A Comprehensive Review about the Molecular Structure of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Insights into Natural Products against COVID-19
Abstract
:1. Introduction
2. Taxonomy and Structure of SARS-CoV-2 Virus
3. COVID-19 Detection Methods
4. Molecular Structure and Functional Determinant of SARS-CoV-2
4.1. SARS-CoV-2 Proteases
4.1.1. Main Protease (Mpro)
4.1.2. Papain-like Protease (PLpro)
4.2. Spike Glycoprotein (S)
4.3. RNA-Dependent RNA Polymerase (RdRp)
4.4. SARS-CoV-2 Nucleocapsid N Protein
5. Therapeutic Approaches for COVID-19
5.1. Antiviral Strategies against SARS-CoV-2
5.2. Immuno-Modulators
5.3. Antibody and Convalescent Plasma Therapy
5.4. COVID-19 Vaccines
5.4.1. BNT162B1 (Pfizer-BioNTech)
5.4.2. CoronaVac (SinoVac)
5.4.3. mRNA1273 (Moderna)
5.4.4. Ad26COVS1 (Jansseen Vaccine)
5.4.5. ChAdOx1 nCoV-19 (AstraZeneca)
5.4.6. SputnikV (Gam-COVID-Vac)
5.4.7. BBIBP-CorV (Sinopharm)
5.4.8. NVX-CoV2373 (Novavax)
5.4.9. BBV152 (Covaxin)
5.4.10. Ad5-nCoV (CanSino)
6. Natural Products and SARS-CoV-2
6.1. Antiviral Natural Products from Fungi
6.2. Natural Products from Algae and SARS-CoV-2
6.3. Antiviral Peptides Derived from Scorpion Venoms
6.4. Natural Compounds from Plants and SARS-CoV-2
6.5. Antiviral Natural Products under Clinical Investigations against SARS-CoV-2
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kooraki, S.; Hosseiny, M.; Myers, L.; Gholamrezanezhad, A. Coronavirus (COVID-19) Outbreak: What the Department of Radiology Should Know. J. Am. Coll. Radiol. 2020, 17, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Ather, A.; Patel, B.; Ruparel, N.B.; Diogenes, A.; Hargreaves, K.M. Coronavirus disease 19 (COVID-19):Implications for clinical dental care. J. Endod. 2020, 46, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E.; Ledford, H.; Mallapaty, S. Six months of Coronavirus: The mysteries scientists are still racing to solve. Nature 2020, 583, 178–179. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- WorldOmeter, Last updated: July 31, 2021, 10:22 GMT. Available online: https://www.worldometers.info/coronavirus/ (accessed on 25 August 2021).
- Bennett, N.J.; Finkbeiner, E.M.; Ban, N.C.; Belhabib, D.; Jupiter, S.D.; Kittinger, J.N.; Mangubhai, S.; Scholtens, J.; Gillk, D.; Christiel, P. The COVID-19 Pandemic, Small-Scale Fisheries and Coastal Fishing Communities. Coast. Manag. 2020, 48, 336–347. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses 2015, 1282, 1–23. [Google Scholar]
- Ray, S.; Maunsell, J.H.R. Different origins of gamma rhythm and high-gamma activity in macaque visual cortex. PLoS Biol. 2011, 9, e1000610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020, 55, 9–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Palasca, O.; Santos, A.; Stolte, C.; Gorodkin, J.; Jensen, L.J. TISSUES 2.0: An integrative web resource on mammalian tissue expression. Database 2018, 2018, bay003. [Google Scholar] [CrossRef]
- Baig, A.M.; Khaleeq, A.; Ali, U.; Syeda, H. Evidence of the COVID-19 Virus Targeting the CNS: Tissue Distribution, Host-Virus Interaction, and Proposed Neurotropic Mechanisms. ACS Chem. Neurosci. 2020, 11, 995–998. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.M.; Venters, C.C.; Di, C.; Pinto, A.M.; Wan, L.; Younis, I.; Cai, Z.; Arai, C.; So, B.R.; Duan, J.; et al. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat. Commun. 2020, 11, 3232. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.; Brierley, S.; Gandhi, M.J.; Cohen, M.A.; Moschella, P.C.; Declan, A.B.L. Repurposing therapeutics for potential treatment of SARS-CoV-2: A review. Viruses 2020, 12, 705. [Google Scholar] [CrossRef]
- Contini, A. Virtual screening of an FDA approved drugs database on two COVID-19 coronavirus proteins. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Singhal, T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87, 281. [Google Scholar] [CrossRef] [Green Version]
- Vivekanandhan, K.; Shanmugam, P.; Barabadi, H.; Arumugam, V.; Daniel Raj Daniel Paul Raj, D.; Sivasubramanian, M.; Ramasamy, S.; Anand, K.; Boomi, P.; Chandrasekaran, B.; et al. Emerging Therapeutic Approaches to Combat COVID-19: Present Status and Future Perspectives. Front. Mol. Biosci. 2021, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Suwannarach, N.; Kumla, J.; Sujarit, K.; Pattananandecha, T.; Saenjum, C.; Lumyong, S. Natural Bioactive Compounds from Fungi as Potential Candidates for Protease Inhibitors and Immunomodulators to Apply for Coronaviruses. Molecules 2020, 25, 1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangsinth, P.; Sillapachaiyaporn, C.; Nilkhet, S.; Tencomnao, T.; Ung, A.T.; Chuchawankul, S. Mushroom-derived bioactive compounds potentially serve as the inhibitors of SARS-CoV-2 main protease: An in silico approach. J. Tradit. Complement. Med. 2021, 11, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, F.R.; Howlader, S.; Raihan, T.; Hasan, M. Plants Metabolites: Possibility of Natural Therapeutics Against the COVID-19 Pandemic. Front. Med. 2020, 7, 444. [Google Scholar] [CrossRef]
- Kasozi, K.I.; Niedbała, G.; Alqarni, M.; Zirintunda, G.; Ssempijja, F.; Musinguzi, S.P.; Usman, I.M.; Matama, K.; Hetta, H.F.; Mbiydzenyuy, N.E.; et al. Bee Venom—A Potential Complementary Medicine Candidate for SARS-CoV-2 Infections. Front. Public Health 2020, 8, 755. [Google Scholar] [CrossRef]
- El-Rayes, R.A.; Mohallal, M.E.; El-Shahat, Y.M.; Ebaid, H.M.; Miller, K.; Strong, P.N.; Abdel-Rahman, M.A. Cytotoxic effects of Smp24 and Smp43 Scorpion Venom antimicrobial peptides on tumour and non-tumour cell lines. Int. J. Pept. Res. Ther. 2019, 26, 1409–1415. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Rahman, M.A.; Harrison, P.L.; Strong, P.N. Snapshots of scorpion venomics. J. Arid Environ. 2015, 112, 170–176. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Quintero-Hernández, V.; Possani, L.D. Venom proteomic and venomous glands transcriptomic analysis of the Egyptian scorpion Scorpio maurus palmatus (Arachnida: Scorpionidae). Toxicon 2013, 74, 193–207. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Van Regenmortel, M.H.V.; Fauquet, C.M.; Bishop, D.H.L.; Carstens, E.B.; Estes, M.K.; Lemon, S.M.; Maniloff, J.; Mayo, M.A.; McGeoch, D.J.; Pringle, C.R.; et al. Virus Taxonomy: Classification and Nomenclature of Viruses. Seventh Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2000. [Google Scholar]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structural biology: Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Wu, K.; Li, W.; Peng, G.; Li, F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA 2009, 106, 19970–19974. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Epand, R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta Biomembr. 2003, 1614, 116–121. [Google Scholar] [CrossRef] [Green Version]
- Abraham, S.; Kienzle, T.E.; Lapps, W.; Brian, D.A. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology 1990, 176, 296–301. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Bárcena, M.; Oostergetel, G.T.; Bartelink, W. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. USA 2009, 106, 582–587. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta. Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, C.; Goldsmith, J.A.; Schaub, J.M.; Divenere, A.M.; Kuo, H.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Peng, R.; Wu, L.-A.; Wang, Q.; Qi, J.; Gao, G.F. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860. [Google Scholar] [CrossRef] [PubMed]
- El-Fakharany, E.M.; El-Maradny, Y.A.; Othman, A.M.; Gerges, M.M.; Belal, F.H.; Behery, E.A. COVID-19 coronavirus: Pathogenesis, clinical features, diagnostics, epidemiology, prevention and control. Microb. Biosyst. 2020, 5, 7–20. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, D.; Zhang, Y.; Ma, J.; Qi, J.; Wang, Q.; Lu, G.; Wu, Y.; Yan, J.; Shi, Y.; et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017, 8, 15092. [Google Scholar] [CrossRef] [PubMed]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93, e01815-18. [Google Scholar] [CrossRef] [Green Version]
- Gierer, S.; Bertram, S.; Kaup, F.; Wrensch, F.; Heurich, A.; Kramer-Kuhl, A.; Welsch, K.; Winkler, M.; Meyer, B.; Drosten, C.; et al. The Spike Protein of the Emerging Betacoronavirus EMC Uses a Novel Coronavirus Receptor for Entry, Can Be Activated by TMPRSS2, and Is Targeted by Neutralizing Antibodies. J. Virol. 2013, 87, 5502–5511. [Google Scholar] [CrossRef] [Green Version]
- Glowacka, I.; Bertram, S.; Muller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [Green Version]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Fiorillo, B.; Marchianò, S.; Sepe, V.; Biagioli, M.; Finamore, C.; Bozza, S.; Francisci, D.; Distrutti, E.; Carino, A.; Moraca, F. Hijacking SARS-CoV-2/ACE2 Receptor Interaction by Natural and Semi-synthetic Steroidal Agents Acting on Functional Pockets on the Receptor Binding Domain. Front. Chem. 2020, 8, 572885. [Google Scholar]
- Singh, A.V. Potential of amentoflavone with antiviral properties in COVID-19 treatment. Asian Biomed. 2021, 15, 153–159. [Google Scholar] [CrossRef]
- Kleine-Weber, H.; Elzayat, M.T.; Hoffmann, M.; Pöhlmann, S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018, 8, 16597. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Le Infez. Med. 2020, 28, 174–184. [Google Scholar]
- Vennema, H.; Godeke, G.J.; Rossen, J.W.; Voorhout, W.F.; Horzinek, M.C.; Opstelten, D.J.; Rottier, P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996, 15, 2020–2028. [Google Scholar] [CrossRef]
- Cong, Y.; Ulasli, M.; Schepers, H. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in Coronaviral life cycle. J. Virol. 2019, 94, e01925-19. [Google Scholar] [CrossRef] [Green Version]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.Q.; Hu, H.C.; Gao, M.N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef]
- Singh, B.; Datta, B.; Ashish, A.; Dutta, G. A comprehensive review on current COVID-19 detection methods: From lab care to point of care diagnosis. Sensors Int. 2021, 2, 100119. [Google Scholar] [CrossRef]
- Needle, D.; Lountos, G.T.; Waugh, D.S. Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1102–1111. [Google Scholar] [CrossRef] [Green Version]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.-H. An Overview of Severe Acute Respiratory Syndrome–Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef]
- Ul Qamar, M.T.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Malik, Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar]
- Centers for Disease Control and Prevention. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020. [Google Scholar]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA J. Am. Med. Assoc. 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Li, D.; Li, J. Immunologic testing for SARS-CoV-2 infection from the antigen perspective. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Sidiq, Z.; Hanif, M.; Dwivedi, K.K.; Chopra, K.K. Benefits and limitations of serological assays in COVID-19 infection. Indian J. Tuberc. 2020, 67, S163. [Google Scholar] [CrossRef]
- Jalandra, R.; Yadav, A.K.; Verma, D.; Dalal, N.; Sharma, M.; Singh, R.; Kumar, A.; Solanki, P.R. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed. Pharmacother. 2020, 129, 110446. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Lists Two COVID-19 Tests for Emergency Use; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Arena, F.; Pollini, S.; Rossolini, G.M.; Margaglione, M. Summary of the available molecular methods for detection of SARS-CoV-2 during the ongoing pandemic. Int. J. Mol. Sci. 2021, 22, 1298. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wu, X.; Zhang, X.; Hou, X.; Liang, T.; Wang, D.; Teng, F.; Dai, J.; Duan, H.; Guo, S.; et al. SARS-CoV-2 Proteome Microarray for Mapping COVID-19 Antibody Interactions at Amino Acid Resolution. ACS Cent. Sci. 2020, 6, 2238–2249. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417–428. [Google Scholar] [CrossRef]
- El-Tholoth, M.; Bau, H.H.; Song, J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Sharar, M.; Saied, E.M.; Rodriguez, M.C.; Arenz, C.; Montes-Bayón, M.; Linscheid, M.W. Elemental Labelling and Mass Spectrometry for the Specific Detection of Sulfenic Acid Groups in Model Peptides: A Proof of Concept. Anal Bioanal Chem 2017, 409, 2015–2027. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Chiu, C.; Rodino, K.G.; Miller, M.B. Point-counterpoint: Should we be performing metagenomic next-generation sequencing for infectious disease diagnosis in the clinical laboratory? J. Clin. Microbiol. 2020, 58, e01739-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzelak, L.; Temmam, S.; Planchais, C.; Demeret, C.; Tondeur, L.; Huon, C.; Guivel-Benhassine, F.; Staropoli, I.; Chazal, M.; Dufloo, J.; et al. A comparison of four serological assays for detecting anti–SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Che, X.Y.; Qiu, L.W.; Liao, Z.Y.; Wang, Y.D.; Wen, K.; Pan, Y.X.; Hao, W.; Mei, Y.B.; Cheng, V.C.C.; Yuen, K.Y. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 2005, 191, 2033–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.W.; Li, Y.; Zhang, H.N.; Wang, W.; Yang, X.; Qi, H.; Li, H.; Men, D.; Zhou, J.; Tao, S.C. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Varnaite, R.; García, M.; Glans, H.; Maleki, K.T.; Sandberg, J.T.; Tynell, J.; Christ, W.; Lagerqvist, N.; Asgeirsson, H.; Ljunggren, H.-G.; et al. Expansion of SARS-CoV-2-Specific Antibody-Secreting Cells and Generation of Neutralizing Antibodies in Hospitalized COVID-19 Patients. J. Immunol. 2020, 2437–2446. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, M.; Yuan, J.; Wang, F.; Wang, Z.; Li, J.; Zhang, M.; Xing, L.; Wei, J.; Peng, L.; et al. Laboratory Diagnosis and Monitoring the Viral Shedding of SARS-CoV-2 Infection. Innovation 2020, 1, 100061. [Google Scholar] [CrossRef]
- De Assis, R.R.; Jain, A.; Nakajima, R.; Jasinskas, A.; Felgner, J.; Obiero, J.M. Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent blood using a coronavirus antigen microarray. Nat. Commun. 2020, 12, 6. [Google Scholar]
- Weitzel, T.; Legarraga, P.; Iruretagoyena, M.; Pizarro, G.; Vollrath, V.; Araos, R.; Munita, J.M.; Porte, L. Head-to-head comparison of four antigen-based rapid detection tests for the diagnosis of SARS-CoV-2 in respiratory samples. bioRxiv 2020, arXiv:2020.05.27.119255. [Google Scholar] [CrossRef]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef] [PubMed]
- Hilgenfeld, R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014, 281, 4085–4096. [Google Scholar] [CrossRef] [Green Version]
- Zumla, A.; Chan, J.F.W.; Azhar, E.I.; Hui, D.S.C.; Yuen, K.-Y. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandeel, M.; Al-Taher, A.; Li, H.; Schwingenschlogl, U.; Al-Nazawi, M. Molecular dynamics of Middle East Respiratory Syndrome Coronavirus (MERS CoV) fusion heptad repeat trimers. Comput. Biol. Chem. 2018, 75, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tomlinson, A.C.A.; Wong, A.H.M.; Zhou, D.; Desforges, M.; Talbot, P.J.; Benlekbir, S.; Rubinstein, J.L.; Rini, J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. eLife 2019, 8, e51230. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Al-Nazawi, M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020, 251, 117627. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Palm, J.; Mesters, J.; Siddell, S.G. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO 2002, 21, 3213–3224. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yu, H.; Yang, H.; Xue, F.; Wu, Z.; Shen, W.; Li, J.; Zhou, Z.; Ding, Y.; Zhao, Q.; et al. Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. J. Virol. 2008, 82, 2515–2527. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.; Yan, L.; Zhang, N.; Guo, Y.; Yang, C.; Lou, Z.; Rao, Z. The newly emerged SARS-like coronavirus HCoV-EMC also has an “Achilles” heel: “Current effective inhibitor targeting a 3C-like protease”. Protein Cell 2013, 4, 248–250. [Google Scholar] [CrossRef]
- Wang, W.; Ma, X.; Han, J.; Zhou, M.; Ren, H.; Pan, Q.; Zheng, C.; Zheng, Q. Neuroprotective Effect of Scutellarin on Ischemic Cerebral Injury by Down-Regulating the Expression of Angiotensin-Converting Enzyme and AT1 Receptor. PLoS ONE 2016, 11, e0146197. [Google Scholar] [CrossRef]
- Yang, H.; Yang, M.; Ding, Y.; Liu, Y.; Lou, Z.; Zhou, Z.; Sun, L.; Mo, L.; Ye, S.; Pang, H.; et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 2003, 100, 13190–13195. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Yao, S.; Zhao, W.; Li, M.; Liu, J.; Shang, W.; Xie, H.; Ke, C.; Hu, H.; Gao, M.; et al. How health anxiety influences responses to viral outbreaks like COVID-19: What all decision-makers, health authorities, and health care professionals need to know. J. Anxiety Disord. 2020, 71, 102211. [Google Scholar] [CrossRef]
- Hattori, S.I.; Higashi-Kuwata, N.; Hayashi, H.; Allu, S.R.; Raghavaiah, J.; Bulut, H.; Das, D.; Anson, B.J.; Lendy, E.K.; Takamatsu, Y.; et al. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021, 12, 668. [Google Scholar] [CrossRef] [PubMed]
- Douangamath, A.; Fearon, D.; Gehrtz, P.; Krojer, T.; Lukacik, P.; Owen, C.D.; Resnick, E.; Strain-Damerell, C.; Aimon, A.; Ábrányi-Balogh, P.; et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020, 11, 5047. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.L.; Kania, R.S.; Brothers, M.A.; Davies, J.F.; Ferre, R.A.; Gajiwala, K.S.; He, M.; Hogan, R.J.; Kozminski, K.; Li, L.Y.; et al. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [Google Scholar] [CrossRef]
- Fu, L.; Ye, F.; Feng, Y.; Yu, F.; Wang, Q.; Wu, Y.; Zhao, C.; Sun, H.; Huang, B.; Niu, P.; et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020, 11, 4417. [Google Scholar] [CrossRef]
- Sacco, M.D.; Ma, C.; Lagarias, P.; Gao, A.; Townsend, J.A.; Meng, X.; Dube, P.; Zhang, X.; Hu, Y.; Kitamura, N.; et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020, 6, eabe0751. [Google Scholar] [CrossRef]
- Fu, L.; Wang, B.; Yuan, T.; Chen, X.; Ao, Y. Clinical characteristics of coronavirus diease 2019 (COVID-19) in china: A systemic review and meta-analysis. J. Infect. 2020, 80, 656–665. [Google Scholar] [CrossRef]
- Adlercreutz, P. Immobilisation and application of lipases in organic media. Chem. Soc. Rev. 2013, 42, 6406–6436. [Google Scholar] [CrossRef] [Green Version]
- Harcourt, B.; Jukneliene, D.; Kanjanahaluethai, A.; Bechill, J. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004, 78, 13600–13612. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.-G.; Bieganowska Bourcier, K.; Freeman, G.J.; Hafler, D.A. Examination of CD8 + T Cell Function in Humans Using MHC Class I Tetramers: Similar Cytotoxicity but Variable Proliferation and Cytokine Production Among Different Clonal CD8 + T Cells Specific to a Single Viral Epitope. J. Immunol. 2000, 165, 6214–6220. [Google Scholar] [CrossRef] [Green Version]
- Xian, X.; Moraghebi, R.; Löfvall, H.; Fasth, A.; Henriksen, K.; Richter, J.; Woods, N.-B.; Moscatelli, I. Generation of gene-corrected functional osteoclasts from osteopetrotic induced pluripotent stem cells. Stem Cell Res. Ther. 2020, 11, 179. [Google Scholar] [CrossRef]
- Ratia, K.; Pegan, S.; Takayama, J.; Sleeman, K.; Coughlin, M.; Baliji, S.; Chaudhuri, R.; Fu, W.; Prabhakar, B.S.; Johnson, M.E.; et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA 2008, 105, 16119–16124. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Aleksandra, J.; Wojdyla, M.W.; Cui, S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B 2020, 11, 237–245. [Google Scholar] [CrossRef]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef] [Green Version]
- Rut, W.; Groborz, K.; Zhang, L.; Sun, X.; Zmudzinski, M.; Pawlik, B.; Wang, X.; Jochmans, D.; Neyts, J.; Młynarski, W.; et al. SARS-CoV-2 M pro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021, 17, 222–228. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Walls, A.C.; Lang, Y.; Wang, C.; Li, Z.; Koerhuis, D.; Boons, G.-J.; Bosch, B.-J.; Rey, F.A.; de Groot, R.J. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019, 26, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Hulswit, R.J.G.; de Haan, C.A.M.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2016; Volume 96. [Google Scholar]
- Kirchdoerfer, R.N.; Wang, N.; Pallesen, J.; Wrapp, D.; Turner, H.L.; Cottrell, C.A.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018, 8, 15701. [Google Scholar] [CrossRef] [Green Version]
- Gui, M.; Song, W.; Zhou, H.; Xu, J.; Chen, S.; Xiang, Y.; Wang, X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017, 27, 119–129. [Google Scholar] [CrossRef]
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Aboagye, J.; Yew, C.W.; Ng, O.W.; Monteil, V.M. Overexpression of the nucleocapsid protein of middle East respiratory syndrome coronavirus up- regulates CXCL10. Biosci. Rep. 2018, 38, BSR20181059. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Saikatendu, K.S.; Joseph, J.S.; Subramanian, V.; Neuman, B.W.; Buchmeier, M.J.; Stevens, R.C.; Kuhn, P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007, 81, 3913–3921. [Google Scholar] [CrossRef] [Green Version]
- Kang, L.; Li, Y.; Hu, S.; Chen, M.; Yang, C.; Yang, B.X.; Wang, Y.; Hu, J.; Lai, J.; Ma, X.; et al. The mental health of medical workers in Wuhan, China dealing with the 2019 novel coronavirus. The Lancet Psychiatry 2020, 7, e14. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.-W.; Yuan, S.; Yuen, K.-S.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef] [Green Version]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation and Treatment Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Senanayake, S.L. Drug repurposing strategies for COVID-19. Futur. Drug Discov. 2020, 2, 6–8. [Google Scholar] [CrossRef]
- National Institutes of Health. A prospective/retrospective, randomized controlled clinical study of antiviral therapy in the 2019-nCoV pneumonia. National Institutes of Health: Bethesda, MD, USA, 2020. [Google Scholar]
- Santoro, M.G.; Carafoli, E. Remdesivir: From Ebola to COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 145–150. [Google Scholar] [CrossRef]
- Trials, N.C. Mild/moderate 2019-nCoV remdesivir RCT. Available online: https://clinicaltrials.gov/ct2/show/NCT04261517 (accessed on 25 August 2021).
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.-X.; et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef]
- Release, N.N. NIH Clinical Trial Shows Remdesivir Accelerates Recovery from Advanced COVID-19. Available online: https://www.nih.gov/news-events/news-releases/nih-clinical-trial-shows-remdesivir-accelerates-recovery-advanced-covid-19 (accessed on 25 August 2021).
- Trials, N.C. Efficacy and safety of hydroxychloroquine for treatment of pneumonia caused by 2019-nCoV (HC-nCoV). Available online: https://www.cochranelibrary.com/central/doi/10.1002/central/CN-02080006/full (accessed on 25 August 2021).
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 449–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, J.; Minko, T. Recent Developments on Therapeutic and Diagnostic Approaches for COVID-19. AAPS J. 2021, 23. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Williamson, P.; Zheng, W. Drug repurposing of synergistic drug combinations as an alternative approach for the treatment of severe infectious diseases. Curr. Opin. Pharmacol. 2019, 48. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, K.; Rarey, M. What is the potential of structure-based target prediction methods?. Future medicinal chemistry. Future Med. Chem. 2014, 6, 1987–1989. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 10, 607–613. [Google Scholar] [CrossRef]
- Kaur, S.; Bansal, Y.; Kumar, R.; Bansal, G. A panoramic review of IL-6: Structure, pathophysiological roles and inhibitors. Bioorganic Med. Chem. 2020, 28, 115327. [Google Scholar] [CrossRef]
- Wu, J.; Shen, J.; Han, Y.; Qiao, Q.; Dai, W.; He, B.; Pang, R.; Zhao, J.; Luo, T.; Guo, Y.; et al. Upregulated IL-6 Indicates a Poor COVID-19 Prognosis: A Call for Tocilizumab and Convalescent Plasma Treatment. Front. Immunol. 2021, 12, 455. [Google Scholar] [CrossRef]
- Saleem, A.; Akhtar, M.F.; Haris, M.; Abdel-Daim, M.M. Recent updates on immunological, pharmacological, and alternative approaches to combat COVID-19. Inflammopharmacology 2021, 30, 1331–1346. [Google Scholar] [CrossRef]
- Russell, C.D.; Millar, J.E.; Baillie, J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020, 395, 473–475. [Google Scholar] [CrossRef] [Green Version]
- Lammers, T.; Sofias, A.; van der Mee, l.R.; Schiffelers, R.; Storm, G.; Tacke, F.; Koschmieder, S.; Brümmendorf, T.H.; Kiessling, F.M.J. Dexamethasone nanomedicines for COVID-19. Nat. Nanotechnol. 2020, 15, 622–624. [Google Scholar] [CrossRef]
- Meyerowitz, E.A.; Vannier, A.G.L.; Friesen, M.G.N.; Schoenfeld, S.; Gelfand, J.A.; Callahan, M.V.; Kim, A.Y.; Reeves, P.M.; Poznansky, M.C. Rethinking the role of hydroxychloroquine in the treatment of COVID-19. FASEB J. 2020, 34, 6027–6037. [Google Scholar] [CrossRef]
- Bekerman, E.; Neveu, G.; Shulla, A.; Brannan, J.; Pu, S.; Wang, S.; Xiao, F.; Barouch-Bentov, R.; Bakken, R.; Mateo, R.; et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Invest. 2017, 127, 1338–1352. [Google Scholar] [CrossRef]
- Sorrell, F.J. Family-wide structural analysis of human numb-associated protein kinases. Structure 2016, 24, 401–411. [Google Scholar] [CrossRef] [Green Version]
- Carbajo-Lozoya, J.; Müller, M.A.; Kallies, S.; Thiel, V.; Drosten, C.; von Brunn, A. Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res. 2012, 165, 112–117. [Google Scholar] [CrossRef]
- Wang, C.H. Adjuvant treatment with a mammalian target of rapamycin inhibitor, sirolimus, and steroids improves outcomes in patients with severe H1N1 pneumonia and acute respiratory failure. Crit. Care Med. 2014, 42, 313–321. [Google Scholar] [CrossRef]
- Tiberghien, P. Collecting and evaluating convalescent plasma for COVID-19 treatment: Why and how. Vox Sang. 2020, 115, 488–494. [Google Scholar] [CrossRef]
- Marando, M.; Tamburello, A. Immunoglobulins or convalescent plasma to tackle COVID-19: Buying time to save lives–current situation and perspectives. Swiss Med. Wkly. 2020, 150, 1718. [Google Scholar] [CrossRef]
- Chen, L. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Kumar, G.V.; Jeyanthi, V.; Ramakrishnan, S. A short review on antibody therapy for COVID-19. New Microbes New Infect. 2020, 100682. [Google Scholar]
- Lin, L.; Lu, L.; Cao, W.; Li, T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020, 9, 727–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, B.L.; McCullough, J. Treatment for emerging viruses: Convalescent plasma and COVID-19. Transfus. Apher. Sci. Off. J. World Apher. Assoc. Off. J. Eur. Soc. Haemapheresis 2020, 59, 102790. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.C.; Goh, H.P.; Koh, D. Treatment of COVID-19: Old tricks for new challenges. Crit. Care 2020, 24, 91. [Google Scholar] [CrossRef] [Green Version]
- Lurie, N.; Saville, M.; Hatchett, R.; Halton, J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020, 382, 1969–1973. [Google Scholar] [CrossRef]
- Won, J.H.; Lee, H. The Current Status of Drug Repositioning and Vaccine Developments for the COVID-19 Pandemic. Int. J. Mol. Sci. 2020, 21, 9775. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Jansen, K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Zeng, G.; Pan, H.X. Immunogenicity and safety of a SARS-CoV-2 inactivated vaccine in healthy adults aged 18–59 years: Report of the randomized, double-blind, and placebo-controlled phase 2 clinical trial. Medrxiv 2020, 25, 102. [Google Scholar]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G. An mRNA vaccine against SARS-CoV-2—preliminary report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Chu, L.; McPhee, R.; Huang, W.; Bennett, H.; Pajon, R.; Nestorova, B.; Leav, B.; mRNA-1273 Study Group. A preliminary report of a randomized controlled phase 2 trial of the safety and immunogenicity of mRNA-1273 SARS-CoV-2 vaccine. Vaccine 2021, 39, 2791–2799. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A. Safety and efficacy of single-dose Ad26. COV2. S vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef] [PubMed]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Jones, I.; Roy, P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet 2021, 397, 642–643. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef]
- Delrue, I.; Verzele, D.; Madder, A.; Nauwynck, H.J. Inactivated virus vaccines from chemistry to prophylaxis: Merits, risks and challenges. Expert Rev. Vaccines 2012, 11, 695–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, S.; Zhang, Y.; Wang, Y. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef]
- Crasto, A.M. BBIBP-CorV, Sinopharm COVID-19 Vaccine. New Drug Approvals. Available online: https://newdrugapprovals.org/2021/03/23/bbibp-corv-sinopharm-covid-19-vaccine/ (accessed on 25 August 2021).
- Pollet, J.; Chen, W.H.; Strych, U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv. Drug Deliv. Rev. 2021, 170, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E.; Mallapaty, S. Novavax Covid vaccine protects people against variants. Nature 2021, 590, 17. [Google Scholar] [CrossRef]
- Ella, R.; Vadrevu, K.M.; Jogdand, H.; Prasad, S.; Reddy, S.; Sarangi, V.; Ganneru, B.; Sapkal, G.; Yadav, P.; Abraham, P.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: A double-blind, randomised, phase 1 trial. Lancet Infect. Dis. 2021, 21, 637–646. [Google Scholar] [CrossRef]
- Sharma, O.; Sultan, A.A.; Ding, H.; Triggle, C.R. A review of the progress and challenges of developing a vaccine for COVID-19. Front. Immunol. 2020, 11, 2413. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, J.; Zhang, Z.; Wu, J.; Zhang, J.; Hu, H.; Zhu, T.; Zhang, J.; Luo, L.; Fan, P.; et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: Preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect. Dis. 2021, 3099, 1–11. [Google Scholar] [CrossRef]
- Abdel-Azeem, A.M.; Zaki, S.M.; Khalil, W.F.; Makhlouf, N.A.; Farghaly, L.M. Anti-rheumatoid Activity of Secondary Metabolites Produced by Endophytic Chaetomium globosum. Front. Microbiol. 2016, 7, 1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, Z.; Collado, J.; Singh, S.; Jayasuriya, H.; Dewey, R.; Polishook, J.D.; Dombrowski, A.W.; Zink, D.L.; Platas, G.; Pelaez, F. Isolation, structure, and HIV-1-integrase inhibitory activity of structurally diverse fungal metabolites. J. Ind. Microbiol. Biotechnol. 2003, 30, 721–731. [Google Scholar] [CrossRef]
- Isaka, M.; Berkaew, P.; Intereya, K.; Komwijit, S.; Sathitkunanon, T. Antiplasmodial and antiviral cyclohexadepsipeptides from the endophytic fungus Pullularia sp. BCC 8613. Tetrahedron 2007, 63, 6855–6860. [Google Scholar] [CrossRef]
- Roy, B.G. Potential of small-molecule fungal metabolites in antiviral chemotherapy. Antivir. Chem. Chemother. 2017, 25, 20–52. [Google Scholar] [CrossRef]
- Fernández-Montero, J.V.; Barreiro, P.; Soriano, V. HIV protease inhibitors: Recent clinical trials and recommendations on use. Expert Opin. Pharmacother. 2009, 10, 1615–1629. [Google Scholar] [CrossRef]
- Mielech, A.M.; Kilianski, A.; Baez-Santos, Y.M.; Mesecar, A.D.; Baker, S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 2014, 450, 64–70. [Google Scholar] [CrossRef]
- Rao, P.; Patel, R.; Shukla, A.; Parmar, P.; Rawal, R.M.; Saraf, M.; Goswami, D. Identifying structural–functional analogue of GRL0617, the only well-established inhibitor for papain-like protease (PLpro) of SARS-CoV2 from the pool of fungal metabolites using docking and molecular dynamics simulation. Mol. Divers. 2021, 1–21. [Google Scholar] [CrossRef]
- Rao, P.; Shukla, A.; Parmar, P.; Rawal, R.M.; Patel, B.; Saraf, M.; Goswami, D. Reckoning a fungal metabolite, Pyranonigrin A as a potential Main protease (Mpro) inhibitor of novel SARS-CoV-2 virus identified using docking and molecular dynamics simulation. Biophys. Chem. 2020, 264, 106425. [Google Scholar] [CrossRef]
- Patel, R.S.; Vanzara, A.G.; Patel, N.R.; Vasava, A.M.; Patil, S.M.; Rajput, K.S. In-silico Discovery of Fungal Metabolites Bergenin, Quercitrin and Dihydroartemisinin as Potential Inhibitors against Main Protease of SARS-CoV-2. Coronaviruses 2020, 1, 1–23. [Google Scholar] [CrossRef]
- Patel, D.K.; Patel, K.; Kumar, R.; Gadewar, M.; Tahilyani, V. Pharmacological and analytical aspects of bergenin: A concise report. Asian Pacific J. Trop. Dis. 2012, 2, 163–167. [Google Scholar] [CrossRef]
- Piacente, S.; Pizza, C.; De Tommasi, N.; Mahmood, N. Constituents of Ardisia japonica and their in vitro anti-HIV activity. J. Nat. Prod. 1996, 59, 565–569. [Google Scholar] [CrossRef]
- Zuo, G.Y.; Li, Z.Q.; Chen, L.R.; Xu, X.J. In vitro anti-HCV activities of Saxifraga melanocentra and its related polyphenolic compounds. Antivir. Chem. Chemother. 2005, 16, 393–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.B.; Nakamura, N.; Miyashiro, H.; Hattori, M.; Jong, C.P. Triterpenoids and flavonoids isolated from the leaves of Alnus firma. Korean J. Pharmacogn. 2007, 38, 76–83. [Google Scholar]
- Aoki, C.; Hartati, S.; Santi, M.R.; Lydwina; Firdaus, R.; Hanafi, M.; Kardono, L.B.S.; Shimizu, Y.; Sudarmono, P.; Hotta, H. Isolation and identification of substances with anti-hepatitis c virus activities from kalanchoe pinnata. Int. J. Pharm. Pharm. Sci. 2014, 6, 211–215. [Google Scholar]
- Chiow, K.H.; Phoon, M.C.; Putti, T.; Tan, B.K.H.; Chow, V.T. Evaluation of antiviral activities of Houttuynia cordata Thunb. extract, quercetin, quercetrin and cinanserin on murine coronavirus and dengue virus infection. Asian Pac. J. Trop. Med. 2016, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Keating, G.M. Dihydroartemisinin/piperaquine: A review of its use in the treatment of uncomplicated plasmodium falciparum malaria. Drugs 2012, 72, 937–961. [Google Scholar] [CrossRef]
- Wicklow, D.T.; Poling, S.M. Antimicrobial activity of pyrrocidines from Acremonium zeae against endophytes and pathogens of maize. Phytopathology 2009, 99, 109–115. [Google Scholar] [CrossRef] [Green Version]
- Duecker, F.L.; Heinze, R.C.; Heretsch, P. Synthesis of Swinhoeisterol A, Dankasterone A and B, and Periconiastone A by Radical Framework Reconstruction. J. Am. Chem. Soc. 2020, 142, 104–108. [Google Scholar] [CrossRef]
- Ebrahimi, K.S.; Ansari, M.; Hosseyni Moghaddam, M.S.; Ebrahimi, Z.; Salehi, Z.; Shahlaei, M.; Moradi, S. In silico investigation on the inhibitory effect of fungal secondary metabolites on RNA dependent RNA polymerase of SARS-CoV-II: A docking and molecular dynamic simulation study. Comput. Biol. Med. 2021, 135, 104613. [Google Scholar] [CrossRef]
- Moore, J.B.; June, C.H. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef] [Green Version]
- Faulds, D.; Goa, K.L.; Benfield, P. Cyclosporin: A Review of its Pharmacodynamic and Pharmacokinetic Properties, and Therapeutic Use in Immunoregulatory Disorders. Drugs 1993, 45, 953–1040. [Google Scholar] [CrossRef]
- Watashi, K.; Hijikata, M.; Hosaka, M.; Yamaji, M.; Shimotohno, K. Cyclosporin A Suppresses Replication of Hepatitis C Virus Genome in Cultured Hepatocytes. Hepatology 2003, 38, 1282–1288. [Google Scholar] [CrossRef]
- Li, H.S.; Kuok, D.I.T.; Cheung, M.C.; Ng, M.M.T.; Ng, K.C.; Hui, K.P.Y.; Peiris, J.S.M.; Chan, M.C.W.; Nicholls, J.M. Effect of interferon alpha and cyclosporine treatment separately and in combination on Middle East Respiratory Syndrome Coronavirus (MERS-CoV) replication in a human in-vitro and ex-vivo culture model. Antiviral Res. 2018, 155, 89–96. [Google Scholar] [CrossRef]
- De Wilde, A.H.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Thiel, V.; Narayanan, K.; Makino, S.; Snijder, E.J.; van Hemert, M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011, 92, 2542–2548. [Google Scholar] [CrossRef]
- Guisado-Vasco, P.; Valderas-Ortega, S.; Carralón-González, M.M.; Roda-Santacruz, A.; González-Cortijo, L.; Sotres-Fernández, G.; Martí-Ballesteros, E.M.; Luque-Pinilla, J.M.; Almagro-Casado, E.; La Coma-Lanuza, F.J.; et al. Clinical characteristics and outcomes among hospitalized adults with severe COVID-19 admitted to a tertiary medical center and receiving antiviral, antimalarials, glucocorticoids, or immunomodulation with tocilizumab or cyclosporine: A retrospective obser. EClinicalMedicine 2020, 28, 100591. [Google Scholar] [CrossRef]
- Mallard, B.; Leach, D.N.; Wohlmuth, H.; Tiralongo, J. Synergistic immuno-modulatory activity in human macrophages of a medicinal mushroom formulation consisting of Reishi, Shiitake and Maitake. PLoS ONE 2019, 14, e0224740. [Google Scholar] [CrossRef] [Green Version]
- El-Maradny, Y.A.; El-Fakharany, E.M.; Abu-Serie, M.M.; Hashish, M.H.; Selim, H.S. Lectins purified from medicinal and edible mushrooms: Insights into their antiviral activity against pathogenic viruses. Int. J. Biol. Macromol. 2021, 179, 239–258. [Google Scholar] [CrossRef]
- Channappanavar, R.; Perlman, S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017, 39, 529–539. [Google Scholar] [CrossRef]
- Vetvicka, V.; Vashishta, A.; Saraswat-Ohri, S.; Vetvickova, J. Immunological Effects of Yeast- and Mushroom-Derived β-Glucans. J. Med. Food 2008, 11, 615–622. [Google Scholar] [CrossRef]
- Ahmad, M.F.; Ahmad, F.A.; Khan, M.I.; Alsayegh, A.A.; Wahab, S.; Alam, M.I.; Ahmed, F. Ganoderma lucidum: A potential source to surmount viral infections through β-glucans immunomodulatory and triterpenoids antiviral properties. Int. J. Biol. Macromol. 2021, 187, 769–779. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, J.; Zhang, T. Immunomodulatory activities of polysaccharides from Ganoderma on immune effector cells. Food Chem. 2021, 340, 127933. [Google Scholar] [CrossRef]
- Cai, S.; Xiao, H.; Wang, X.; Lin, S.; Zhong, J.J. Bioconversion of a ganoderic acid 3-hydroxy-lanosta-8,24-dien-26-oic acid by a crude enzyme from Ganoderma lucidum. Process Biochem. 2020, 95, 12–16. [Google Scholar] [CrossRef]
- Zhu, Q.; Bang, T.H.; Ohnuki, K.; Sawai, T.; Sawai, K.; Shimizu, K. Inhibition of neuraminidase by Ganoderma triterpenoids and implications for neuraminidase inhibitor design. Sci. Rep. 2015, 5, 13194. [Google Scholar] [CrossRef]
- Li, Q.Z.; Zheng, Y.Z.; Zhou, X.W. Fungal immunomodulatory proteins: Characteristic, potential antitumor activities and their molecular mechanisms. Drug Discov. Today 2019, 24, 307–314. [Google Scholar] [CrossRef]
- Elaya Perumal, U.; Sundararaj, R. Algae: A potential source to prevent and cure the novel coronavirus – A review. Int. J. Emerg. Technol. 2020, 479–483. [Google Scholar]
- Alam, M.A.; Parra-Saldivar, R.; Bilal, M.; Afroze, C.A.; Ahmed, M.N.; Iqbal, H.M.N.; Xu, J. Algae-derived bioactive molecules for the potential treatment of sars-cov-2. Molecules 2021, 26, 2134. [Google Scholar] [CrossRef]
- O’Keefe, B.R.; Giomarelli, B.; Barnard, D.L.; Shenoy, S.R.; Chan, P.K.S.; McMahon, J.B.; Palmer, K.E.; Barnett, B.W.; Meyerholz, D.K.; Wohlford-Lenane, C.L.; et al. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010, 84, 2511–2521. [Google Scholar] [CrossRef] [Green Version]
- Barre, A.; Simplicien, M.; Benoist, H.; Van Damme, E.J.M.; Rouge, P. Mannose specific lectins from marine algae: Diverse structural scaffolds associated to common virucidal and anti-cancer properties. Mar. Drugs 2019, 17, 440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa, G.P.; Tavares, W.R.; Sousa, P.M.C.; Pagès, A.K.; Seca, A.M.L.; Pinto, D.C.G.A. Seaweed Secondary Metabolites with Beneficial Health Effects: An Overview of Successes in In Vivo Studies and Clinical Trials. Mar. Drugs 2019, 18, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L. Antiviral activity of seaweeds and their extracts. In Therapeutic and Nutritional Uses of Algae; CRC Press: Boca Raton, FL, USA, 2018; pp. 175–211. [Google Scholar]
- Shi, Q.; Wang, A.; Lu, Z.; Qin, C.; Hu, J.; Yin, J. Overview on the antiviral activities and mechanisms of marine polysaccharides from seaweeds. Carbohydr. Res. 2017, 453–454, 1–9. [Google Scholar] [CrossRef]
- Gerber, P.; Dutcher, J.D.; Adams, E.V.; Sherman, J.H. Protective effect of seaweed extracts for chicken embryos infected with influenza B or mumps virus. Exp. Biol. Med. 1958, 99, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Deig, E.F.; Ehresmann, D.W.; Hatch, M.T.; Riedlinger, D.J. Inhibition of herpesvirus replication bymarine algae extracts. Antimicrob. Agents Chemother. 1974, 6, 524–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehresmann, D.W.; Deig, E.F.; Hatch, M.T.; DiSalvo, L.H.; Vedros, N.A. Antiviral substances from California marine algae. J. Phycol. 1977, 13, 37–40. [Google Scholar] [CrossRef]
- Richards, J.T.; Kern, E.R.; Glasgow, L.A.; Overall, J.C.; Deign, E.F.; Hatch, M.T. Antiviral activity of extracts from marine algae. Antimicrob. Agents Chemother. 1978, 14, 24–30. [Google Scholar] [CrossRef] [Green Version]
- Neushul, M. Antiviral Carbohydrates from Marine Red Algae; Lindstrom, S.C., Gabrielson, P.W., Eds.; Springer: Amsterdam, The Netherlands, 1990; pp. 99–104. [Google Scholar]
- Park, J.; Hoon, J.; Min, J.; Kwon, H.; Jae, H.; Min, Y.; Kim, D.; Song, W.; Bae, Y. Dieckol, a SARS-CoV 3CL(pro) inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg. Med. Chem. 2013, 21, 3730–3737. [Google Scholar] [CrossRef]
- Millet, J.K.; Séron, K.; Labitt, R.N.; Danneels, A.; Palmer, K.E.; Whittaker, G.R.; Dubuisson, J.; Belouzard, S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antiviral Res. 2016, 133, 1–8. [Google Scholar] [CrossRef]
- Lee, C. Griffithsin, a highly potent broad-spectrum antiviral lectin from red algae: From discovery to clinical application. Mar. Drugs 2019, 17, 267. [Google Scholar] [CrossRef] [Green Version]
- Elshabrawy, H.A. SARS-CoV-2: An update on potential antivirals in light of SARS-CoV antiviral drug discoveries. Vaccines 2020, 8, 335. [Google Scholar] [CrossRef]
- Morokutti-Kurz, M.; Fröba, M.; Graf, P.; Große, M.; Grassauer, A.; Auth, J.; Schubert, U.; Prieschl-Grassauer, E. Iota-carrageenan neutralizes SARS-CoV-2 and inhibits viral replication in vitro. PLoS ONE 2021, 16, e0237480. [Google Scholar] [CrossRef]
- Kwon, P.S.; Oh, H.; Kwon, S.J.; Jin, W.; Zhang, F.; Fraser, K.; Hong, J.J.; Linhardt, R.J.; Dordick, J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020, 6, 4–7. [Google Scholar] [CrossRef]
- Grassauer, A.; Weinmuellner, R.; Meier, C.; Pretsch, A.; Prieschl-Grassauer, E.; Unger, H. Iota-Carrageenan is a potent inhibitor of rhinovirus infection. Virol. J. 2008, 5, 107. [Google Scholar] [CrossRef] [Green Version]
- Buck, C.B.; Thompson, C.D.; Roberts, J.N.; Müller, M.; Lowy, D.R.; Schiller, J.T. Carrageenan is a potent inhibitor of papillomavirus infection. PLOS Pathog. 2006, 2, e69. [Google Scholar] [CrossRef] [Green Version]
- Alsaidi, S.; Cornejal, N.; Mahoney, O.; Melo, C.; Verma, N.; Bonnaire, T.; Chang, T.; O’Keefe, B.R.; Sailer, J.; Zydowsky, T.M.; et al. Griffithsin and Carrageenan Combination Results in Antiviral Synergy against SARS-CoV-1 and 2 in a Pseudoviral Model. Mar. Drugs 2021, 19, 418. [Google Scholar] [CrossRef]
- Joseph, J.; Karthika, T.; Ajay, A.; Das, V.R.A.; Raj, V.S. Green tea and Spirulina extracts inhibit SARS, MERS, and SARS-2 spike pseudotyped virus entry in vitro. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hayashi, T.; Hayashi, K.; Maeda, M.; Kojima, I. Calcium spirulan, an inhibitor of enveloped virus replication, from a blue-green alga Spirulina platensis. J. Nat. Prod. 1996, 59, 83–87. [Google Scholar] [CrossRef]
- Witvrouw, M.; Este, J.A.; Mateu, M.Q.; Reymen, D.; Andrei, G.; Snoeck, R.; Ikeda, S.; Pauwels, R.; Bianchini, N.V.; Desmyter, J. Activity of a sulfated polysaccharide extracted from the red seaweed Aghardhiella tenera against human immunodeficiency virus and other enveloped viruses. Antivir. Chem. Chemother. 1994, 5, 297–303. [Google Scholar] [CrossRef] [Green Version]
- Gustafson, K.R.; Sowder, R.C.; Henderson, L.E.; Cardellina, J.H.; McMahon, J.B.; Rajamani, U.; Pannell, L.K.; Boyd, M.R. Isolation, primary sequence determination, and disulfide bond structure of cyanovirin-N, an anti-HIV (Human Immunodeficiency Virus) protein from the cyanobacterium Nostoc ellipsosporum. Biochem. Biophys. Res. Commun. 1997, 238, 223–228. [Google Scholar] [CrossRef]
- Dey, B.; Lerner, D.L.; Lusso, P.; Boyd, M.R.; Elder, J.H.; Berger, E.A. Multiple Antiviral Activities of Cyanovirin-N: Blocking of Human Immunodeficiency Virus Type 1 gp120 Interaction with CD4 and Coreceptor and Inhibition of Diverse Enveloped Viruses. J. Virol. 2000, 74, 4562–4569. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Hayashi, K.; Hirata, M.; Kuroda, E.; Suzuki, E.; Kubo, Y.; Hayashi, T. Antiviral sulfated polysaccharide from Navicula directa, a diatom collected from deep-sea water in Toyama Bay. Biol. Pharm. Bull. 2006, 29, 2135–2139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ale, M.T.; Meyer, A.S. Fucoidans from brown seaweeds: An update on structures, extraction techniques and use of enzymes as tools for structural elucidation. RSC Adv. 2013, 3, 8131–8141. [Google Scholar] [CrossRef] [Green Version]
- Shanura Fernando, I.P.; Asanka Sanjeewa, K.K.; Samarakoon, K.W.; Lee, W.W.; Kim, H.S.; Ranasinghe, P.; Gunasekara, U.K.D.S.S.; Jeon, Y.J. Antioxidant and anti-inflammatory functionality of ten Sri Lankan seaweed extracts obtained by carbohydrase assisted extraction. Food Sci. Biotechnol. 2018, 27, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Fernando, I.P.S.; Ryu, B.M.; Ahn, G.; Yeo, I.K.; Jeon, Y.J. Therapeutic potential of algal natural products against metabolic syndrome: A review of recent developments. Trends Food Sci. Technol. 2020, 97, 286–299. [Google Scholar] [CrossRef]
- Irhimeh, M.R.; Fitton, J.H.; Lowenthal, R.M. Pilot clinical study to evaluate the anticoagulant activity of fucoidan. Blood Coagul. Fibrinolysis 2009, 20, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Krylova, N.V.; Ermakova, S.P.; Lavrov, V.F.; Leneva, I.A.; Kompanets, G.G.; Iunikhina, O.V.; Nosik, M.N.; Ebralidze, L.K.; Falynskova, I.N.; Silchenko, A.S.; et al. The comparative analysis of antiviral activity of native and modified fucoidans from brown algae fucus evanescens in Vitro and in Vivo. Mar. Drugs 2020, 18, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pozharitskaya, O.; Obluchinskaya, E.; Shikov, A. Mechanisms of Bioactivities of Fucoidan from the Brown Seaweed Fucus vesiculosus L. of the Barents Sea. Mar. Drugs 2020, 18, 275. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Sun, F.; Wang, L.; Gao, M.; Xie, Y.; Sun, Y.; Liu, H.; Yuan, Y.; Yi, W.; Huang, Z.; et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranotics 2020, 10, 12223–12240. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernandez, V.; Aoki-Utsubo, C.; Moustafa, M.A.; Possani, L.D.; Hotta, H.; El-Bitar, A.M.; Sarhan, M.; Abdel-Rahman, M.A. Smp76, a scorpine-like peptide isolated from the venom of the scorpion Scorpio maurus palmatus, with a potent antiviral activity against hepatitis C virus and dengue virus. Int. J. Pept. Res. Ther. 2020, 26, 811–821. [Google Scholar]
- Hong, W.; Li, T.; Song, Y.; Zhang, R.; Zeng, Z.; Han, S.; Zhang, X.; Wu, Y.; Li, W.; Cao, Z. Inhibitory activity and mechanism of two scorpion venom peptides against herpes simplex virus type 1. Antiviral Res. 2014, 102, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Hong, W.; Zeng, Z.; Wu, Y.; Hu, K.; Tian, X.; Li, W.; Cao, Z. Mucroporin-M1 Inhibits Hepatitis B Virus Replication by Activating the Mitogen-activated Protein Kinase (MAPK) Pathway and Down-regulating HNF4α in Vitro and in Vivo. J. Biol. Chem. 2012, 287, 30181–30190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Z.; Li, F.; Xia, Z.; Guo, X.; Gao, M.; Sun, F.; Cheng, Y.; Wu, Y.; Li, W.; Ali, S.A.; et al. The Scorpion Venom Peptide Smp76 Inhibits Viral Infection by Regulating Type-I Interferon Response. Virol. Sin. 2018, 33, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.F.; Rateb, M.E.; El Kassem, L.T.A.; Hawas, U.W. Anti-HCV protease of diketopiperazinApplied es produced by the red sea sponge-saaociated fungus Aspergillus versicolor. Appl. Biochem. Microbiol. 2017, 53, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhao, Z.; He, Y.; Wu, L.; Cai, D.; Hong, W.; Wu, Y.; Cao, Z.; Zheng, C.; Li, W. A new natural α-helical peptide from the venom of the scorpion Heterometrus petersii kills HCV. Peptides 2011, 32, 11–19. [Google Scholar] [CrossRef]
- Hong, W.; Zhang, R.; Di, Z.; He, Y.; Zhao, Z.; Hu, J.; Wu, Y.; Li, W.; Cao, Z. Design of histidine-rich peptides with enhanced bioavailability and inhibitory activity against hepatitis C virus. Biomaterials 2013, 34, 3511–3522. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, L.; Zhong, M.; Zhang, Y.; Han, C.; Li, Q.; Yang, J.; Zhou, D.; Shi, W.; He, B.; et al. Anti-HIV-1 activity of a new scorpion venom peptide derivative Kn2-7. PLoS ONE 2012, 7, e34947. [Google Scholar] [CrossRef] [Green Version]
- Grimes, J.M.; Grimes, K.V. p38 MAPK inhibition: A promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020, 144, 63–65. [Google Scholar] [CrossRef]
- Cao, R.; Hu, H.; Li, Y.; Wang, X.; Xu, M.; Liu, J.; Zhang, H.; Yan, Y.; Zhao, L.; Li, W. Anti-SARS-CoV-2 Potential of Artemisinins In Vitro. ACS Infect. Dis. 2020, 6, 2524–2531. [Google Scholar] [CrossRef]
- Bettuzzi, S.; Gabba, L.; Cataldo, S. Efficacy of a Polyphenolic, Standardized Green Tea Extract for the Treatment of COVID-19 Syndrome: A Proof-of-Principle Study. Covid 2021, 1, 2–12. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, X.; Hu, N.; Zhang, X.; Sun, T.; Xu, J.; Bian, X. Baicalin attenuates angiotensin II-induced endothelial dysfunction. Biochem. Biophys. Res. Commun. 2015, 465, 101–107. [Google Scholar] [CrossRef]
- Zandi, K.; Musall, K.; Oo, A.; Cao, D.; Liang, B.; Hassandarvish, P.; Lan, S.; Slack, R.L.; Kirby, K.A.; Bassit, L.; et al. Baicalein and baicalin inhibit sars-cov-2 rna-dependent-rna polymerase. Microorganisms 2021, 9, 893. [Google Scholar] [CrossRef]
- Chen, F.; Chan, K.H.; Jiang, Y.; Kao, R.Y.T.; Lu, H.T.; Fan, K.W.; Cheng, V.C.C.; Tsui, W.H.W.; Hung, I.F.N.; Lee, T.S.W.; et al. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Li, S.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzyme Inhib. Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef]
- Senthil Kumar, K.J.; Vani, M.G.; Wang, C.S.; Chen, C.C.; Chen, Y.C.; Lu, L.P.; Huang, C.H.; Lai, C.S.; Wang, S.Y. Geranium and lemon essential oils and their active compounds downregulate angiotensin-converting enzyme 2 (ACE2), a SARS-CoV-2 spike receptor-binding domain, in epithelial cells. Plants 2020, 9, 770. [Google Scholar] [CrossRef]
- Xiao, T.; Cui, M.; Zheng, C.; Wang, M.; Sun, R.; Gao, D.; Bao, J.; Ren, S.; Yang, B.; Lin, J.; et al. Myricetin Inhibits SARS-CoV-2 Viral Replication by Targeting Mpro and Ameliorates Pulmonary Inflammation. Front. Pharmacol. 2021, 12, 1012. [Google Scholar] [CrossRef]
- Benarba, B.; Pandiella, A. Medicinal Plants as Sources of Active Molecules Against COVID-19. Front. Pharmacol. 2020, 11, 1189. [Google Scholar] [CrossRef]
- Górniak, I.; Bartoszewski, R.; Króliczewski, J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 2019, 18, 241–272. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, M.; Jiang, H.; Suzuki, Y.; Li, X.; Xiao, P.; Tanaka, T.; Ling, H.; Yang, B.; Saitoh, H.; Zhang, L.; et al. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antiviral Res. 2009, 82, 73–81. [Google Scholar] [CrossRef]
- Ghosh, D. A cinnamon - derived procyanidin type—A compound: A potential candidate molecule against coronaviruses including COVID - 19. J. Ayurveda Case Rep. 2020, 3, 122. [Google Scholar] [CrossRef]
- Jo, S.; Kim, S.; Shin, D.H.; Kim, M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020, 35, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltane, R.; Chrouda, A.; Mostafa, A.; Al-Karmalawy, A.A.; Chouaïb, K.; Dhahri, A.; Pashameah, R.A.; Alasiri, A.; Kutkat, O.; Shehata, M.; et al. Strong inhibitory activity and action modes of synthetic maslinic acid derivative on highly pathogenic coronaviruses: Covid-19 drug candidate. Pathogens 2021, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.C.; Kuo, Y.H.; Jan, J.T.; Liang, P.H.; Wang, S.Y.; Liu, H.G.; Lee, C.K.; Chang, S.T.; Kuo, C.J.; Lee, S.S.; et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007, 50, 4087–4095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Wang, S.; Zhao, J.; Yu, C.; Hu, Y.; Tu, Y.; Yang, Z.; Zheng, J.; Wang, Y.; Gao, Y. Naringenin ameliorates renovascular hypertensive renal damage by normalizing the balance of renin-angiotensin system components in rats. Int. J. Med. Sci. 2019, 16, 644–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abubakar, M.B.; Usman, D.; El-Saber Batiha, G.; Cruz-Martins, N.; Malami, I.; Ibrahim, K.G.; Abubakar, B.; Bello, M.B.; Muhammad, A.; Gan, S.H.; et al. Natural Products Modulating Angiotensin Converting Enzyme 2 (ACE2) as Potential COVID-19 Therapies. Front. Pharmacol. 2021, 12, 629935. [Google Scholar] [CrossRef]
- Kotwal, G.J.; Kaczmarek, J.N.; Leivers, S.; Ghebremariam, Y.T.; Kulkarni, A.P.; Bauer, G.; De Beer, C.; Preiser, W.; Mohamed, A.R. Anti-HIV, anti-poxvirus, and anti-SARS activity of a nontoxic, acidic plant extract from the Trifollium species Secomet-V/anti-Vac suggests that it contains a novel broad-spectrum antiviral. Ann. N. Y. Acad. Sci. 2005, 1056, 293–302. [Google Scholar] [CrossRef]
- Orhan, I.E.; Senol Deniz, F.S. Natural Products as Potential Leads Against Coronaviruses: Could They be Encouraging Structural Models Against SARS-CoV-2? Nat. Products Bioprospect. 2020, 10, 171–186. [Google Scholar] [CrossRef]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, L.E.; Lin, C.N.; Su, B.L.; Jan, T.R.; Chen, C.M.; Wang, C.H.; Lin, D.S.; Lin, C.T.; Chueh, L.L. Synergistic antiviral effect of Galanthus nivalis agglutinin and nelfinavir against feline coronavirus. Antiviral Res. 2010, 88, 25–30. [Google Scholar] [CrossRef]
- Keyaerts, E.; Vijgen, L.; Pannecouque, C.; Van Damme, E.; Peumans, W.; Egberink, H.; Balzarini, J.; Van Ranst, M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 2007, 75, 179–187. [Google Scholar] [CrossRef]
- Wink, M. Potential of DNA Intercalating Alkaloids and Other Plant Secondary Metabolites against SARS-CoV-2 Causing COVID-19. Diversity 2020, 12, 175. [Google Scholar] [CrossRef]
- Schnitzler, P.; Schuhmacher, A.; Astani, A.; Reichling, J. Melissa officinalis oil affects infectivity of enveloped herpesviruses. Phytomedicine Int. J. Phyther. Phytopharm. 2008, 15, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Abdizadeh, R.; Hadizadeh, F.; Abdizadeh, T. In silico analysis and identification of antiviral coumarin derivatives against 3 - chymotrypsin - like main protease of the novel coronavirus SARS - CoV - 2. Mol. Divers. 2021, 1–24. [Google Scholar] [CrossRef]
- Ryu, Y.B.; Jeong, H.J.; Kim, J.H.; Kim, Y.M.; Park, J.-Y.; Kim, D.; Nguyen, T.T.H.; Park, S.-J.; Chang, J.S.; Park, K.H.; et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 2010, 18, 7940–7947. [Google Scholar] [CrossRef]
- Ho, T.-Y.; Wu, S.-L.; Chen, J.-C.; Li, C.-C.; Hsiang, C.-Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007, 74, 92–101. [Google Scholar] [CrossRef]
- Park, J.-Y.; Yuk, H.J.; Ryu, H.W.; Lim, S.H.; Kim, K.S.; Park, K.H.; Ryu, Y.B.; Lee, W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 2017, 32, 504–512. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.S.; Lee, J.; Lee, J.M.; Kim, Y.; Chin, Y.W.; Jee, J.G.; Keum, Y.S.; Jeong, Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorganic Med. Chem. Lett. 2012, 22, 4049–4054. [Google Scholar] [CrossRef]
- Cho, J.K.; Curtis-Long, M.J.; Lee, K.H.; Kim, D.W.; Ryu, H.W. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013, 21, 3051–3057. [Google Scholar] [CrossRef]
- Roh, C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomed. 2012, 2173. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.W.; Seo, K.H.; Curtis-Long, M.J.; Oh, K.Y.; Oh, J.W.; Cho, J.K.; Lee, K.H.; Park, K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzyme Inhib. Med. Chem. 2014, 29, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. [Google Scholar] [CrossRef]
- Güler, H.I.; Tatar, G.; Yildiz, O.; Belduz, A.O.; Kolayli, S. An investigation of ethanolic propolis extracts: Their potential inhibitor properties against ACE-II receptors for COVID-19 treatment by Molecular Docking Study. Sci. Prepr. 2020, 26, 1695–1703. [Google Scholar]
- Vardhan, S.; Sahoo, S.K. Searching inhibitors for three important proteins of COVID-19 through molecular docking studies. arXiv 2020, arXiv:2004.08095. [Google Scholar]
- Roshdy, W.H.; Rashed, H.A.; Kandeil, A.; Mostafa, A.; Moatasim, Y.; Kutkat, O.; AboShama, N.M.; Gomaa, M.R.; El-Sayed, I.H.; El Guindy, N.M.; et al. EGYVIR: An immunomodulatory herbal extract with potent antiviral activity against SARS-CoV-2. PLoS ONE 2020, 15, e0241739. [Google Scholar] [CrossRef]
- Kumar, A.; Choudhir, G.; Shukla, S.K.; Sharma, M.; Tyagi, P.; Bhushan, A.; Rathore, M. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J. Biomol. Struct. Dyn. 2020, 39, 3760–3770. [Google Scholar] [CrossRef]
- Sahlan, M.; Irdiani, R.; Flamandita, D.; Aditama, R.; Alfarraj, S.; Ansari, M.J.; Khayrani, A.C.; Pratami, D.K.; Lischer, K. Molecular interaction analysis of Sulawesi propolis compounds with SARS-CoV-2 main protease as preliminary study for COVID-19 drug discovery. J. King Saud Univ. Sci. 2021, 33, 101234. [Google Scholar] [CrossRef]
- Salehzadeh, F.; Pourfarzi, F.; Ataei, S. The Impact of Colchicine on The COVID-19 Patients; A Clinical Trial Study. Res. Sq. 2020, 1–11. [Google Scholar] [CrossRef]
- Huang, F.; Li, Y.; Leung, E.L.H.; Liu, X.; Liu, K.; Wang, Q.; Lan, Y.; Li, X.; Yu, H.; Cui, L.; et al. A review of therapeutic agents and Chinese herbal medicines against SARS-COV-2 (COVID-19). Pharmacol. Res. 2020, 158, 104929. [Google Scholar] [CrossRef]
- Tallei, T.E.; Tumilaar, S.G.; Niode, N.J.; Fatimawali; Kepel, B.J.; Idroes, R.; Effendi, Y.; Sakib, S.A.; Emran, T. Bin Potential of Plant Bioactive Compounds as SARS-CoV-2 Main Protease (Mpro) and Spike (S) Glycoprotein Inhibitors: A Molecular Docking Study. Scientifica 2020, 2020. [Google Scholar] [CrossRef]
- Di Pierro, F.; Iqtadar, S.; Khan, A.; Ullah Mumtaz, S.; Masud Chaudhry, M.; Bertuccioli, A.; Derosa, G.; Maffioli, P.; Togni, S.; Riva, A.; et al. Potential clinical benefits of quercetin in the early stage of COVID-19: Results of a second, pilot, randomized, controlled and open-label clinical trial. Int. J. Gen. Med. 2021, 14, 2807–2816. [Google Scholar] [CrossRef]
- Jang, W.D.; Jeon, S.; Kim, S.; Lee, S.Y. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc. Natl. Acad. Sci. USA 2021, 118, e2024302118. [Google Scholar] [CrossRef]
- Gligorijević, N.; Stanić-Vučinić, D.; Radomirović, M.; Stojadinović, M.; Khulal, U.; Nedić, O.; Ćirković Veličković, T. Role of resveratrol in prevention and control of cardiovascular disorders and cardiovascular complications related to COVID-19 disease: Mode of action and approaches explored to increase its bioavailability. Molecules 2021, 26, 2834. [Google Scholar] [CrossRef]
- Huang, Y.; Hongt, C. Tetrandrine. Cardiovasc. Drug Rev. 1998, 16, 1–15. [Google Scholar] [CrossRef]
- Rauchensteiner, F.; Matsumura, Y.; Yamamoto, Y.; Yamaji, S.; Tani, T. Analysis and comparison of Radix Glycyrrhizae (licorice) from Europe and China by capillary-zone electrophoresis (CZE). J. Pharm. Biomed. Anal. 2005, 38, 594–600. [Google Scholar] [CrossRef]
- Seki, H.; Sawai, S.; Ohyama, K.; Mizutani, M.; Ohnishi, T.; Sudo, H.; Fukushima, E.O.; Akashi, T.; Aoki, T.; Saito, K.; et al. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 2011, 23, 4112–4123. [Google Scholar] [CrossRef] [Green Version]
- Van de Sand, L.; Bormann, M.; Alt, M.; Schipper, L.; Heilingloh, C.S.; Steinmann, E.; Todt, D.; Dittmer, U.; Elsner, C.; Witzke, O.; et al. Glycyrrhizin effectively inhibits sars-cov-2 replication by inhibiting the viral main protease. Viruses 2021, 13, 609. [Google Scholar] [CrossRef]
- Ding, H.; Deng, W.; Ding, L.; Ye, X.; Yin, S.; Huang, W. Glycyrrhetinic acid and its derivatives as potential alternative medicine to relieve symptoms in nonhospitalized COVID-19 patients. J. Med. Virol. 2020, 92, 2200–2204. [Google Scholar] [CrossRef]
- Ang, L.; Lee, H.W.; Choi, J.Y.; Zhang, J.; Soo Lee, M. Herbal medicine and pattern identification for treating COVID-19: A rapid review of guidelines. Integr. Med. Res. 2020, 9, 100407. [Google Scholar] [CrossRef]
- Niu, F.; Xu, X.; Zhang, R.; Sun, L.; Gan, N.; Wang, A. Ursodeoxycholic acid stimulates alveolar fluid clearance in LPS-induced pulmonary edema via ALX/cAMP/PI3K pathway. J. Cell. Physiol. 2019, 234, 20057–20065. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, M.; Liu, H.; Liu, S. A critical review: Traditional uses, phytochemistry, pharmacology and toxicology of Stephania tetrandra S. Moore (Fen Fang Ji). Phytochem. Rev. 2020, 19, 449–489. [Google Scholar] [CrossRef]
- Kaserer, T.; Rigo, R.; Schuster, P.; Alcaro, S.; Sissi, C.; Schuster, D. Optimized Virtual Screening Workflow for the Identification of Novel G-Quadruplex Ligands. J. Chem. Inf. Model. 2016, 56, 484–500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, M.; Chen, X.; Cao, K.; You, Y.; Qian, Y.; Yu, W. Berberine reduces circulating inflammatory mediators in patients with severe COVID-19. Br. J. Surg. 2021, 108, e9–e11. [Google Scholar] [CrossRef] [PubMed]
- Varghese, F.S.; van Woudenbergh, E.; Overheul, G.J.; Eleveld, M.J.; Kurver, L.; van Heerbeek, N.; van Laarhoven, A.; Miesen, P.; den Hartog, G.; de Jonge, M.I. Berberine and obatoclax inhibit SARS-CoV-2 replication in primary human nasal epithelial cells in vitro. bioRxiv 2020, 13, 282. [Google Scholar]
- Krupanidhi, S.; Abraham Peele, K.; Venkateswarulu, T.; Ayyagari, V.S.; Nazneen Bobby, M.; John Babu, D.; Venkata Narayana, A.; Aishwarya, G. Screening of phytochemical compounds of Tinospora cordifolia for their inhibitory activity on SARS-CoV-2: An in silico study. J. Biomol. Struct. Dyn. 2020, 39, 5799–5803. [Google Scholar] [CrossRef]
- Debnath, B.; Singh, W.S.; Das, M.; Goswami, S.; Singh, M.K.; Maiti, D.; Manna, K. Role of plant alkaloids on human health: A review of biological activities. Mater. Today Chem. 2018, 9, 56–72. [Google Scholar] [CrossRef]
- Borquaye, L.S.; Gasu, E.N.; Ampomah, G.B.; Kyei, L.K.; Amarh, M.A.; Mensah, C.N.; Nartey, D.; Commodore, M.; Adomako, A.K.; Acheampong, P. Alkaloids from Cryptolepis sanguinolenta as Potential Inhibitors of SARS-CoV-2 Viral Proteins: An In Silico Study. Biomed Res. Int. 2020, 2020, 5324560. [Google Scholar] [CrossRef]
- Gendelman, O.; Amital, H.; Bragazzi, N.L.; Watad, A.; Chodick, G. Continuous hydroxychloroquine or colchicine therapy does not prevent infection with SARS-CoV-2: Insights from a large healthcare database analysis. Autoimmun. Rev. 2020, 19, 102566. [Google Scholar] [CrossRef]
- Rocca, R.; Moraca, F.; Costa, G.; Alcaro, S.; Distinto, S.; Maccioni, E.; Ortuso, F.; Artese, A.; Parrotta, L. Structure-Based Virtual Screening of Novel Natural Alkaloid Derivatives as Potential Binders of h-telo and c-myc DNA G-Quadruplex Conformations. Molecules 2014, 20, 206–223. [Google Scholar] [CrossRef] [Green Version]
- Shaldam, M.A.; Yahya, G.; Mohamed, N.H.; Abdel-Daim, M.M.; Al Naggar, Y. In silico screening of potent bioactive compounds from honey bee products against COVID-19 target enzymes. Environ. Sci. Pollut. Res. 2020, 28, 40507–40514. [Google Scholar] [CrossRef]
- Muhseen, Z.T.; Hameed, A.R.; Al-Hasani, H.M.; Qamar, M.T.U.; Li, G. Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: Integrated computational approach. J. Mol. Liq. 2020, 320, 114493. [Google Scholar] [CrossRef]
- Luo, P.; Liu, D.; Li, J. Pharmacological perspective: Glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105995. [Google Scholar] [CrossRef]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; De Jesus, P.D.; Teriete, P.; Hull, M.V. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020, 586, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Campione, E.; Cosio, T.; Rosa, L.; Lanna, C.; Di Girolamo, S.; Gaziano, R.; Valenti, P.; Bianchi, L. Lactoferrin as Protective Natural Barrier of Respiratory and Intestinal Mucosa against Coronavirus Infection and Inflammation. Int. J. Mol. Sci. 2020, 21, 4903. [Google Scholar] [CrossRef] [PubMed]
- Bagetta, D.; Maruca, A.; Lupia, A.; Mesiti, F.; Catalano, R.; Romeo, I.; Moraca, F.; Ambrosio, F.A.; Costa, G.; Artese, A. Mediterranean products as promising source of multi-target agents in the treatment of metabolic syndrome. J. Biomol. Struct. Dyn. 2020, 186, 111903. [Google Scholar] [CrossRef]
- Abian, O.; Ortega-Alarcon, D.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Vega, S.; Reyburn, H.T.; Rizzuti, B.; Velázquez-Campoy, A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020, 164, 1693–1703. [Google Scholar] [CrossRef]
- Thomson, R. Naturally Occurring Quinones; Academic Press: London, UK, 2012. [Google Scholar]
- Ter-Ellen, B.M.; Kumar, N.D.; Bouma, E.M.; Troost, B.; van De Pol, D.P.I.; van Der-Metselaar, H.H.E.; Apperloo, L.; Van, D.G.; van Den Berge, M.; Nawijn, M.C.; et al. Resveratrol And Pterostilbene Potently Inhibit SARS-CoV-2 Infection In Vitro. bioRxiv 2020, 285940. [Google Scholar] [CrossRef]
- Gaber, A.; Alsanie, W.F.; Kumar, D.N.; Refat, M.S.; Saied, E.M. Novel Papaverine Metal Complexes with Potential Anticancer Activities. Molecules 2020, 25, 5447. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; Kim, H. Mini-Review on the Roles of Vitamin C, Vitamin D, and Selenium in the Immune System against COVID-19. Molecules 2020, 25, 5346. [Google Scholar] [CrossRef]
- Carr, A.C.; Rowe, S. The Emerging Role of Vitamin C in the Prevention and Treatment of COVID-19. Nutrients 2020, 12, 3286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Rao, X.; Li, Y.; Zhu, Y.; Liu, F.; Guo, G.; Luo, G.; Meng, Z.; De Backer, D.; Xiang, H. Pilot trial of high-dose vitamin C in critically ill COVID-19 patients. Ann. Intensive Care 2021, 11, 5. [Google Scholar] [CrossRef]
- Hemilä, H.; Chalker, E. Vitamin C may reduce the duration of mechanical ventilation in critically ill patients: A meta-regression analysis. J. Intensive Care 2020, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemilä, H.; Chalker, E. Vitamin C Can Shorten the Length of Stay in the ICU: A Meta-Analysis. Nutrients 2019, 11, 708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemilä, H.; Chalker, E. Vitamin C as a Possible Therapy for COVID-19. IC Infect. Chemother. 2020, 52, 222–223. [Google Scholar] [CrossRef] [PubMed]
- Hemilä, H.; Chalker, E. Vitamin C for preventing and treating the common cold. Cochrane Database Syst. Rev. 2013, 1, 31. [Google Scholar] [CrossRef] [Green Version]
- Cheng, R.Z. Can early and high intravenous dose of vitamin C prevent and treat coronavirus disease 2019 (COVID-19)? Med. Drug Discov. 2020, 5, 100028. [Google Scholar] [CrossRef]
- Torjesen, I. Evidence does not support vitamin D for reducing respiratory infections, reviews conclude. BMJ 2020, 369, m2629. [Google Scholar] [CrossRef]
- Kow, C.S.; Hadi, M.A.; Hasan, S.S. Vitamin D Supplementation in Influenza and COVID-19 Infections Comment on: “Evidence that Vitamin D Supplementation Could Reduce Risk of Influenza and COVID-19 Infections and Deaths”. Nutrients 2020, 12, 1626. [Google Scholar] [CrossRef]
- Ali, N. Role of vitamin D in preventing of COVID-19 infection, progression and severity. J. Infect. Public Health 2020, 13, 1373–1380. [Google Scholar] [CrossRef]
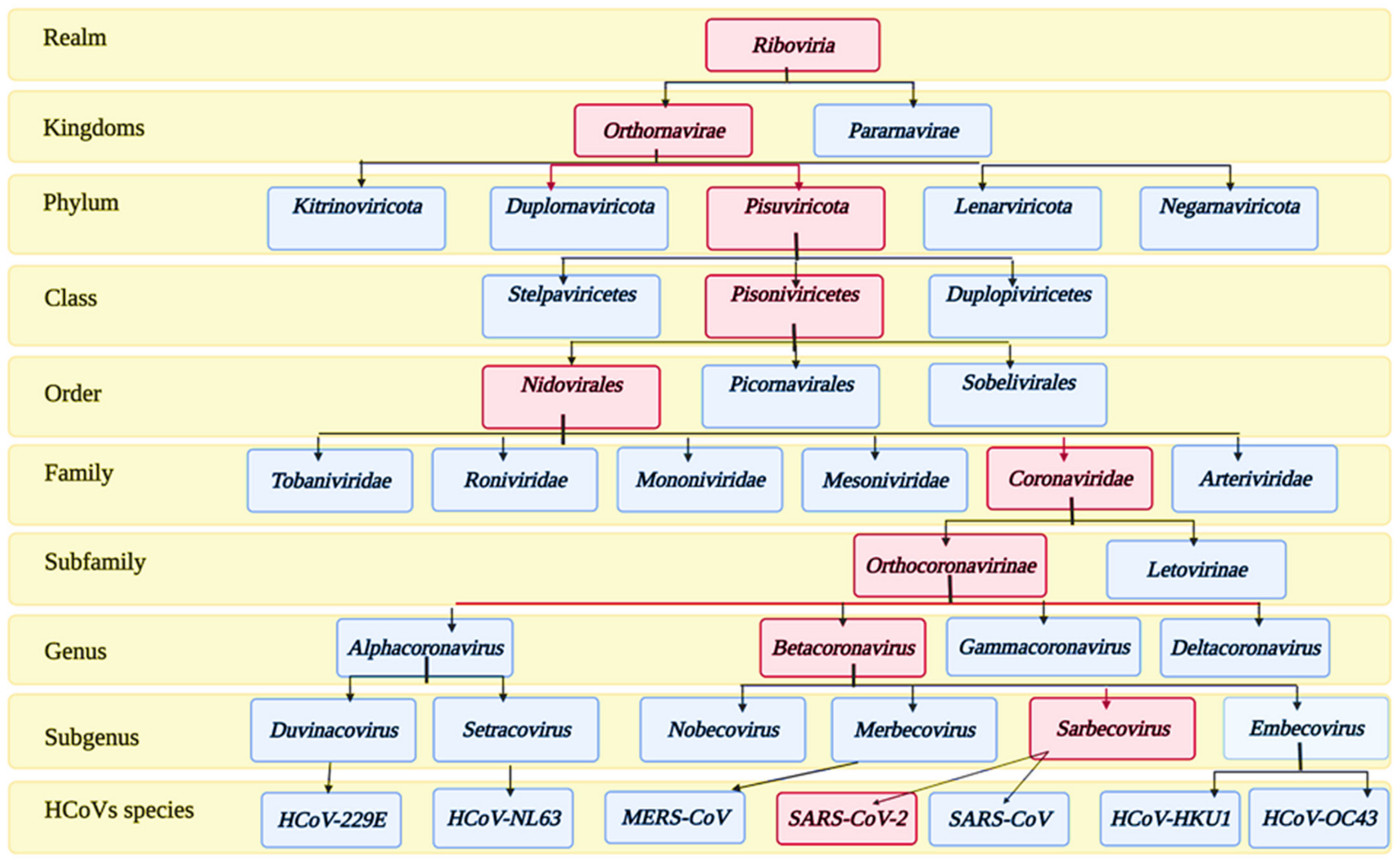



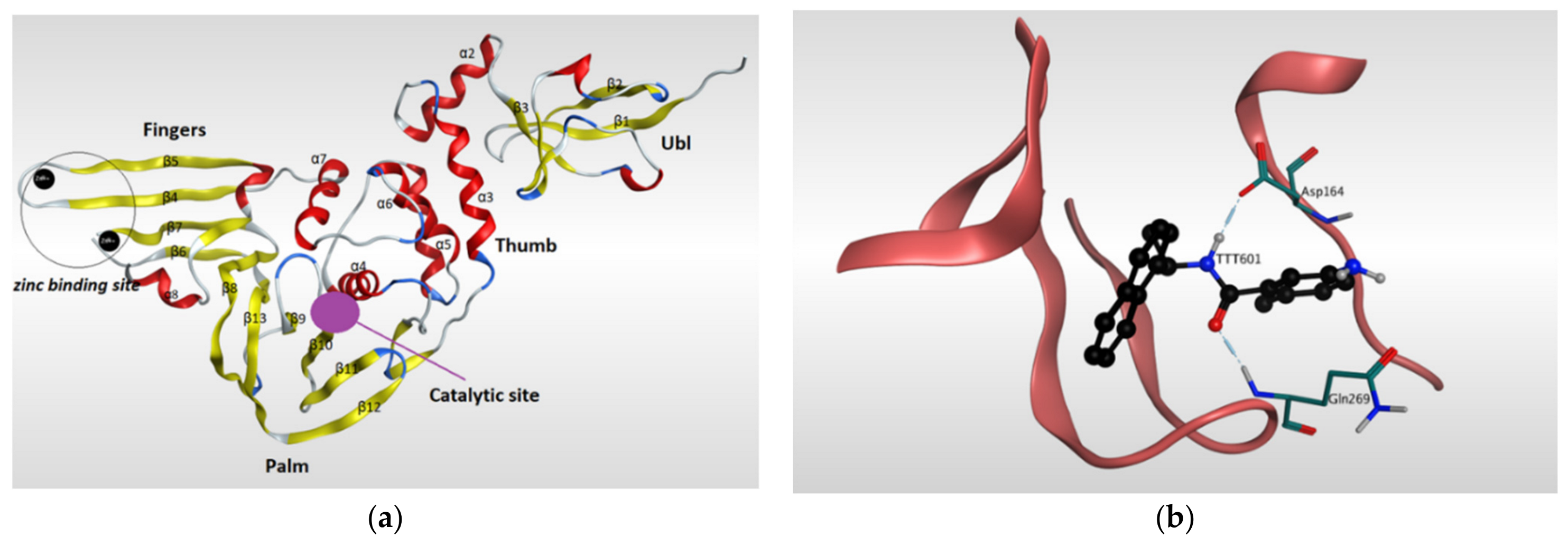


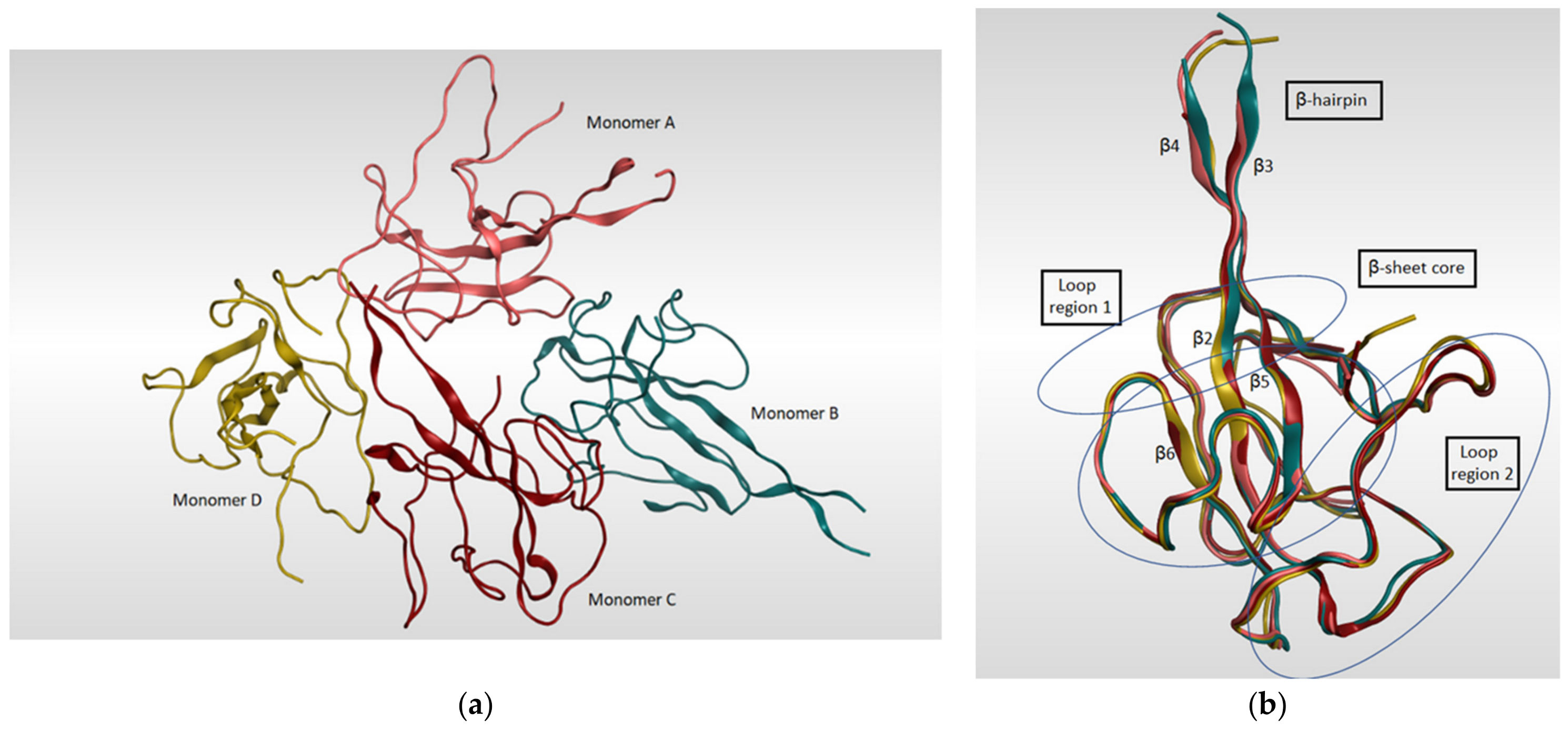
| PDB ID | Resolution | Macromolecule | Ligand | Reference |
|---|---|---|---|---|
| 6lU7 | 2.16 | - SARS-CoV-2 main protease - synthetic construct | N3 inhibitor | [95] |
| 6M2N | 2.20 | - 3CLpro - SARS-CoV-2 main protease | Baicalein | [96] |
| 7K3T | 1.2 | - 3C-like protease (3CL pro) | - | |
| 6YB7 | 1.25 | - SARS-CoV-2 main protease - ORF1ab polyprotein | - | |
| 7JKV | 1.25 | -3CLpro | N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | [97] |
| 5R8T | 1.27 | - 3CLpro - SARS-CoV-2 main protease | - | [98] |
| 6XKH | 1.28 | - 3CLpro | - | |
| 5R82 | 1.31 | - 3CLpro - SARS-CoV-2 main protease. | 6-(ethylamino)pyridine-3-carbonitrile | |
| 5RH4 | 1.34 | - 3CLpro - SARS-CoV-2 main protease | (2R)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid | |
| 5RGJ | 1.34 | - 3CLpro - SARS-CoV-2 main protease | (5S)-7-(pyrazin-2-yl)-2-oxa-7-azaspiro[4 .4]nonane | [98] |
| 7K40 | 1.35 | - 3CLpro | Boceprevir (bound form) | |
| 6XHN | 1.377 | - 3CLpro | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate | [99] |
| 7BRR | 1.4 | - 3C-like proteinase - 3CL-PRO - 3CLp | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid | [100] |
| 6XHM | 1.41 | - 3CLpro | N-[(2S)-1-({(2S)-4-hydroxy-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | [99] |
| 5RFD | 1.41 | - 3CLpro - SARS-CoV-2 main protease | 2-[(methylsulfonyl)methyl]-1H-benzimidazole | [98] |
| 5RGR | 1.41 | - 3CLpro - SARS-CoV-2 main protease | N,1-dimethyl-N-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | [98] |
| 5RF9 | 1.43 | - 3CLpro - SARS-CoV-2 main protease | 1-[(2~{S})-2-methylmorpholin-4-yl]-2-pyrazol-1-yl-ethanone | [98] |
| 5RFW | 1.43 | - 3CLpro - SARS-CoV-2 main protease | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one | [98] |
| 5RHB | 1.43 | - 3CLpro - SARS-CoV-2 main protease | (E)-1-(pyrimidin-2-yl)methanimine | [98] |
| 5RGW | 1.43 | - 3CLpro - SARS-CoV-2 main protease | 2-(5-cyanopyridin-3-yl)-N-(pyridin-3-yl)acetamide | |
| 5RF8 | 1.44 | - 3CLpro - SARS-CoV-2 main protease | 4-amino-N-(pyridin-2-yl)benzenesulfonamide | [98] |
| 6WNP | 1.443 | -3CLpro - ORF1ab polyprotein | Boceprevir (bound form) | |
| 6XHO | 1.446 | - 3CLpro | Ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | [99] |
| 5RF6 | 1.45 | - 3CLpro - SARS-CoV-2 main protease | 5-(1,4-oxazepan-4-yl)pyridine-2-carbonitrile | [98] |
| 6XR3 | 1.45 | - 3CLpro | N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | |
| 6XBG | 1.45 | - SARS-CoV-2 main protease - 3CLpro | UAW246 inhibitor | [101] |
| 6W79 | 1.46 | - SARS-CoV-2 main protease | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | |
| 5RFE | 1.46 | - 3CLpro - SARS-CoV-2 main protease | N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | [98] |
| 5RED | 1.47 | - 3CLpro - SARS-CoV-2 main protease | 4-[2-(phenylsulfanyl)ethyl]morpholine | [98] |
| 6XHL | 1.471 | - 3CLpro | N-[(2S)-1-({(2S)-4-hydroxy-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | [99] |
| 6WCO | 1.48 | - SARS-CoV-2 main protease | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclopentylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | |
| 5RFB | 1.48 | - 3CLpro - SARS-CoV-2 main protease | N-[(1-methyl-1H-1,2,3-triazol-4-yl)methyl]ethanamine | [98] |
| 5RFV | 1.48 | - 3CLpro - SARS-CoV-2 main protease | 1-[4-(thiophene-2-carbonyl)piperazin-1-yl]ethan-1-one | [98] |
| 5C5O | 1.5 | - 3CLpro | (2S)-3-(1H-imidazol-5-yl)-2-({[(3S,4aR,8aS)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal | |
| 5RF3 | 1.5 | - 3CLpro - SARS-CoV-2 main protease. | pyrimidin-5-amine | [98] |
| 5RGK | 1.43 | - 3CLpro - SARS-CoV-2 main protease | 2-fluoro-N-[2-(pyridin-4-yl)ethyl]benzamide | [98] |
| PDB ID | Resolution | Source Organism | Macromolecule | Ligand | Reference |
|---|---|---|---|---|---|
| 7CMD | 2.59 | - SARS-CoV-2 | - PLpro | GRL0617 | [107] |
| 6WX4 | 1.655 | - SARS-CoV-2 - Saltans group | - Nonstructural protein 3 - PLpro - pp1ab - ORF1ab polyprotein | VIR251 | [110] |
| 6WUU | 2.79 | - SARS-CoV-2 - Synthetic construct | - PLpro - pp1ab - ORF1ab polyprotein | VIR250 | [110] |
| 6YVA | 3.18 | - SARS-CoV-2 - Mus musculus | - Replicase polyprotein 1a - pp1ab - ORF1ab polyprotein - Ubiquitin-like protein ISG15 - Interferon-induced 15 kDa protein - Interferon-induced 17 kDa protein - IP17 - Ubiquitin cross-reactive protein | mISG15 | [40] |
| 7JRN | 2.48 | - SARS-CoV-2 | - PLpro | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | - |
| 6W9C | 2.7 | - SARS-CoV-2 | - Papain-like proteinase | - | |
| 7CJM | 3.2 | - SARS-CoV-2 | - Nonstructural protein 3 - PLpro | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | - |
| 6XA9 | 2.9 | - SARS-CoV-2 - Homo sapiens | - PLpro - Nonstructural - CTD-propargylamide - Interferon-induced 15 kDa protein - Interferon-induced 17 kDa protein - IP17 - Ubiquitin cross-reactive protein - hUCRP ein 3 | ISG15 | - |
| 7CMD | 2.59 | - SARS-CoV-2 | - Replicase polyprotein 1ab - pp1ab - ORF1ab polyprotein | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide | [108] |
| 6XAA | 2.7 | - SARS-CoV-2 | - PLpro - Nonstructural protein 3 - Ubiquitin-propargylamide | ubiquitin propargylamide | - |
| 7CJD | 2.501 | - SARS-CoV-2 | - Replicase polyprotein 1ab | - | [108] |
| PDB ID | Resolution | Source Organism | Macromolecule | Reference |
|---|---|---|---|---|
| 6M1V | 1.5 | - SARS-CoV-2 | - spike protein | - |
| 7JMP | 1.712 | - SARS-CoV-2 - Homo sapiens | - Spike protein S1 - COVA2-39 heavy chain - COVA2-39 light chain | - |
| 6YZ5 | 1.8 | - SARS-CoV-2 - Lama glama | - Spike glycoprotein - Nanobody H11-D4 | - |
| 7BZ5 | 1.84 | - SARS-CoV-2 - Homo sapiens | - Spike protein S1 - Heavy chain of B38 - Light chain of B38 | [115] |
| 6ZBP | 1.85 | - SARS-CoV-2 - Lama glama | - Spike glycoprotein - H11-H4 | |
| 7C8V | 2.15 | - Synthetic construct - SARS-CoV-2 | - Synthetic nanobody SR4 - Spike glycoprotein | - |
| 6WAQ | 2.2 | - Lama glama - SARS-CoV | - nanobody SARS VHH-72 - Spike glycoprotein | - |
| 6XC4 | 2.341 | - SARS-CoV-2 | - Spike protein S1 - CC12.3 heavy chain - CC12.3 light chain | [34] |
| 7JMO | 2.359 | - SARS-CoV-2 - Homo sapiens | - Spike protein S1 - COVA2-04 heavy chain - COVA2-04 light chain | |
| 6XLU | 2.4 | - SARS-CoV-2 | - Spike glycoprotein | [15] |
| 7CHB | 2.4 | - Homo sapiens - SARS-CoV-2 | - BD-236 Fab heavy chain - BD-236 Fab light chain - SARS-CoV-2 receptor binding domain | - |
| 6YLA | 2.42 | - SARS-CoV-2 - Homo sapiens | - Spike glycoprotein - Heavy Chain - Light chain | - |
| 6M0J | 2.45 | - Homo sapiens - SARS-CoV-2 | - Angiotensin-converting enzyme 2 - Spike receptor binding domain | [116] |
| 6VYB | 3.20 | - Homo sapiens - SARS-CoV-2 | - Spike glycoprotein | [116] |
| PDB ID | Resolution | Source Organism | Macromolecule Name | Reference |
|---|---|---|---|---|
| 7BTF | 2.95 | - SARS-CoV-2 | - NSP12 - pp1ab - ORF1ab polyprotein | [108] |
| - SARS-CoV-2 | - NSP7 - pp1ab - ORF1ab polyprotein | |||
| - SARS-CoV-2 | - NSP8 - pp1ab - ORF1ab polyprotein | |||
| 6M71 | 2.9 | - SARS-CoV-2 | - NSP 12 - pp1ab - ORF1ab polyprotein | [108] |
| - SARS-CoV-2 | - NSP 7 - pp1ab - ORF1ab polyprotein | |||
| - SARS-CoV-2 | - NSP 8 - pp1ab - ORF1ab polyprotein | |||
| 7BZF | 3.26 | - SARS-CoV-2 | - NSP 8-1 | [72,102] |
| - SARS-CoV-2 | - NSP7 | |||
| - SARS-CoV-2 | - RNA-directed RNA polymerase - NSP12 | |||
| 7C2K | 2.93 | - SARS-CoV-2 | - RNA-directed RNA polymerase - NSP12 | [102] |
| - SARS-CoV-2 | - NSP 8-1 | |||
| - SARS-CoV-2 | - NSP7 | |||
| 6YYT | 2.90 | - SARS-CoV-2 - Synthetic construct | - NSP12 | [117] |
| - SARS-CoV-2 - Synthetic construct | - NSP 8 | |||
| - SARS-CoV-2 - Synthetic construct | - NSP7 |
| PDB ID | Resolution | Source of Organism | Macromolecules Name | Reference |
|---|---|---|---|---|
| 6M3M | 2.70 | - SARS-CoV-2 | - Nucleoprotein | [123] |
| 6WZO | 1.42 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | [123] |
| 6WZQ | 1.45 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | [123] |
| 6M3M | 2.7 | - SARS-CoV-2 | - Nucleocapsid protein | [122] |
| 6WKP | 2.67 | - SARS-CoV-2 | - Nucleocapsid protein - Nucleoprotein - Protein N | - |
| 6YUN | 1.44 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | - |
| 7CE0 | 1.5 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | - |
| 7CDZ | 1.8 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | - |
| 6YI3 | (NMR) | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | - |
| 6VYO | 1.7 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | - |
| 6WJI | 2.05 | - SARS-CoV-2 | - Nucleocapsid protein - Nucleoprotein - Protein N | - |
| 7C22 | 2.00 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | [124] |
| 6ZCO | 1.36 | - SARS-CoV-2 | - Nucleoprotein - Nucleocapsid protein - Protein N | - |
| Vaccine | Classification | Efficiency | Required Dose | Postvaccination Symptom | Vaccine Producer |
|---|---|---|---|---|---|
| Moderna mRNA1273 | mRNA vaccine | 94% | 2 doses/3 weeks apart | Local and systemic reaction | Moderna, and National institute of allergy and infectious diseases, Cambridge, MA, USA |
| Pfizer-BioNTech (BNT162B1) | mRNA vaccine | 95% | 2 doses/3 weeks apart | Pain, redness, joint pain, muscle pain, paroxysmal ventricular | Pfizer (New York, NY, USA), and BioNTech, Mainz, Germany |
| Covaxin (BBV152) | Inactivated virus vaccine | 81% | 2 doses/4 weeks apart | Pain at site of injection | Bharat Biotech, Genome Valley, Hyderabad, India |
| Sinopharm (BBIBPCorV) | Inactivated virus vaccine | 79% | 2 doses/3 weeks apart | Mild cellulitis | Beijing Bio-Institute, Beijing, China |
| CanSino (Ad5-nCoV) | Vector vaccine | 66% | 1 dose | Pain at site of injection | CanSino Biologics, Tianjin, China |
| AstraZeneca (ChAdOx1/ AZD1222) | Vector vaccine | 70% | 2 doses/12 weeks apart | A pathogenic PF4-dependent syndrome may develop | AstraZeneca, Oxford, England |
| Jansseen (Ad26COVS1) | Vector vaccine | 73% | 2 doses/3 weeks apart | Irritation | Janssen Pharmaceutical Companies, Leiden, The Netherlands |
| SputnikV (Gram Covid Vac) | Vector vaccine | 91% | 2 doses/3 weeks apart | Flu-like illness, headache, asthenia, renal colic, deep vein thrombosis | Gamaleya Research Institute, Moscow, Russia |
| SinoVac (CoronaVac) | Virus vaccine inactivated with aluminum hydroxide | 50% | 2 doses/2 weeks apart | Pain, fever | Wuhan Institute, and Sinovac Biotech, Beijing, China |
| Novavax (NVXCoV2373) | Virus resemble vaccine particles (adjuvanted recombinant protein nanoparticle) | 96% | 2 doses/3 weeks apart | Fatigue, headache, pain | Novavax Biotechnology company, Gaithersburg, MD, USA |
| Medicinal Plants | Antiviral Compound(s) | Virus | Structure | Reference |
|---|---|---|---|---|
| Umbelliferae | Glycycoumarin | Inhibit SARS-CoV-2 3CLpro by hydrogen bonding interactions |  | [267,277] |
| Torreya nucifera | Amentoflavone | Inhibit SARS-CoV-2 3CLpro by blocking the S-protein and ACE2 interaction |  | [51,278] |
| Rheum officinale Baill | Emodin | Inhibit SARS-CoV by blocking the S protein and ACE2 interaction |  | [279] |
| Broussonetia papyrifera | Broussochalcone B | CoV cysteine proteases inhibitor |  | [280] |
| Aglaia perviridis | Myricetin | Inhibit the SARS-CoV helicase |  | [281] |
| Cinnamomi cortex | Procyanidin A2 | wtSARS-CoV and SARS-CoVS pseudovirus |  | [263] |
| Paulownia tomentosa | Tomentin C3 | Inhibit SARS-CoV |  | [282] |
| Camellia sinensis | Catechin gallate | Inhibit SARS-CoV N protein |  | [283] |
| Psoralea corylifolia | Bavachinin | Inhibit SARS-CoV PLpro |  | [284] |
| Panax ginseng | Ginsenoside-Rb1 | Inhibit SARS-CoV replication |  | [285] |
| Artemisia annua | Arteannuin B | Inhibit SARS-CoV-2 |  | [253] |
| Scutellaria baicalensis | Baicalin | Inhibit SARS-CoV-2 by blocking the interaction with ACE2 |  | [286] |
| Origanum vulgare | Carvacrol | Inhibit SARS-CoV-2 by blocking the interaction with ACE2 |  | [50] |
| Cymbopogonwinterianus, Geranium | Citronellol | Inhibit SARS-CoV-2 proteins |  | [287] |
| EGYVIR | Curcumin-piperine | Inhibit SARS-CoV-2 |  | [288] |
| Essential oils | Geraniol | Inhibit SARS-CoV-2 by blocking the interaction with ACE2 |  | [50] |
| Bile acids | Glyco-ursodeoxycholic acid | Inhibit SARS-CoV-2 main protease |  | [289] |
| Rhodiola rosea | Herbacetin | Inhibit SARS-CoV-2 main protease |  | [290] |
| Essential oils | Limonene | Inhibit SARS-CoV-2 proteins |  | [287] |
| Essential oils | Linalool | Inhibit SARS-CoV-2 proteins |  | [287] |
| Citrus oils | Neryl acetate | Inhibit SARS-CoV-2 proteins |  | [287] |
| Olea europaea | Oleanolic acid | Inhibit SARS-CoV-2 main protease |  | [289] |
| Cirsium setidens | Pectolinarin | Inhibit SARS-CoV-2 main protease |  | [290] |
| Bile Acids | Ursodeoxycholic acid | Inhibit SARS-CoV-2 main protease |  | [289] |
| Bile Acids | Ursolic acid | Inhibit SARS-CoV-2 main protease |  | [289] |
| Natural Source | Antiviral Compound(s) | Chemical Structure | Reference |
|---|---|---|---|
| Artemisia annua | Artemisinin |  | [253,304] |
| Berberis vulgaris | Berberine |  | [305,306,307,308,309,310] |
| Bile acids | Chenodeoxycholic acid |  | [289] |
| Colchicum autumnale | Colchicine |  | [311,312] |
| Rheum officinale and Reynoutria multiflora | Emodin |  | [313] |
| Glycyrrhiza glabra | Glycyrrhizin |  | [314,315] |
| Stephania tetrandra | Hanfangchin A |  | [316] |
| Mammalian and Animal milk | Lactoferrin |  | [317] |
| Onion, Garlic, Peppermint, Fenugreek, Origan Capers, Onions, Elderberries, Kale, Okra, Apple Peels, Aronia Berries, Cranberries, Asparagus, Goji Berries | Quercetin |  | [280,318,319,320] |
| Red wine, Red grape juice, Peanuts, Fresh grapes, Pistachios, Peanut butter, Cocoa powder, Dark chocolate, Milk chocolate, Strawberries, Jackfruit skin, Blueberries, Bilberries, Red currants, Cranberries, Lingonberries, Mulberries | Resveratrol |  | [321,322] |
| Citrus Fruit, Tropical Fruit, Peppers, Cruciferous Vegetables, Leafy Vegetables | Vitamin C |  | [323,324,325,326,327,328,329,330] |
| Human and Jay Jasmine plant | Vitamin D |  | [331] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saied, E.M.; El-Maradny, Y.A.; Osman, A.A.; Darwish, A.M.G.; Abo Nahas, H.H.; Niedbała, G.; Piekutowska, M.; Abdel-Rahman, M.A.; Balbool, B.A.; Abdel-Azeem, A.M. A Comprehensive Review about the Molecular Structure of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Insights into Natural Products against COVID-19. Pharmaceutics 2021, 13, 1759. https://doi.org/10.3390/pharmaceutics13111759
Saied EM, El-Maradny YA, Osman AA, Darwish AMG, Abo Nahas HH, Niedbała G, Piekutowska M, Abdel-Rahman MA, Balbool BA, Abdel-Azeem AM. A Comprehensive Review about the Molecular Structure of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Insights into Natural Products against COVID-19. Pharmaceutics. 2021; 13(11):1759. https://doi.org/10.3390/pharmaceutics13111759
Chicago/Turabian StyleSaied, Essa M., Yousra A. El-Maradny, Alaa A. Osman, Amira M. G. Darwish, Hebatallah H. Abo Nahas, Gniewko Niedbała, Magdalena Piekutowska, Mohamed A. Abdel-Rahman, Bassem A. Balbool, and Ahmed M. Abdel-Azeem. 2021. "A Comprehensive Review about the Molecular Structure of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Insights into Natural Products against COVID-19" Pharmaceutics 13, no. 11: 1759. https://doi.org/10.3390/pharmaceutics13111759
APA StyleSaied, E. M., El-Maradny, Y. A., Osman, A. A., Darwish, A. M. G., Abo Nahas, H. H., Niedbała, G., Piekutowska, M., Abdel-Rahman, M. A., Balbool, B. A., & Abdel-Azeem, A. M. (2021). A Comprehensive Review about the Molecular Structure of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Insights into Natural Products against COVID-19. Pharmaceutics, 13(11), 1759. https://doi.org/10.3390/pharmaceutics13111759