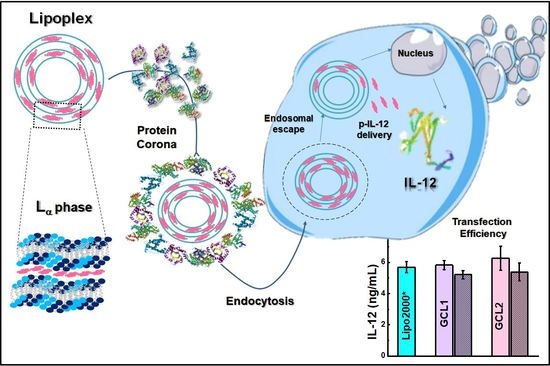
Gemini Cationic Lipid-Type Nanovectors Suitable for the Transfection of Therapeutic Plasmid DNA Encoding for Pro-Inflammatory Cytokine Interleukin-12
Abstract
1. Introduction
2. Materials and Methods
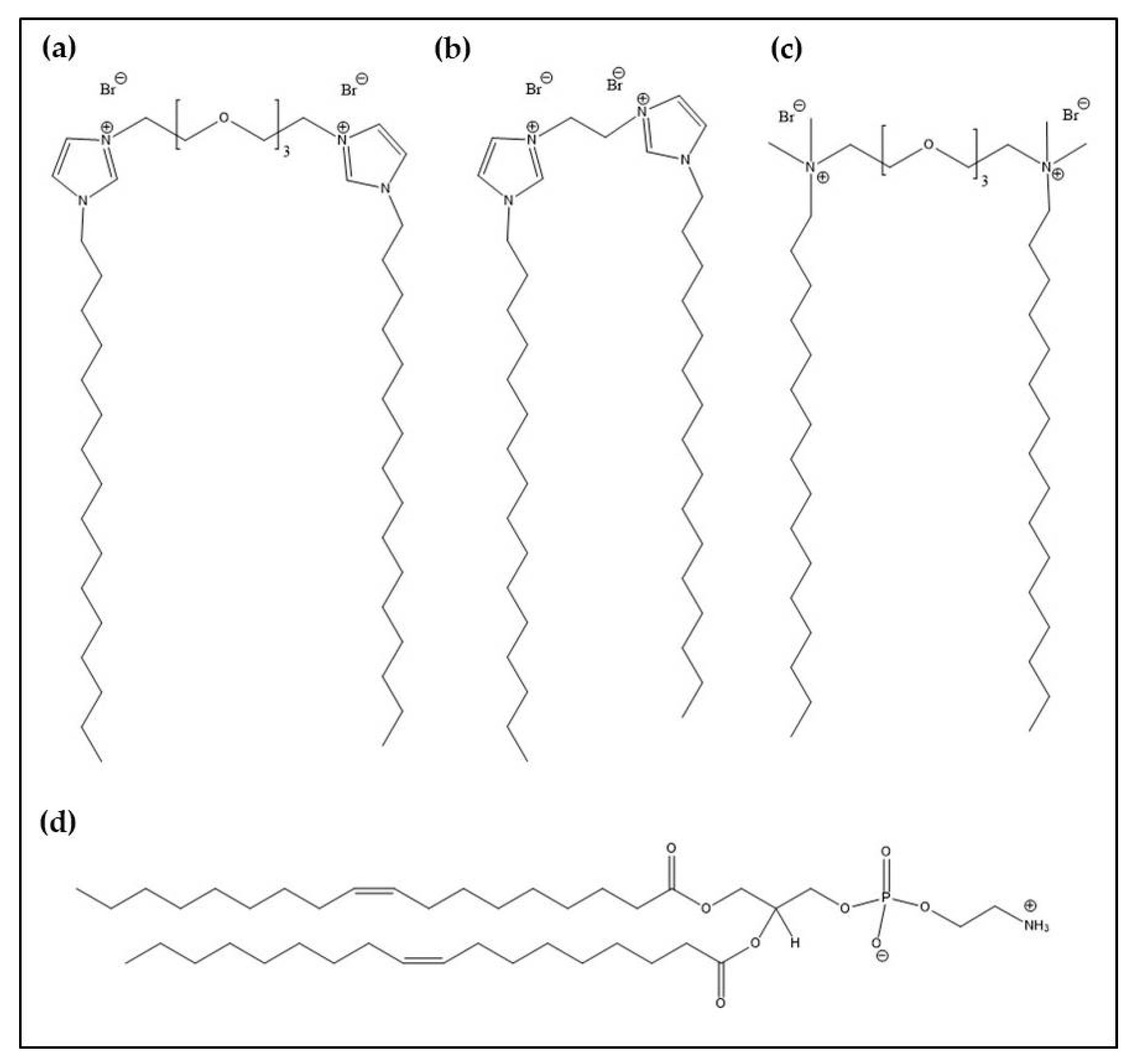
2.1. Materials
2.2. Preparation of Lipoplexes
2.3. Physicochemical Characterization Methods
2.4. Protein Corona Studies
2.5. In Vitro Studies
2.5.1. Luminometry
2.5.2. Enzyme-Linked Immunosorbent Assay
3. Results and Discussion
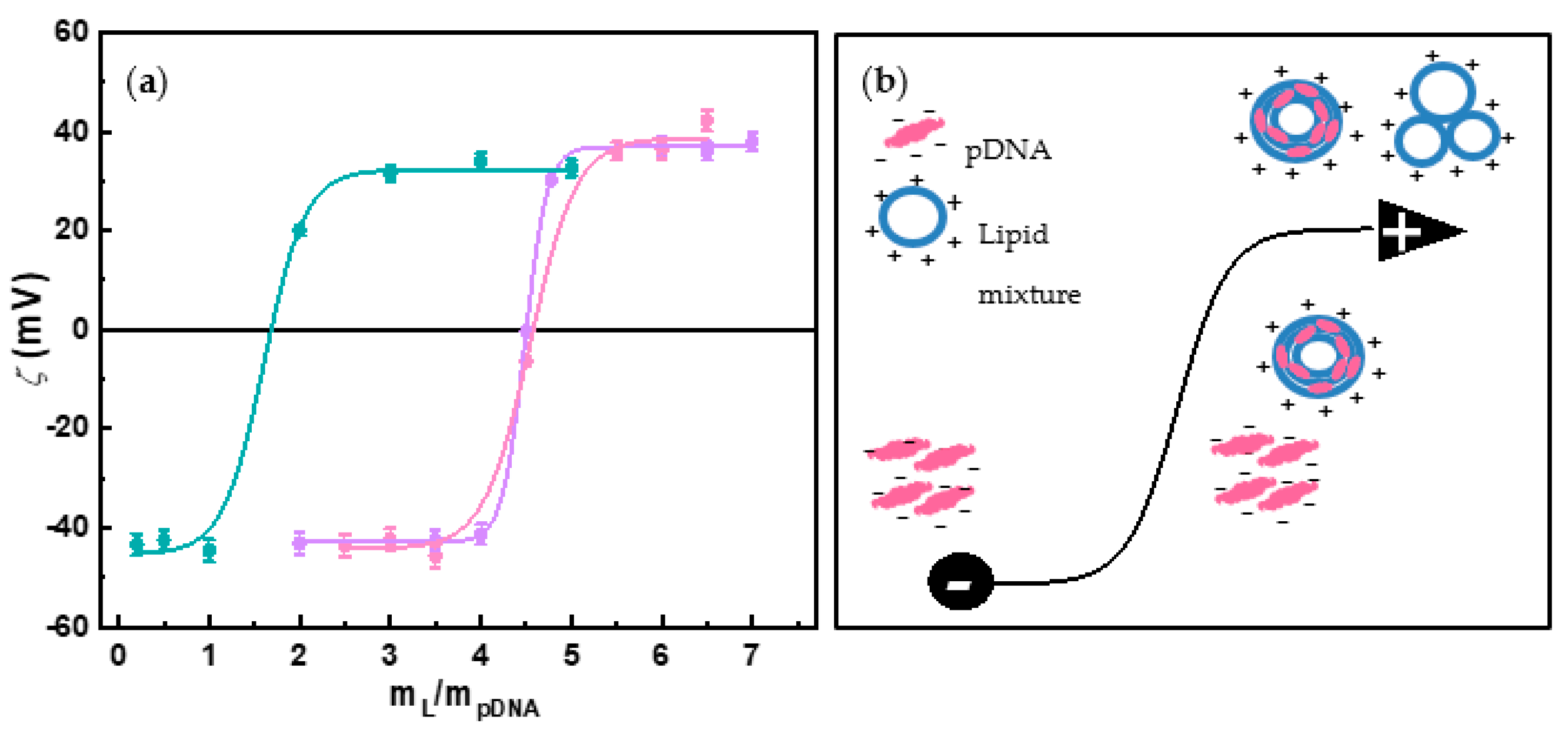
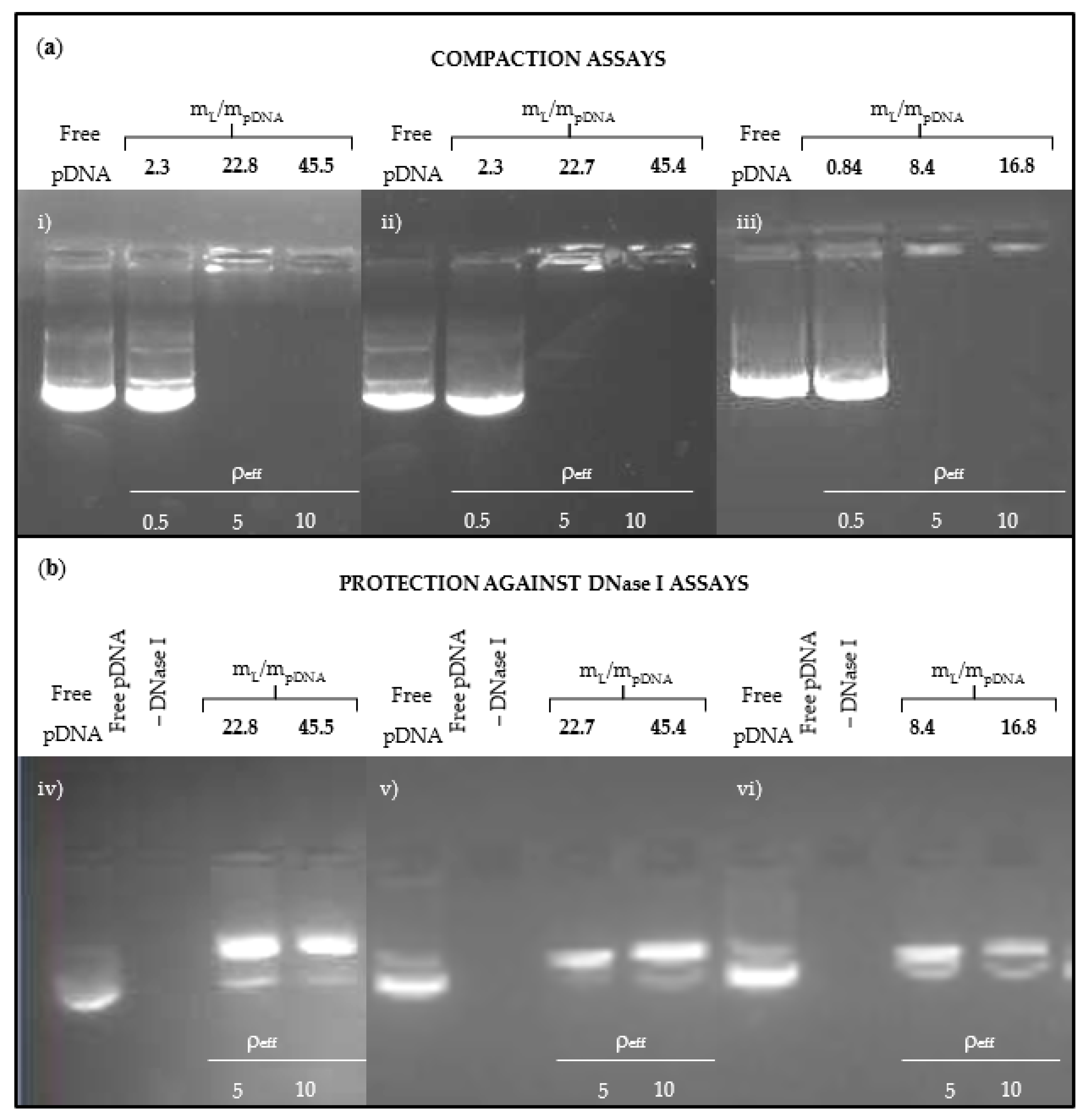
3.1. Physicochemical Characterization
3.2. Protein Corona Characterization
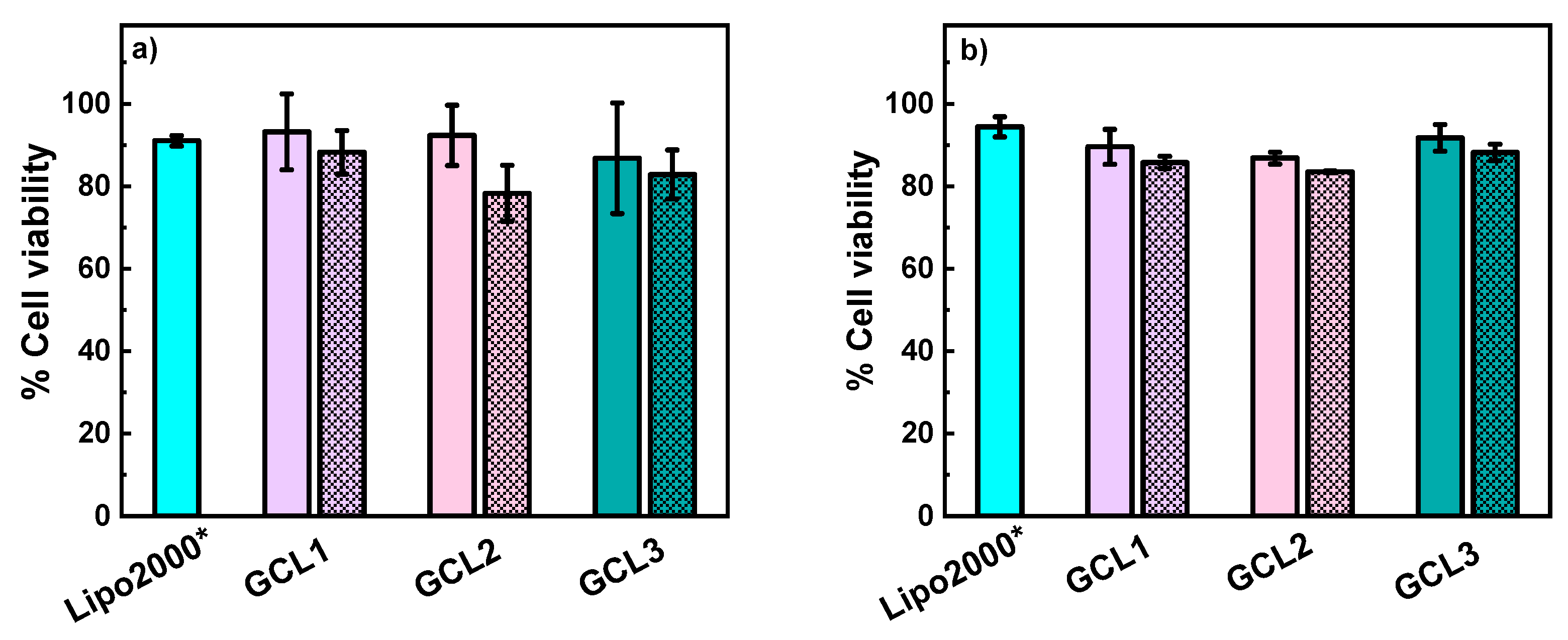
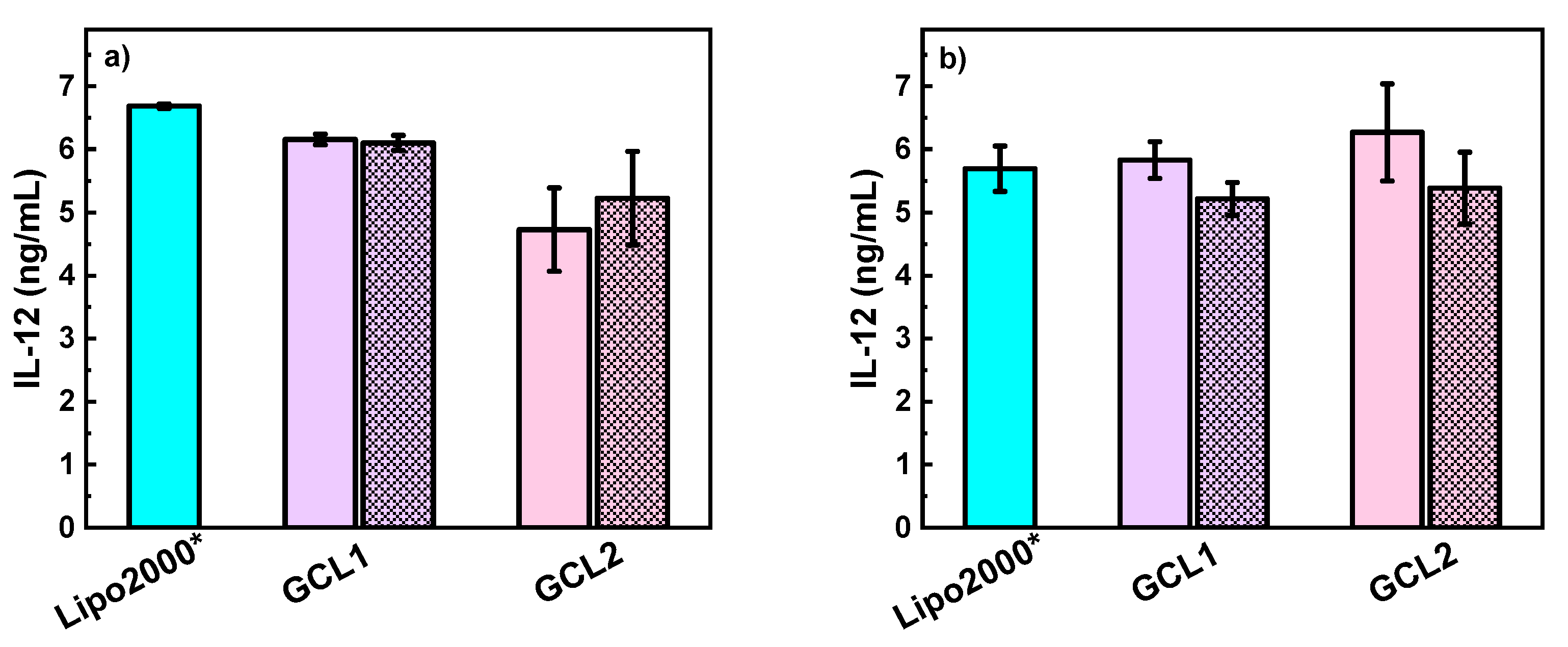
3.3. In Vitro Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kobayashi, M.; Fitz, L.; Ryan, M.; Hewick, R.M.; Clark, S.C.; Chan, S.; Loudon, R.; Sherman, F.; Perussia, B.; Trinchieri, G. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J. Exp. Med. 1989, 170, 827–845. [Google Scholar] [CrossRef]
- Stern, A.S.; Podlaski, F.J.; Hulmes, J.D.; Pan, Y.C.E.; Quinn, P.M.; Wolitzky, A.G.; Familletti, P.C.; Stremlo, D.L.; Truitt, T.; Chizzonite, R.; et al. Purification to homogeneity and partial characterization of cytotoxic lymphocyte maturation factor from human B-lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 1990, 87, 6808–6812. [Google Scholar] [CrossRef]
- Del Vecchio, M.; Bajetta, E.; Canova, S.; Lotze, M.T.; Wesa, A.; Parmiani, G.; Anichini, A. Interleukin-12: Biological properties and clinical application. Clin. Cancer Res. 2007, 13, 4677–4685. [Google Scholar] [CrossRef] [PubMed]
- Trinchieri, G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 2003, 3, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Otani, T.; Nakamura, S.; Toki, M.; Motoda, R.; Kurimoto, M.; Orita, K. Identification of IFN-gamma-producing cells in IL-12/IL-18-treated mice. Cell. Immunol. 1999, 198, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.S.; Macatonia, S.E.; Tripp, C.S.; Wolf, S.F.; Ogarra, A.; Murphy, K.M. Development of Th1 CD4+ T-cells through IL-12 produced by listeria-induced macrophages. Science 1993, 260, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Hamza, T.; Barnett, J.B.; Li, B. Interleukin 12 a key immunoregulatory cytokine in infection applications. Int. J. Mol. Sci. 2010, 11, 789–806. [Google Scholar] [CrossRef]
- Seder, R.A.; Kelsall, B.L.; Jankovic, D. Differential roles for IL-12 in the maintenance of immune responses in infectious versus autoimmune disease. J. Immunol. 1996, 157, 2745–2748. [Google Scholar] [PubMed]
- Guo, Y.; Cao, W.; Zhu, Y. Immunoregulatory functions of the IL-12 family of cytokines in antiviral systems. Viruses 2019, 11, 772. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, X.R.; Ju, Z.Y.; He, W.F. Advances in the research of mechanism and related immunotherapy on the cytokine storm induced by coronavirus disease 2019. Zhonghua Shao Shang Za Zhi 2020, 36, 471–475. [Google Scholar] [CrossRef]
- Chen, H.-W.; Chen, H.-Y.; Wang, L.-T.; Wang, F.-H.; Fang, L.-W.; Lai, H.-Y.; Chen, H.-H.; Lu, J.; Hung, M.-S.; Cheng, Y.; et al. Mesenchymal stem cells tune the development of monocyte-derived dendritic cells toward a myeloid-derived suppressive phenotype through growth-regulated oncogene chemokines. J. Immunol. 2013, 190, 5065–5077. [Google Scholar] [CrossRef]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodriguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef]
- Hussain, S.; Xie, Y.J.; Li, D.; Malik, S.I.; Hou, J.C.; Leung, E.L.H.; Fan, X.X. Current strategies against COVID-19. Chin. Med. 2020, 15, 12. [Google Scholar] [CrossRef]
- Xu, Z.; Patel, A.; Tursi, N.J.; Zhu, X.; Muthumani, K.; Kulp, D.W.; Weiner, D.B. Harnessing recent advances in synthetic DNA and electroporation technologies for rapid vaccine development against COVID-19 and other emerging infectious diseases. Front. Med. Technol. 2020, 2. [Google Scholar] [CrossRef]
- Jensen, S.; Twitty, C.; Paustian, C.; Laws, M.; McDonnell, G.; Wegmann, K.; Moudgil, T.; Afentoulis, M.; Han, M.; Foerter, K.M.; et al. 480 Preliminary evaluation of a novel coronavirus vaccine (CORVax) using electroporation of plasmid DNA encoding a stabilized prefusion SARS-CoV-2 spike protein alone or with transfection of plasmid IL-12. J. Immunother. Cancer 2020, 8, A296. [Google Scholar] [CrossRef]
- Colombo, M.P.; Trinchieri, G. Interleukin-12 in anti-tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002, 13, 155–168. [Google Scholar] [CrossRef]
- Voest, E.E.; Kenyon, B.B.; Oreilly, M.S.; Truitt, G.; Damato, R.J.; Folkman, J. Inhibition of angiogenesis in vivo by interleukin-12. J. Natl. Cancer Inst. 1995, 87, 581–586. [Google Scholar] [CrossRef]
- Lasek, W.; Zagożdżon, R.; Jakobisiak, M. Interleukin 12: Still a promising candidate for tumor immunotherapy? Cancer Immunol. Immunother. 2014, 63, 419–435. [Google Scholar] [CrossRef]
- Nguyen, K.G.; Vrabel, M.R.; Mantooth, S.M.; Hopkins, J.J.; Wagner, E.S.; Gabaldon, T.A.; Zaharoff, D.A. Localized interleukin-12 for cancer immunotherapy. Front. Immunol. 2020, 11, 2510. [Google Scholar] [CrossRef]
- Jenks, S. After initial setback, IL-12 regaining popularity. J. Natl. Cancer Inst. 1996, 88, 576–577. [Google Scholar] [CrossRef]
- Mazzolini, G.; Alfaro, C.; Sangro, B.; Feijoo, E.; Ruiz, J.; Benito, A.; Tirapu, M.; Arina, A.; Sola, J.; Herraiz, M.; et al. Intratumoral injection of dendritic cells engineered to secrete interleukin-12 by recombinant adenovirus in patients with metastatic gastrointestinal carcinomas. J. Clin. Oncol. 2005, 23, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Tahara, H.; Zitvogel, L.; Storkus, W.J.; Zeh, H.J.; McKinney, T.G.; Schreiber, R.D.; Gubler, U.; Robbins, P.D.; Lotze, M.T. Effective eradication of established murine tumors with IL-12 gene therapy using a polycistronic retroviral vector. J. Immunol. 1995, 154, 6466–6474. [Google Scholar]
- Quetglas, J.I.; Rodriguez-Madoz, J.R.; Bezunartea, J.; Ruiz-Guillen, M.; Casales, E.; Medina-Echeverz, J.; Prieto, J.; Berraondo, P.; Hervas-Stubbs, S.; Smerdou, C. Eradication of liver-implanted tumors by semliki forest virus expressing IL-12 requires efficient long-term immune responses. J. Immunol. 2013, 190, 2994–3004. [Google Scholar] [CrossRef] [PubMed]
- Fewell, J.G.; Matar, M.M.; Rice, J.S.; Brunhoeber, E.; Slobodkin, G.; Pence, C.; Worker, M.; Lewis, D.H.; Anwer, K. Treatment of disseminated ovarian cancer using nonviral interleukin-12 gene therapy delivered intraperitoneally. J. Gene Med. 2009, 11, 718–728. [Google Scholar] [CrossRef]
- Bunuales, M.; Düzgünes, N.; Zalba, S.; Garrido, M.J.; Tros de Ilarduya, C. Efficient gene delivery by EGF-lipoplexes in vitro and in vivo. Nanomedicine 2011, 6, 89–98. [Google Scholar] [CrossRef]
- Anwer, K.; Barnes, M.N.; Fewell, J.; Lewis, D.H.; Alvarez, R.D. Phase-I clinical trial of IL-12 plasmid/lipopolymer complexes for the treatment of recurrent ovarian cancer. Gene Ther. 2010, 17, 360–369. [Google Scholar] [CrossRef]
- Lucas, M.L.; Heller, L.; Coppola, D.; Heller, R. IL-12 plasmid delivery by in vivo electroporation for the successful treatment of established subcutaneous B16.F10 melanoma. Mol. Ther. 2002, 5, 668–675. [Google Scholar] [CrossRef]
- Charoensit, P.; Kawakami, S.; Higuchi, Y.; Yamashita, F.; Hashida, M. Enhanced growth inhibition of metastatic lung tumors by intravenous injection of ATRA-cationic liposome/IL-12 pDNA complexes in mice. Cancer Gene Ther. 2010, 17, 512–522. [Google Scholar] [CrossRef]
- Janeway, C.A.J.; Travers, P.; Walport, M.; Shlomchik, M.J. The complement system and innate immunity. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Barbero, F.; Russo, L.; Vitali, M.; Piella, J.; Salvo, I.; Borrajo, M.L.; Busquets-Fite, M.; Grandori, R.; Bastus, N.G.; Casals, E.; et al. Formation of the protein corona: The interface between nanoparticles and the immune system. Semin. Immunol. 2017, 34, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Farokhzad, O.C.; Mahmoudi, M. Biological Identity of Nanoparticles In Vivo: Clinical Implications of the Protein Corona. Trends Biotechnol. 2017, 35, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Gao, H.L. The Impact of Protein Corona on the Behavior and Targeting Capability of Nanoparticle-Based Delivery System. Int. J. Pharm. 2018, 552, 328–339. [Google Scholar] [CrossRef]
- Nierenberg, D.; Khaled, A.R.; Flores, O. Formation of a Protein Corona Influences the Biological Identity of Nanomaterials. Rep. Pract. Oncol. Radiother. 2018, 23, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Lynch, I.; Cedervall, T.; Lundqvist, M.; Cabaleiro-Lago, C.; Linse, S.; Dawson, K.A. The nanoparticle—Protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv. Colloid Interface Sci. 2007, 134, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Molinaro, R.; Parodi, A.; Furman, N.E.T.; Salvatore, F.; Tasciotti, E. The Impact of Nanoparticle Protein Corona on Cytotoxicity, Immunotoxicity and Target Drug Delivery. Nanomedicine 2016, 11, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Molinaro, R.; Taraballi, F.; Furman, N.E.T.; Sherman, M.B.; Parodi, A.; Salvatore, F.; Tasciotti, E. Effects of the protein corona on liposome-liposome and liposome-cell interactions. Int. J. Nanomed. 2016, 11, 3049–3063. [Google Scholar] [CrossRef]
- Kumar, K.; Barrán-Berdón, A.L.; Datta, S.; Muñoz-Úbeda, M.; Aicart-Ramos, C.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. A delocalizable cationic headgroup together with an oligo-oxyethylene spacer in gemini cationic lipids improves their biological activity as vectors of plasmid DNA. J. Mater. Chem. B 2015, 3, 1495–1506. [Google Scholar] [CrossRef]
- Misra, S.K.; Muñoz-Úbeda, M.; Datta, S.; Barrán-Berdón, A.L.; Aicart-Ramos, C.; Castro-Hartmann, P.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Effects of a delocalizable cation on the headgroup of gemini lipids on the lipoplex-type nano-aggregates directly formed from plasmid DNA. Biomacromolecules 2013, 14, 3951–3963. [Google Scholar] [CrossRef]
- Barrán-Berdón, A.L.; Misra, S.K.; Datta, S.; Muñoz-Úbeda, M.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Cationic gemini lipids containing polyoxyethylene spacers as improved transfecting agents of plasmid DNA in cancer cells. J. Mater. Chem. B 2014, 2, 4640–4652. [Google Scholar] [CrossRef]
- Safinya, C.R.; Ewert, K.K.; Majzoub, R.N.; Leal, C. Cationic liposome-nucleic acid complexes for gene delivery and gene silencing. New J. Chem. 2014, 38, 5164–5172. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Llizo, A.; Li, P.; Wang, C.X.; Guo, Y.Y.; Ao, M.Q.; Bai, L.L.; Wang, C.; Yang, Y.L.; Xu, G.Y. High transfection efficiency of homogeneous DNA nanoparticles induced by imidazolium gemini surfactant as nonviral vector. J. Phys. Chem. C 2013, 117, 26573–26581. [Google Scholar] [CrossRef]
- Junquera, E.; Aicart, E. Recent progress in gene therapy to deliver nucleic acids with multivalent cationic vectors. Adv. Colloid Interface Sci. 2016, 233, 161–175. [Google Scholar] [CrossRef]
- Sharma, V.D.; Ilies, M.A. Heterocyclic cationic gemini surfactants: A comparative overview of their synthesis, self-assembling, physicochemical, and biological properties. Med. Res. Rev. 2014, 34, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Kamel, A.O.; Wettig, S.D. Interactions between DNA and gemini surfactant: Impact on gene therapy: Part I. Nanomedicine 2016, 11, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Kamel, A.O.; Wettig, S.D. Interactions between DNA and gemini surfactant: Impact on gene therapy: Part II. Nanomedicine 2016, 11, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pulido, A.; Aicart, E.; Llorca, O.; Junquera, E. Compaction process of calf thymus DNA by mixed cationic-zwitterionic liposomes: A physicochemical study. J. Phys. Chem. B 2008, 112, 2187–2197. [Google Scholar] [CrossRef]
- Muñoz-Úbeda, M.; Misra, S.K.; Barrán-Berdón, A.L.; Datta, S.; Aicart-Ramos, C.; Castro-Hartmann, P.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. How does the spacer length of cationic gemini lipids influence the lipoplex formation with plasmid DNA? Physicochemical and biochemical characterizations and their relevance in gene therapy. Biomacromolecules 2012, 13, 3926–3937. [Google Scholar] [CrossRef]
- Muñoz-Úbeda, M.; Misra, S.K.; Barrán-Berdón, A.L.; Aicart-Ramos, C.; Sierra, M.B.; Biswas, J.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Why is less cationic lipid required to prepare lipoplexes from plasmid DNA than linear DNA in gene therapy? J. Am. Chem. Soc. 2011, 133, 18014–18017. [Google Scholar] [CrossRef]
- Bednar, J.; Woodcock, C.L. Chromatin; Academic Press Inc.: San Diego, CA, USA, 1999; Volume 304, pp. 191–213. [Google Scholar]
- Llorca, O.; McCormack, E.; Hynes, G.; Grantham, J.; Cordell, J.; Carrascosa, J.L.; Willison, K.R.; Fernández, J.J.; Valpuesta, J.M. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 1999, 402, 693–696. [Google Scholar] [CrossRef]
- Dubochet, J.; Adrian, M.; Chang, J.J.; Homo, J.C.; Lepault, J.; McDowall, A.W.; Schultz, P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988, 21, 129–228. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Arribas, N.; Martínez-Negro, M.; Villar, E.M.; Pérez, L.; Aicart, E.; Taboada, P.; Guerrero-Martínez, A.; Junquera, E. Biocompatible nanovector of siRNA consisting of arginine-based cationic lipid for gene knockdown in cancer cells. ACS Appl. Mater. Interfaces 2020, 12, 34536–34547. [Google Scholar] [CrossRef]
- Martínez-Negro, M.; Blanco-Fernández, L.; Tentori, P.M.; Pérez, L.; Pinazo, A.; de Ilarduya, C.T.; Aicart, E.; Junquera, E. A gemini cationic lipid with histidine residues as a novel lipid-based gene nanocarrier: A biophysical and biochemical study. Nanomaterials 2018, 8, 1061. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Negro, M.; Sánchez-Arribas, N.; Guerrero-Martínez, A.; Moyá, M.L.; de Ilarduya, C.T.; Mendicuti, F.; Aicart, E.; Junquera, E. A non-viral plasmid DNA delivery system consisting on a lysine-derived cationic lipid mixed with a fusogenic lipid. Pharmaceutics 2019, 11, 632. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Negro, M.; Guerrero-Martínez, A.; García-Rio, L.; Domènech, O.; Aicart, E.; de Ilarduya, C.T.; Junquera, E. Multidisciplinary approach to the transfection of plasmid DNA by a nonviral nanocarrier based on a gemini-bolaamphiphilic hybrid lipid. ACS Omega 2018, 3, 208–217. [Google Scholar] [CrossRef]
- Liu, D.X.; Mori, A.; Huang, L. Role of liposome size and RES blockade in controlling biodistribution and tumor uptake of GM1-containing liposomes. Biochim. Biophys. Acta 1992, 1104, 95–101. [Google Scholar] [CrossRef]
- Gabizon, A.; Papahadjopoulos, D. The role of surface-charge and hydrophilic groups on liposome clearance in vivo. Biochim. Biophys. Acta 1992, 1103, 94–100. [Google Scholar] [CrossRef]
- Pozzi, D.; Colapicchioni, V.; Caracciolo, G.; Piovesana, S.; Capriotti, A.L.; Palchetti, S.; De Grossi, S.; Riccioli, A.; Amenitsch, H.; Lagana, A. Effect of polyethyleneglycol (PEG) chain length on the bio-nano-interactions between PEGylated lipid nanoparticles and biological fluids: From nanostructure to uptake in cancer cells. Nanoscale 2014, 6, 2782–2792. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Arribas, N.; Martínez-Negro, M.; Villar, E.M.; Pérez, L.; Osío Barcina, J.; Aicart, E.; Taboada, P.; Guerrero-Martínez, A.; Junquera, E. Protein expression knockdown in cancer cells induced by a gemini cationic lipid nanovector with histidine-based polar heads. Pharmaceutics 2020, 12, 791. [Google Scholar] [CrossRef]
- Martinez-Negro, M.; Gonzalez-Rubio, G.; Aicart, E.; Landfester, K.; Guerrero-Martinez, A.; Junquera, E. Insights into colloidal nanoparticle-protein corona interactions for nanomedicine applications. Adv. Colloid Interface Sci. 2021, 289. [Google Scholar] [CrossRef]
- Kreuter, J.; Shamenkov, D.; Petrov, V.; Ramge, P.; Cychutek, K.; Koch-Brandt, C.; Alyautdin, R. Apolipoprotein-mediated transport of nanoparticle-bound drugs across the blood-brain barrier. J. Drug Target. 2002, 10, 317–325. [Google Scholar] [CrossRef]
- Furumoto, K.; Yokoe, J.-I.; Ogawara, K.-i.; Amano, S.; Takaguchi, M.; Higaki, K.; Kai, T.; Kimura, T. Effect of coupling of albumin onto surface of PEG liposome on its in vivo disposition. Int. J. Pharm. 2007, 329, 110–116. [Google Scholar] [CrossRef]
- Caracciolo, G.; Cardarelli, F.; Pozzi, D.; Salomone, F.; Maccari, G.; Bardi, G.; Capriotti, A.L.; Cavaliere, C.; Papi, M.; Lagana, A. Selective targeting capability acquired with a protein corona adsorbed on the surface of 1,2-dioleoyl-3-trimethylammonium propane/DNA nanoparticles. ACS Appl. Mater. Interfaces 2013, 5, 13171–13179. [Google Scholar] [CrossRef]
- Mochizuki, S.; Kanegae, N.; Nishina, K.; Kamikawa, Y.; Koiwai, K.; Masunaga, H.; Sakurai, K. The role of the helper lipid dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine. Biochim. Biophys. Acta 2013, 1828, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.X.; Munye, M.M.; Tagalakis, A.D.; Manunta, M.D.I.; Hart, S.L. The role of the helper lipid on the DNA transfection efficiency of lipopolyplex formulations. Sci. Rep. 2014, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Carbajo-Gordillo, A.I.; Rodriguez-Lavado, J.; Blanco, J.L.J.; Benito, J.M.; Di Giorgio, C.; Velaz, I.; de Ilarduya, C.T.; Mellet, C.O.; Fernandez, J.M.G. Trehalose-based siamese twin amphiphiles with tunable self-assembling, DNA nanocomplexing and gene delivery properties. Chem. Commun. 2019, 55, 8227–8230. [Google Scholar] [CrossRef] [PubMed]



| Lipoplexes | ρeff = 5 | ρeff = 10 | ||
|---|---|---|---|---|
| Dh (nm) | PDI | Dh (nm) | PDI | |
| GCL1/DOPE-pCMV-Luc | 265 | 0.39 | 181 | 0.36 |
| GCL2/DOPE-pCMV-Luc | 205 | 0.30 | 152 | 0.24 |
| GCL3/DOPE-pCMV-Luc | 202 | 0.36 | 185 | 0.39 |
| Proteins | % | |||
|---|---|---|---|---|
| GCL1 | GCL2 | GCL3 | ||
| Alpha-1-antitrypsin SV = 1 | 11 | |||
| Alpha-1-antitrypsin SV = 3 | ||||
| APOC4-APOC2 | ||||
| Apolipoprotein A-I | ||||
| Apolipoprotein A-II | ||||
| Apolipoprotein A-IV | 9 | |||
| Apolipoprotein B-100 | ||||
| Apolipoprotein C-I | ||||
| Apolipoprotein C-III | ||||
| Apolipoprotein D | ||||
| Apolipoprotein E | 7 | |||
| Apolipoprotein M | ||||
| C4b-binding protein alpha chain | ||||
| Complement C3 | ||||
| Haptoglobin | ||||
| Haptoglobin-related protein | 5 | |||
| Hyaluronan-binding protein 2 | ||||
| Ig heavy constant gamma 1 (Fragment) | ||||
| Ig kappa constant | ||||
| Ig lambda constant 3 | ||||
| Inter-alpha-trypsin inhibitor heavy chain H1 | 3 | |||
| Isoform 2 of Ig heavy constant mu | ||||
| Protein AMBP | ||||
| Prothrombin | ||||
| SAA2-SAA4 readthrough | ||||
| Serum albumin | 1 | |||
| Serum amyloid A-1 protein | ||||
| Trypsin | ||||
| Vitronectin | 0 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Arribas, N.; Martínez-Negro, M.; Aicart-Ramos, C.; Tros de Ilarduya, C.; Aicart, E.; Guerrero-Martínez, A.; Junquera, E. Gemini Cationic Lipid-Type Nanovectors Suitable for the Transfection of Therapeutic Plasmid DNA Encoding for Pro-Inflammatory Cytokine Interleukin-12. Pharmaceutics 2021, 13, 729. https://doi.org/10.3390/pharmaceutics13050729
Sánchez-Arribas N, Martínez-Negro M, Aicart-Ramos C, Tros de Ilarduya C, Aicart E, Guerrero-Martínez A, Junquera E. Gemini Cationic Lipid-Type Nanovectors Suitable for the Transfection of Therapeutic Plasmid DNA Encoding for Pro-Inflammatory Cytokine Interleukin-12. Pharmaceutics. 2021; 13(5):729. https://doi.org/10.3390/pharmaceutics13050729
Chicago/Turabian StyleSánchez-Arribas, Natalia, María Martínez-Negro, Clara Aicart-Ramos, Conchita Tros de Ilarduya, Emilio Aicart, Andrés Guerrero-Martínez, and Elena Junquera. 2021. "Gemini Cationic Lipid-Type Nanovectors Suitable for the Transfection of Therapeutic Plasmid DNA Encoding for Pro-Inflammatory Cytokine Interleukin-12" Pharmaceutics 13, no. 5: 729. https://doi.org/10.3390/pharmaceutics13050729
APA StyleSánchez-Arribas, N., Martínez-Negro, M., Aicart-Ramos, C., Tros de Ilarduya, C., Aicart, E., Guerrero-Martínez, A., & Junquera, E. (2021). Gemini Cationic Lipid-Type Nanovectors Suitable for the Transfection of Therapeutic Plasmid DNA Encoding for Pro-Inflammatory Cytokine Interleukin-12. Pharmaceutics, 13(5), 729. https://doi.org/10.3390/pharmaceutics13050729