Characterization of Liposomes Using Quantitative Phase Microscopy (QPM)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Liposome Preparation
2.3. Liposome Size Reduction
2.4. Liposome Characterization: Size and ζ-Potential
2.5. Liposome Immobilization for Imaging Purpose
2.6. Imaging
2.7. Image Processing and Analysis
2.7.1. Phase Retrieval Algorithm
2.7.2. Size Distribution of Liposomes
- Recording of the phase-shifted interferograms of liposome samples.
- High-resolution phase recovery by employing the AIA algorithm.
- Removal of any background information from the recovered phase images either physically (through a reference/sample free interferogram) or numerically.
- Convert the phase map into a height map by using Equation (4). The value of is assumed to be equal to 0.04.
- Count the number of liposomes present in the recovered height map.
- Find the maximum height values of all liposomes using the image-processing toolbox in MATLAB and utilize these values to draw a histogram plot.
3. Results
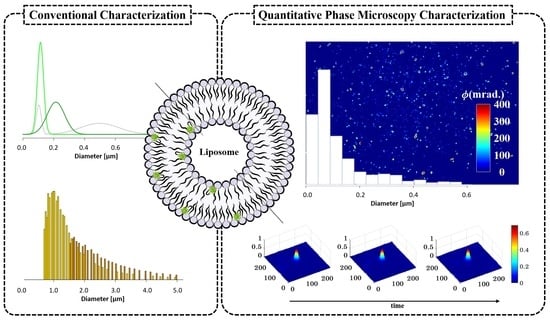
3.1. Conventional Characterization of Labeled Liposomes
3.2. QPM Label-Free Characterization of Liposomes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Gadekar, V.; Borade, Y.; Kannaujia, S.; Rajpoot, K.; Anup, N.; Tambe, V.; Kalia, K.; Tekade, R.K. Nanomedicines accessible in the market for clinical interventions. J. Control Release 2021, 330, 372–397. [Google Scholar] [CrossRef]
- Cao, J.; Huang, D.; Peppas, N.A. Advanced engineered nanoparticulate platforms to address key biological barriers for delivering chemotherapeutic agents to target sites. Adv. Drug Deliv. Rev. 2020, 167, 170–188. [Google Scholar] [CrossRef]
- Fontana, F.; Figueiredo, P.; Martins, J.P.; Santos, H.A. Requirements for Animal Experiments: Problems and Challenges. Small 2020, 17, e2004182. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, S.T.; Faria, M.; Crampin, E.J. An analytical approach for quantifying the influence of nanoparticle polydispersity on cellular delivered dose. J. R. Soc. Interface 2018, 15, 20180364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.H.; Jang, W.Y.; Ko, Y.T. The Effect of Surface Charges on the Cellular Uptake of Liposomes Investigated by Live Cell Imaging. Pharm. Res. 2017, 34, 704–717. [Google Scholar] [CrossRef]
- Varenne, F.; Makky, A.; Gaucher-Delmas, M.; Violleau, F.; Vauthier, C. Multimodal Dispersion of Nanoparticles: A Comprehensive Evaluation of Size Distribution with 9 Size Measurement Methods. Pharm. Res. 2016, 33, 1220–1234. [Google Scholar] [CrossRef] [Green Version]
- Eleamen Oliveira, E.; Barendji, M.; Vauthier, C. Understanding Nanomedicine Size and Biological Response Dependency: What Is the Relevance of Previous Relationships Established on Only Batch-Mode DLS-Measured Sizes? Pharm. Res. 2020, 37, 161. [Google Scholar] [CrossRef]
- Robson, A.L.; Dastoor, P.C.; Flynn, J.; Palmer, W.; Martin, A.; Smith, D.W.; Woldu, A.; Hua, S. Advantages and Limitations of Current Imaging Techniques for Characterizing Liposome Morphology. Front. Pharmacol. 2018, 9, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducat, E.; Evrard, B.; Peulen, O.; Piel, G. Cellular uptake of liposomes monitored by confocal microscopy and flow cytometry. J. Drug Deliv. Sci. Technol. 2011, 21, 469–477. [Google Scholar] [CrossRef]
- Snipstad, S.; Hak, S.; Baghirov, H.; Sulheim, E.; Morch, Y.; Lelu, S.; von Haartman, E.; Back, M.; Nilsson, K.P.R.; Klymchenko, A.S.; et al. Labeling nanoparticles: Dye leakage and altered cellular uptake. Cytom. A 2017, 91, 760–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qhattal, H.S.; Hye, T.; Alali, A.; Liu, X. Hyaluronan polymer length, grafting density, and surface poly(ethylene glycol) coating influence in vivo circulation and tumor targeting of hyaluronan-grafted liposomes. ACS Nano 2014, 8, 5423–5440. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, K.I.; Tassone, C.; Ehrlich, N.; Andresen, T.L.; Flyvbjerg, H. How to Characterize Individual Nanosize Liposomes with Simple Self-Calibrating Fluorescence Microscopy. Nano Lett. 2018, 18, 2844–2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonsen, J.B.; Larsen, J.B.; Hempel, C.; Eng, N.; Fossum, A.; Andresen, T.L. Unique Calibrators Derived from Fluorescence-Activated Nanoparticle Sorting for Flow Cytometric Size Estimation of Artificial Vesicles: Possibilities and Limitations. Cytom. A 2019, 95, 917–924. [Google Scholar] [CrossRef]
- Cauzzo, J.; Nystad, M.; Holsaeter, A.M.; Basnet, P.; Skalko-Basnet, N. Following the Fate of Dye-Containing Liposomes In Vitro. Int. J. Mol. Sci. 2020, 21, 4847. [Google Scholar] [CrossRef]
- Rodriguez-Lorenzo, L.; Fytianos, K.; Blank, F.; Von Garnier, C.; Rothen-Rutishauser, B.; Petri-Fink, A. Fluorescence-Encoded Gold Nanoparticles: Library Design and Modulation of Cellular Uptake into Dendritic Cells. Small 2014, 10, 1341–1350. [Google Scholar] [CrossRef] [Green Version]
- Munter, R.; Kristensen, K.; Pedersbaek, D.; Larsen, J.B.; Simonsen, J.B.; Andresen, T.L. Dissociation of fluorescently labeled lipids from liposomes in biological environments challenges the interpretation of uptake studies. Nanoscale 2018, 10, 22720–22724. [Google Scholar] [CrossRef]
- Suutari, T.; Silen, T.; Saari, H.O.; Desai, D.; Kerkelä, E.; Laitinen, S.; Yliperttula, M.; Karaman, D.S.; Rosenholm, J.M.; Viitala, T.; et al. Real-Time Label-Free Monitoring of Nanoparticle Cell Uptake. Small 2016, 12, 6289–6300. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Ng, W.B.; Bernt, W.; Cho, N.-J. Validation of size estimation of nanoparticle tracking analysis on polydisperse macromolecule assembly. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kong, C.; Pilger, C.; Hachmeister, H.; Wei, X.; Cheung, T.H.; Lai, C.S.W.; Lee, N.P.; Tsia, K.K.; Wong, K.K.Y.; Huser, T. High-contrast, fast chemical imaging by coherent Raman scattering using a self-synchronized two-colour fibre laser. Light. Sci. Appl. 2020, 9, 25. [Google Scholar] [CrossRef] [Green Version]
- Patel, M.; Feith, M.; Janicke, B.; Alm, K.; El-Schich, Z. Evaluation of the Impact of Imprinted Polymer Particles on Morphology and Motility of Breast Cancer Cells by Using Digital Holographic Cytometry. Appl. Sci. 2020, 10, 750. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.; Dubey, V.; Butola, A.; Tinguely, J.-C.; Ahluwalia, B.S.; Mehta, D.S. Sub-nanometer height sensitivity by phase shifting interference microscopy under environmental fluctuations. Opt. Express 2020, 28, 9340–9358. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Depeursinge, C.; Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 2018, 12, 578–589. [Google Scholar] [CrossRef]
- Popescu, G. Quantitative Phase Imaging of Cells and Tissues; McGraw-Hill Education: New York, NY, USA, 2011. [Google Scholar]
- Bhaduri, B.; Edwards, C.; Pham, H.; Zhou, R.; Nguyen, T.H.; Goddard, L.L.; Popescu, G. Diffraction phase microscopy: Principles and applications in materials and life sciences. Adv. Opt. Photonics 2014, 6, 57–119. [Google Scholar] [CrossRef]
- Ahmad, A.; Dubey, V.; Singh, G.; Singh, V.; Mehta, D.S. Quantitative phase imaging of biological cells using spatially low and temporally high coherent light source. Opt. Lett. 2016, 41, 1554–1557. [Google Scholar] [CrossRef]
- Dubois, A.; Grieve, K.; Moneron, G.; Lecaque, R.; Vabre, L.; Boccara, C. Ultrahigh-resolution full-field optical coherence tomography. Appl. Opt. 2004, 43, 2874–2883. [Google Scholar] [CrossRef]
- Ahmad, A.; Dubey, V.; Singh, V.R.; Tinguely, J.-C.; Øie, C.I.; Wolfson, D.L.; Mehta, D.S.; So, P.T.; Ahluwalia, B.S. Quantitative phase microscopy of red blood cells during planar trapping and propulsion. Lab. Chip. 2018, 18, 3025–3036. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.; Mahanty, T.; Dubey, V.; Butola, A.; Ahluwalia, B.S.; Mehta, D.S. Effect on the longitudinal coherence properties of a pseudothermal light source as a function of source size and temporal coherence. Opt. Lett. 2019, 44, 1817–1820. [Google Scholar] [CrossRef] [PubMed]
- Dubey, V.; Popova, D.; Ahmad, A.; Acharya, G.; Basnet, P.; Mehta, D.S.; Ahluwalia, B.S. Partially spatially coherent digital holographic microscopy and machine learning for quantitative analysis of human spermatozoa under oxidative stress condition. Sci. Rep. 2019, 9, 3564. [Google Scholar] [CrossRef]
- Greish, K.; Mathur, A.; Bakhiet, M.; Taurin, S. Nanomedicine: Is it lost in translation? Ther. Deliv. 2018, 9, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchillin, V.P. Recent Advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, S.; Huang, T.; Wang, S.; Yan, X. Analytical techniques for single-liposome characterization. Anal. Methods 2013, 5, 2150–2157. [Google Scholar] [CrossRef]
- Kay, J.G.; Koivusalo, M.; Ma, X.; Wohland, T.; Grinstein, S. Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 2012, 23, 2198–2212. [Google Scholar] [CrossRef]
- Ternullo, S.; Basnet, P.; Holsaeter, A.M.; Flaten, G.E.; de Weerd, L.; Skalko-Basnet, N. Deformable liposomes for skin therapy with human epidermal growth factor: The effect of liposomal surface charge. Eur. J. Pharm. Sci. 2018, 125, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Martins, S.; Tho, I.; Ferreira, D.C.; Souto, E.B.; Brandl, M. Physicochemical properties of lipid nanoparticles: Effect of lipid and surfactant composition. Drug Dev. Ind. Pharm. 2011, 37, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, I.L.; Carl, D.; Langehanenberg, P.; Valenzuela, S.M.; Battle, A.R.; Al Khazaaly, S.; Killingsworth, M.; Kemper, B.; von Bally, G.; Martin, D.K. Structural properties of liposomes from digital holographic microscopy. BioMEMS Nanotechnol. II 2016, 6030, 60361R-1. [Google Scholar] [CrossRef] [Green Version]
- Ban, S.; Min, E.; Ahn, Y.; Popescu, G.; Jung, W. Effect of tissue staining in quantitative phase imaging. J. Biophotonics 2018, 11, e201700402. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Han, B. Advanced iterative algorithm for phase extraction of randomly phase-shifted interferograms. Opt. Lett. 2004, 29, 1671–1673. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential-What they are and what they are not? J. Control Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Joraholmen, M.W.; Basnet, P.; Acharya, G.; Skalko-Basnet, N. PEGylated liposomes for topical vaginal therapy improve delivery of interferon alpha. Eur. J. Pharm. Biopharm. 2017, 113, 132–139. [Google Scholar] [CrossRef] [Green Version]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bangham, A.D.; Horne, R. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef]
- Zhang, H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. In Liposomes: Methods and Protocols; D’Souza, G.G.M., Ed.; Springer: New York, NY, USA, 2017; pp. 17–22. [Google Scholar] [CrossRef]
- Lacroix, A.L.; Vengut-Climent, E.; de Rochambeau, D.; Sleiman, H.F. Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale. ACS Cent. Sci. 2019, 5, 882–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venable, R.M.; Zhang, Y.; Hardy, B.J.; Pastor, R.W. Molecular dynamics simulations of a lipid bilayer and of hexadecane: An investigation of membrane fluidity. Science 1993, 262, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Khandelia, H.; Mouritsen, O.G. Lipid gymnastics: Evidence of complete acyl chain reversal in oxidized phospholipids from molecular simulations. Biophys. J. 2009, 96, 2734–2743. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Millet, L.; Mir, M.; Ding, H.; Unarunotai, S.; Rogers, J.; Gillette, M.U.; Popescu, G. Spatial light interference microscopy (SLIM). Opt. Express 2011, 19, 1016–1026. [Google Scholar] [CrossRef]
- Lee, K.; Kim, K.; Jung, J.; Heo, J.; Cho, S.; Lee, S.; Chang, G.; Jo, Y.; Park, H.; Park, Y. Quantitative phase imaging techniques for the study of cell pathophysiology: From principles to applications. Sensors 2013, 13, 4170–4191. [Google Scholar] [CrossRef]
- Bhardwaj, U.; Burgess, D.J. Physicochemical properties of extruded and non-extruded liposomes containing the hydrophobic drug dexamethasone. Int. J. Pharm. 2010, 388, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, B.; Khlebtsov, N. On the measurement of gold nanoparticle sizes by the dynamic light scattering method. Colloid J. 2011, 73, 118–127. [Google Scholar] [CrossRef]
- Butola, A.; Popova, D.; Prasad, D.K.; Ahmad, A.; Habib, A.; Tinguely, J.C.; Basnet, P.; Acharya, G.; Senthilkumaran, P.; Mehta, D.S. High spatially sensitive quantitative phase imaging assisted with deep neural network for classification of human spermatozoa under stressed condition. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Manca, M.L.; Zaru, M.; Manconi, M.; Lai, F.; Valenti, D.; Sinico, C.; Fadda, A.M. Glycerosomes: A new tool for effective dermal and transdermal drug delivery. Int. J. Pharm. 2013, 455, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Manca, M.L.; Castangia, I.; Zaru, M.; Nácher, A.; Valenti, D.; Fernàndez-Busquets, X.; Fadda, A.M.; Manconi, M. Development of curcumin loaded sodium hyaluronate immobilized vesicles (hyalurosomes) and their potential on skin inflammation and wound restoring. Biomaterials 2015, 71, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Luther, E.; Mendes, L.P.; Pan, J.; Costa, D.F.; Torchilin, V.P. Applications of label-free, quantitative phase holographic imaging cytometry to the development of multi-specific nanoscale pharmaceutical formulations. Cytom. A 2017, 91, 412–423. [Google Scholar] [CrossRef] [Green Version]
- Sternbæk, L.; Kimani Wamaitha, M.; Gawlitza, K.; Janicke, B.; Alm, K.; Wingren Gjörloff, A. Digital holographic microscopy: Macrophage uptake of nanoprobes. Imaging Microsc. 2019, 1, 21–23. [Google Scholar]






| Formulation | Extrusion |
|---|---|
| N1 | 1 × 800 nm 1 |
| N2 | 4 × 800 nm |
| N3 | 4 × 800 nm, 4 × 400 nm |
| N4 | 4 × 800 nm, 4 × 400 nm, 4 × 200 nm |
| Lip | Measured Values | Expected Values | ||||
|---|---|---|---|---|---|---|
| Size [nm] | PdI | ζ-Potential [mV] | Size [nm] | PdI | ζ-Potential [mV] | |
| N1 | 1040 1 | 0.85 ± 0.08 | −55.7 ± 6.3 | >>1000 | 1 | [−5, +5] |
| N2 | 499 ± 124 (74.5%) 2 103 ± 16 (25.5%) | 0.47 ± 0.04 | −59.8 ± 5.1 | 600–800 | <0.250 | [−5, +5] |
| N3 | 214 ± 57 | 0.24 ± 0.02 | −57.1 ± 6.7 | 300–500 | <0.250 | [−5, +5] |
| N4 | 114 ± 20 | 0.11 ± 0.01 | −55.4 ± 6.6 | 150–350 | <0.250 | [−5, +5] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cauzzo, J.; Jayakumar, N.; Ahluwalia, B.S.; Ahmad, A.; Škalko-Basnet, N. Characterization of Liposomes Using Quantitative Phase Microscopy (QPM). Pharmaceutics 2021, 13, 590. https://doi.org/10.3390/pharmaceutics13050590
Cauzzo J, Jayakumar N, Ahluwalia BS, Ahmad A, Škalko-Basnet N. Characterization of Liposomes Using Quantitative Phase Microscopy (QPM). Pharmaceutics. 2021; 13(5):590. https://doi.org/10.3390/pharmaceutics13050590
Chicago/Turabian StyleCauzzo, Jennifer, Nikhil Jayakumar, Balpreet Singh Ahluwalia, Azeem Ahmad, and Nataša Škalko-Basnet. 2021. "Characterization of Liposomes Using Quantitative Phase Microscopy (QPM)" Pharmaceutics 13, no. 5: 590. https://doi.org/10.3390/pharmaceutics13050590
APA StyleCauzzo, J., Jayakumar, N., Ahluwalia, B. S., Ahmad, A., & Škalko-Basnet, N. (2021). Characterization of Liposomes Using Quantitative Phase Microscopy (QPM). Pharmaceutics, 13(5), 590. https://doi.org/10.3390/pharmaceutics13050590