H3PO4, NaCl (0.6–0.7 g/100 mL plasma) | |||||||
| Urine | Urine 0.5–2 L in 24 h | 5.6–7 | erythrocytes 0–3/visual field | 150 mg | 12–20 mg/dL urea, 0.25–0.75 g/24 h uric acid, 1.5 g creatinine, 0–0.14 g/24 h, <0.25 g/24 h glucose, 40–220 mmol/24 h Na+, 25–125 mmol/24 h K+, 2.5–7.5 mmol/24 h Ca2+, 110–250 mmol/24 h Cl−, 13–42 mmol/24 h H3PO4 | [5,7,9] | |||
| thrombocytes 0–4/visual field | |||||||||
| Brain, Spinal cord | CSF ~ 150mL | 7.28–7.32 | lymphocytes and monocytes < 5/mm3; no erythrocytes | lumbar CSF: 0.1––0.4g/L; ventricular CSF: 0.07–0.25 g/L | Na+ (135–147 mmol)/L, K+ (3.5–5.3 mmol)/L, Cl− (95–110 mmol)/L, Ca+ (2.10–2.60 mmol)/L, Mg+ (0.8–1.1 mmol)/L, H3PO4 (0.81–1.45 mmol)/L, Fe (0.2–0.4 mmol)/L, creatinine (50–110 mmol)/L, urea (3.0–6.5 mmol)/L, lactate (1.1–2.4 mmol)/L | 44–50 mmHg | 40–44 mmHg | glucose (2.8–4.4 mmol)/L | [10,11] |
| Lung | ELF | / | Bronchial wash macrophages: 7.2 (5.2–12.3) × 104 cells/mL; neutrophils: 0.7 (0.3–1.0) × 104 cells/mL; lymphocytes: 0.3 (0.2–0.7) × 104 cells/mL; eosinophils: 0.0 (0.0–0.1) × 104 cells/mL; mast cells: 0.01 (0.00–0.02) × 104 cells/mL | total protein, in mg/mL: 14.3 (11.8–24.6) | alveolar (49.2% albumin, 6.3% surfactant protein A) bronchial (63.67% albumin, 3.35% surfactant protein A) | / | / | / | [12] |
| Bronchoalveolar lavage Macrophages: 13.9 (8.9–17.6) × 104 cells/mL; neutrophils: 0.1 (0.0–0.2) × 104 cells/mL; lymphocytes: 0.7 (0.5–1.0) × 104 cells/mL; eosinophils: 0.0 (0.0–0.1) × 104 cells/mL; mast cells: 0.01 (0.00–0.03) × 104 cells/mL | total protein, in mg/mL: 40.4 (28.9–51.4) | ||||||||
| Bile | Gallbladder | 6.89–7.00 | / | total protein: 4.5 g/L | Carbohydrates: 2.4 g/L; chloride: 66.2 mmol/L; H3PO4: 45 mmol/L; K+: 12.8–24.6 mmol/L; Na+: 179–209 mmol/L; Ca+: 3.7–10.8 mmol/L; Ku: 87.5 µmol/L; zinc: 5.4 µmol/L | [13] | |||
| Liver bile | 7.15 | / | total protein: 1.8–7 g/L; albumin: 634 mg/L | Cl−: 105 mmol/L; H3PO4: 4.78 mmol/L; K: 4.8 mmol/L; Na+: 146–149 mmol/L; Ca+: 4 mmol/L; Fu: 15 µmol/L; Ku: 5.8–25.3 µmol/L | [13] | ||||
| Pancreas | Pancreatic juice: ~17–20 mL/kg/day | 7.7 | / | total protein: 6.6 g/L | Cl−: 56 mmol/L; H3PO4: 0.8 mmol/kg; K: 7.6 mmol/L; sodium: 125 mmol/L; Ca+: 0.6 mmol/L; zinc: 18.5 µmol/L; chymotrypsin; trypsin’ carboxypeptidase A and B; lipase | [13] | |||
| Breast milk | Colostrum → mature milk | 7.01–7.29 | / | Total protein: 10.6–22.9 g/L; Whey protein (lactalbumin): 3.6–7.8 g/L | Na+: 0.172–0.501 g/L; K: 0.512–0.745 g/L; Ca+: 0.344–0.481 g/L; Mg+: 0.035–0.042 g/L; Fu: 0.50–1 mg/L; Ku: 0.51–1.34 mg/L; Zn: 1.18–5.59 mg/L; N: 324–479 mg/L; lactose: 57–71 g/L; total fat: 29.5–45.4 g/L | / | / | / | [13] |
| Adapted Media | Incorporated Bacteria | In Vitro Test | Antibiotics | Year | Ref |
|---|---|---|---|---|---|
| MHB spiked with 50% human erythrocytes | ATCC-25922 E. coli, ATCC-29213 S. aureus, ATCC-27853 P. aeruginosa | MIC, growth assay, TKC | Ciprofloxacin, meropenem, tigecycline | 2019 | [14] |
| MHB spiked with thrombocyte concentrates | ATCC-25922 E. coli, ATCC-29213 S. aureus, ATCC-27853 P. aeruginosa | MIC, growth assay, TKC | Ciprofloxacin, meropenem, tigecycline | 2019 | [15] |
| MHB spiked with 50% human serum or NaCl | 209 clinical isolates of Enterobactericae carrying blaKPC | MIC | Cefepime–tazobactam | 2017 | [16] |
| MHB with 12% albumin pH 6 and pH 7.4 at 32 °C, 37 °C, and 42 °C | 20 clinical isolates of S. aureus; ATCC-29213 S. aureus | MIC, growth assay, TKC | Telavancin, vancomycin, teicoplanin | 2018 | [17] |
| MHB with 4%, 8%, 12%, and 16% human albumin MHB with 20%, 50%, and 70% human serum | ATCC-29213 S. aureus | MIC, growth assay, TKC | Clindamycin | 2010 | [18] |
| Pure MHB and 100% serum MHB with 4%, 8%, 12%, and 16% human albumin MHB with 20% and 70% human serum | ATCC-29213 S. aureus, ATCC-27853 P. aeruginosa | Ultrafiltration, MIC, growth assay, TKC, | Moxifloxacin, trovafloxacin | 2007 | [19] |
| Pure MHB and MHB with 50% plasma | 40 methicillin-susceptible S. aureus (MSSA); 38 methicillin-resistant S. aureus (MRSA) | MIC | Vancomycin, fusidic-acid, teicoplanin, iclaprim | 2007 | [20] |
| MHB with 40 mg/L human albumin | ATCC-29213 S. aureus | MIC, growth assay, TKC, | Ampicillin, fosfomycinosfomycin, oxacillin, moxifloxacin | 2004 | [21] |
| Pure MHB and MHB with 65% defibrinated human blood | 17 E. faecium clinical isolates, E. faecalis ATCC29212, S. aureus ATCC25923 | MIC, MBC, TKC | Ampicillin, ciprofloxacin, co-trimoxazole, doxycycline, fusidic-acid, imipenem, gentamicin, mupirocin, novobiocin, rifampin, streptomycin, taurolidine, teicoplanin, trimethoprim-sulfamethoxazole, vancomycin | 1993 | [22] |
| Pure MHB and MHB with 65% defibrinated human blood | Six MDR S. aureus isolates, S. aureus ATCC25923 | MIC, MBC | Amikacin, cefamandole, chloramphenicol, ciprofloxacin, clindamycin, coumermycin, teicoplanin, fusidic acid, gentamycin, imipenem, netilmicin, novobiocin, ofloxacin, oxacillin, rifampicin, tobramycin, trimethoprim-sulfamethoxazole, vancomycin | 1991 | [23] |
| MHB and MHB with 90%, 80%, and 50% urine | 16 urogenic clinical isolates of Enterobacteriaceae | MIC | delafloxacin, ciprofloxacin | 2016 | [24] |
| Body Fluid | Incorporated Bacteria | In Vitro Test | Antibiotics | Year | Ref |
|---|---|---|---|---|---|
| Pooled human urine, MHB | Clinical isolates of E. coli, K. pneumoniae, and P. mirabilis | MIC, TKC | Ceftolozane–tazobactam, meropenem | 2019 | [25] |
| Pooled human bile fluid | ATCC-25922 E. coli, ATCC-29212 E. faecalis | MIC, TKC | Linezolid, tigecycline, meropenem, ciprofloxacin | 2017 | [26] |
| MHB, urine, artificial urine medium | E. coli MG1655 wild-type (DA5438); cysB deletion mutant MG1655 (DA28439); MecR clinical E. coli UTI isolates DA14719, DA24682, and DA24686 | MIC (test strips and Etest), growth measurement (Bioscreen C Analyser) | Mecillinam, meropenem, ampicillin, cefotaxime | 2017 | [27] |
| MHB, urine | Six urogenic clinical isolates: E. coli, S. saprophyticus, K. pneumoniae, P. mirabilis, E. faecalis, and S. epidermidis | MIC, disc-diffusion assay | Ciprofloxacin, ofloxacin, levofloxacin, gentamicin, tobramycin, erythromycin, azithromycin, trim/Sulfa, trimethoprim, tetracycline, doxycycline, cefotaxime, cephalothin, cefazolin, ceftazidime, ampicillin, piperacillin, nitrofurantoin | 2014 | [28] |
| MHB, urine | ATCC-25922 E. coli, ATCC-29213 S. aureus, ATCC-700324 K. oxytoca, ATCC-14153 P. mirabilis, ATCC-29212 E. faecalis | MIC, growth assay, TKC | Trimethoprim, fosfomycin, colistin, amikacin, ertapenem | 2012 | [29] |
| MHB, pooled human CSF in 5% CO2, artificial substitute CSF, 0.5 g/L human albumin, sodium OH (0.064 M solution) | ATCC-29213 S. aureus, ATCC-12228 S. epidermidis | MIC, growth assay, TKC | Linezolid | 2012 | [30] |
| MHB, urine | ATCC-25922 E. coli, ATCC-700324 K. oxycota | MIC, growth assay, TKC | Ciprofloxacin, levofloxacin, moxifloxacin | 2010 | [31] |
| MHB, pooled human CSF in ambient air, pooled human CSF in 5% CO2 | ATCC-29213 S. aureus, clinical isolate of S. aureus (MIC 16mg/L) | TKC | Fosfomycin | 2009 | [32] |
| Reference MHB, pooled human CSF in ambient air, pooled human CSF in 5% CO2, pooled human CSF in 5% CO2 | ATCC-29213 S. aureus | TKC | Cefepime, rifampicin | 2008 | [33] |
| Advantages | Limitations | |
|---|---|---|
| MIC/MBC | Easy and quick to perform | Static approach |
| Screening of a high number of isolates | Problems with turbidity in MIC testing (i.e., with blood, urine, surfactant) | |
| Widely used: EUCAST * and CLSI ** | Two-fold dilution steps might not detect small changes in the efficacy of the tested concentration | |
| Guide values for further testing | Kinetics of bacterial killing are not recorded | |
| MIC is determined by visible growth (107 cells/mL); low growth is not considered | ||
| Difficulties with swarming bacteria or bacteria that produce CO2 (e.g., Proteus mirabilis in urine) | ||
| Instability of antibiotics in MHB or adapted MHB | ||
| Components of the culture media might be spent by dividing bacteria | ||
| TKC | Kinetics of bacterial killing can be observed and time-CFU/mL *** curves can be produced | Labor-intensive and time consuming—not suitable for screening |
| A coherent system with killing curves and growth assays at the same time | Amount of the volume of adapted media or body fluid is rather high | |
| Possibility of mimicking multiple dosing and **** T > MIC | Maintaining a homogenous suspension might be difficult | |
| Time points can be chosen individually, and CFU/mL *** can be evaluated at multiple time points | Instability of antibiotics in MHB or adapted MHB | |
| No problems with the turbidity of media | Components of the culture media might be spent by dividing bacteria | |
| Fresh broth and antibiotic in HF systems |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nussbaumer-Pröll, A.; Zeitlinger, M. Use of Supplemented or Human Material to Simulate PD Behavior of Antibiotics at the Target Site In Vitro. Pharmaceutics 2020, 12, 773. https://doi.org/10.3390/pharmaceutics12080773
Nussbaumer-Pröll A, Zeitlinger M. Use of Supplemented or Human Material to Simulate PD Behavior of Antibiotics at the Target Site In Vitro. Pharmaceutics. 2020; 12(8):773. https://doi.org/10.3390/pharmaceutics12080773
Chicago/Turabian StyleNussbaumer-Pröll, Alina, and Markus Zeitlinger. 2020. "Use of Supplemented or Human Material to Simulate PD Behavior of Antibiotics at the Target Site In Vitro" Pharmaceutics 12, no. 8: 773. https://doi.org/10.3390/pharmaceutics12080773