Implementation and Comparison of Two Pharmacometric Tools for Model-Based Therapeutic Drug Monitoring and Precision Dosing of Daptomycin
Abstract
1. Introduction
2. Material and Methods
2.1. Population Data
2.2. Therapeutic Drug Monitoring of Daptomycin
2.3. Building of Daptomycin Nonparametric Population PK Model for BestDose
2.4. Implementation of A Parametric Model in Tucuxi
2.5. External Validation and Comparison of the Two Models
3. Results
3.1. Population Data
3.2. Population Modeling in the Learning Dataset
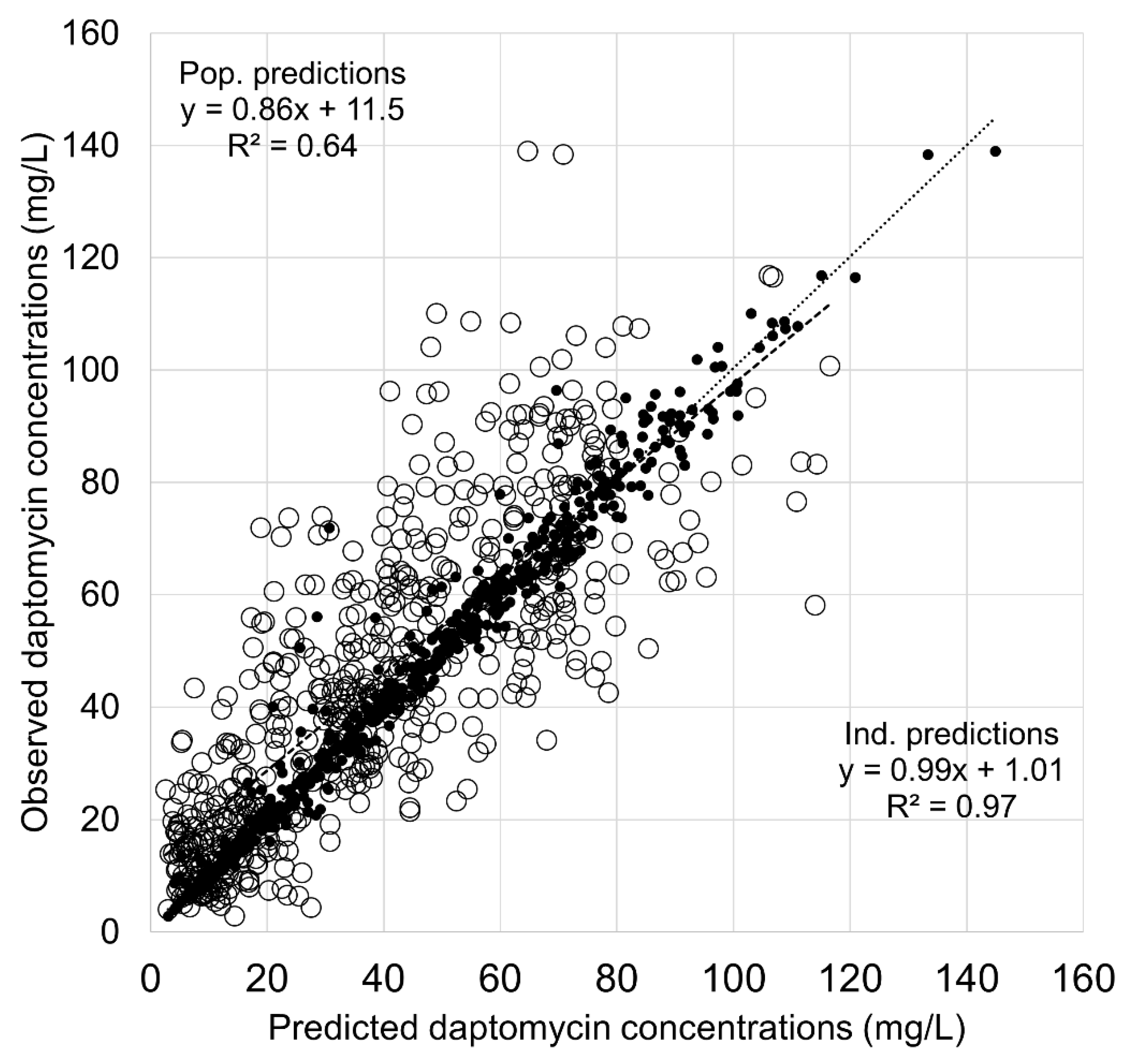
3.3. Comparison of BestDose and Tucuxi Models in the Validation Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gray, D.A.; Wenzel, M. More Than a Pore: A Current Perspective on the In Vivo Mode of Action of the Lipopeptide Antibiotic Daptomycin. Antibiotics 2020, 9, 17. [Google Scholar] [CrossRef]
- Debono, M.; Barnhart, M.; Carrell, C.B.; Hoffmann, J.A.; Occolowitz, J.L.; Abbott, B.J.; Fukuda, D.S.; Hamill, R.L.; Biemann, K.; Herlihy, W.C. A21978C, a complex of new acidic peptide antibiotics: Isolation, chemistry, and mass spectral structure elucidation. J. Antibiot. 1987, 40, 761–777. [Google Scholar] [CrossRef]
- Sauermann, R.; Rothenburger, M.; Graninger, W.; Joukhadar, C. Daptomycin: A review 4 years after first approval. Pharmacology 2008, 81, 79–91. [Google Scholar] [CrossRef]
- Sandoval, N.; Grau, S.; Sorli, L.; Montero, M.; Esteve, E.; Horcajada, J.P. Clinical experience with the use of daptomycin in a tertiary care teaching hospital in Barcelona, Spain. Future Microbiol. 2015, 10, 1145–1154. [Google Scholar] [CrossRef]
- Montange, D.; Berthier, F.; Leclerc, G.; Serre, A.; Jeunet, L.; Berard, M.; Muret, P.; Vettoretti, L.; Leroy, J.; Hoen, B.; et al. Penetration of daptomycin into bone and synovial fluid in joint replacement. Antimicrob. Agents Chemother. 2014, 58, 3991–3996. [Google Scholar] [CrossRef] [PubMed]
- Traunmuller, F.; Schintler, M.V.; Metzler, J.; Spendel, S.; Mauric, O.; Popovic, M.; Konz, K.H.; Scharnagl, E.; Joukhadar, C. Soft tissue and bone penetration abilities of daptomycin in diabetic patients with bacterial foot infections. J. Antimicrob. Chemother. 2010, 65, 1252–1257. [Google Scholar] [CrossRef] [PubMed]
- Osmon, D.R.; Berbari, E.F.; Berendt, A.R.; Lew, D.; Zimmerli, W.; Steckelberg, J.M.; Rao, N.; Hanssen, A.; Wilson, W.R. Diagnosis and management of prosthetic joint infection: Clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect Dis. 2013, 56, e1–e25. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Valour, F.; Karsenty, J.; Gagnieu, M.C.; Perpoint, T.; Lustig, S.; Ader, F.; Martha, B.; Laurent, F.; Chidiac, C.; et al. Daptomycin > 6 mg/kg/day as salvage therapy in patients with complex bone and joint infection: Cohort study in a regional reference center. BMC Infect. Dis. 2016, 16, 83. [Google Scholar] [CrossRef] [PubMed]
- Telles, J.P.; Cieslinski, J.; Tuon, F.F. Daptomycin to bone and joint infections and prosthesis joint infections: A systematic review. Braz. J. Infect. Dis. 2019, 23, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, M.; Jacqueline, C.; Amador, G.; Le Mabecque, V.; Miegeville, A.; Potel, G.; Caillon, J.; Asseray, N. Efficacy of daptomycin combined with rifampicin for the treatment of experimental meticillin-resistant Staphylococcus aureus (MRSA) acute osteomyelitis. Int. J. Antimicrob. Agents 2010, 36, 542–544. [Google Scholar] [CrossRef]
- Figueroa, D.A.; Mangini, E.; Amodio-Groton, M.; Vardianos, B.; Melchert, A.; Fana, C.; Wehbeh, W.; Urban, C.M.; Segal-Maurer, S. Safety of high-dose intravenous daptomycin treatment: Three-year cumulative experience in a clinical program. Clin. Infect Dis. 2009, 49, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Casapao, A.M.; Kullar, R.; Davis, S.L.; Levine, D.P.; Zhao, J.J.; Potoski, B.A.; Goff, D.A.; Crank, C.W.; Segreti, J.; Sakoulas, G.; et al. Multicenter study of high-dose daptomycin for treatment of enterococcal infections. Antimicrob. Agents Chemother. 2013, 57, 4190–4196. [Google Scholar] [CrossRef]
- Kullar, R.; Casapao, A.M.; Davis, S.L.; Levine, D.P.; Zhao, J.J.; Crank, C.W.; Segreti, J.; Sakoulas, G.; Cosgrove, S.E.; Rybak, M.J. A multicentre evaluation of the effectiveness and safety of high-dose daptomycin for the treatment of infective endocarditis. J. Antimicrob. Chemother. 2013, 68, 2921–2926. [Google Scholar] [CrossRef]
- Bhavnani, S.M.; Rubino, C.M.; Ambrose, P.G.; Drusano, G.L. Daptomycin exposure and the probability of elevations in the creatine phosphokinase level: Data from a randomized trial of patients with bacteremia and endocarditis. Clin. Infect Dis. 2010, 50, 1568–1574. [Google Scholar] [CrossRef]
- Safdar, N.; Andes, D.; Craig, W.A. In vivo pharmacodynamic activity of daptomycin. Antimicrob. Agents Chemother. 2004, 48, 63–68. [Google Scholar] [CrossRef]
- Falcone, M.; Russo, A.; Cassetta, M.I.; Lappa, A.; Tritapepe, L.; d’Ettorre, G.; Fallani, S.; Novelli, A.; Venditti, M. Variability of pharmacokinetic parameters in patients receiving different dosages of daptomycin: Is therapeutic drug monitoring necessary? J. Infect Chemother. 2013, 19, 732–739. [Google Scholar] [CrossRef]
- Avent, M.L.; Rogers, B.A. Optimising antimicrobial therapy through the use of Bayesian dosing programs. Int. J. Clin. Pharm. 2019, 41, 1121–1130. [Google Scholar] [CrossRef]
- AntiInfective, I.S.; Wicha, S.G.; Martson, A.G.; Nielsen, E.I.; Koch, B.C.; Friberg, L.E. From Therapeutic Drug Monitoring to Model-Informed Precision Dosing for Antibiotics. Clin. Pharmacol. Ther. 2021, 109, 928–941. [Google Scholar] [CrossRef]
- Kantasiripitak, W.; Van Daele, R.; Gijsen, M.; Ferrante, M.; Spriet, I.; Dreesen, E. Software Tools for Model-Informed Precision Dosing: How Well Do They Satisfy the Needs? Front Pharmacol. 2020, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Bricca, R.; Goutelle, S.; Roux, S.; Gagnieu, M.C.; Becker, A.; Conrad, A.; Valour, F.; Laurent, F.; Triffault-Fillit, C.; Chidiac, C.; et al. Genetic polymorphisms of ABCB1 (P-glycoprotein) as a covariate influencing daptomycin pharmacokinetics: A population analysis in patients with bone and joint infection. J. Antimicrob. Chemother. 2019, 74, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Martens-Lobenhoffer, J.; Kielstein, J.T.; Oye, C.; Bode-Boger, S.M. Validated high performance liquid chromatography-UV detection method for the determination of daptomycin in human plasma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008, 875, 546–550. [Google Scholar] [CrossRef]
- Neely, M.; Philippe, M.; Rushing, T.; Fu, X.; van Guilder, M.; Bayard, D.; Schumitzky, A.; Bleyzac, N.; Goutelle, S. Accurately Achieving Target Busulfan Exposure in Children and Adolescents With Very Limited Sampling and the BestDose Software. Ther. Drug Monit. 2016, 38, 332–342. [Google Scholar] [CrossRef]
- Neely, M.N.; van Guilder, M.G.; Yamada, W.M.; Schumitzky, A.; Jelliffe, R.W. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther. Drug Monit. 2012, 34, 467–476. [Google Scholar] [CrossRef]
- Goutelle, S.; Woillard, J.B.; Neely, M.; Yamada, W.; Bourguignon, L. Nonparametric Methods in Population Pharmacokinetics. J. Clin. Pharmacol. 2020. [Google Scholar] [CrossRef]
- Dvorchik, B.; Arbeit, R.D.; Chung, J.; Liu, S.; Knebel, W.; Kastrissios, H. Population pharmacokinetics of daptomycin. Antimicrob. Agents Chemother. 2004, 48, 2799–2807. [Google Scholar] [CrossRef][Green Version]
- Drennan, P.G.; Thoma, Y.; Barry, L.; Matthey, J.; Sivam, S.; van Hal, S.J. Bayesian Forecasting for Intravenous Tobramycin Dosing in Adults with Cystic Fibrosis Using One versus Two Serum Concentrations in a Dosing Interval. Ther. Drug Monit. 2021, 43, 505–511. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999, 8, 135–160. [Google Scholar] [CrossRef]
- Goedhart, J.; Rishniw, M. BA-plotteR—A web tool for generating Bland-Altman plots and constructing limits of agreement. Res. Vet. Sci. 2021, 137, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Carli, A.V.; Miller, A.O.; Kapadia, M.; Chiu, Y.F.; Westrich, G.H.; Brause, B.D.; Henry, M.W. Assessing the Role of Daptomycin as Antibiotic Therapy for Staphylococcal Prosthetic Joint Infection. J. Bone Jt. Infect 2020, 5, 82–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galar, A.; Munoz, P.; Valerio, M.; Cercenado, E.; Garcia-Gonzalez, X.; Burillo, A.; Sanchez-Somolinos, M.; Juarez, M.; Verde, E.; Bouza, E. Current use of daptomycin and systematic therapeutic drug monitoring: Clinical experience in a tertiary care institution. Int. J. Antimicrob. Agents 2019, 53, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Urakami, T.; Hamada, Y.; Oka, Y.; Okinaka, T.; Yamakuchi, H.; Magarifuchi, H.; Aoki, Y. Clinical pharmacokinetic and pharmacodynamic analysis of daptomycin and the necessity of high-dose regimen in Japanese adult patients. J. Infect. Chemother. 2019, 25, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Sheiner, L.B.; Beal, S.; Rosenberg, B.; Marathe, V.V. Forecasting individual pharmacokinetics. Clin. Pharmacol. Ther. 1979, 26, 294–305. [Google Scholar] [CrossRef]
- Jelliffe, R.; Bayard, D.; Milman, M.; Van Guilder, M.; Schumitzky, A. Achieving target goals most precisely using nonparametric compartmental models and “multiple model” design of dosage regimens. Ther. Drug Monit. 2000, 22, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Tod, M. Individualising aminoglycoside dosage regimens after therapeutic drug monitoring. Simple or complex pharmacokinetic methods ? Clin. Pharm. 2001, 40, 803–814. [Google Scholar] [CrossRef]
- Avery, L.M.; Kuti, J.L.; Weisser, M.; Egli, A.; Rybak, M.J.; Zasowski, E.J.; Arias, C.A.; Contreras, G.A.; Chong, P.P.; Aitken, S.L.; et al. Pharmacodynamic Analysis of Daptomycin-treated Enterococcal Bacteremia: It Is Time to Change the Breakpoint. Clin. Infect. Dis. 2019, 68, 1650–1657. [Google Scholar] [CrossRef] [PubMed]


| Variables | Learning Dataset (n = 81) | Validation Dataset (n = 94) |
|---|---|---|
| Proportion of women/men | 41.9%/58.1% | 42.5%/57.5% |
| Age (years) | 60 ± 18 | 62 ± 17 |
| Body weight (kg) a | 79 ± 20 | 76 ± 18 |
| CLCR (mL/min) a, b | 100 ± 41 | 103 ± 56 |
| Initial dose of daptomycin (mg/kg/24 h) | 8.0 ± 1.9 | 7.6 ± 1.3 |
| Number of TDM occasions per patient | 2.5 ± 7.9 | 2.8 ± 0.5 |
| AUC24h (mg.h/L) | ND | 975 ± 395 |
| AUC24h < 666 mg.h/L (%) | ND | 21.3% |
| Cmin (mg/L) | ND | 21.3 ± 12.9 |
| Cmin > 24.3 mg/L (%) | ND | 31.9% |
| Parameter a | Mean | Median | Variance | Coefficient of Variation |
|---|---|---|---|---|
| V1 (L per 70 kg) | 6.90 | 7.18 | 7.40 | 39.4% |
| Ks (h−1 per 100 mL/min of CLCR) | 0.050 | 0.045 | 0.0020 | 89.6% |
| Ki (h−1) | 0.060 | 0.052 | 0.0025 | 83.2% |
| Kcp (h−1) | 0.693 | 0.287 | 0.669 | 118.0% |
| Kpc (h−1) | 0.667 | 0.449 | 0.424 | 97.7% |
| PK Quantity | BestDose Estimate | Tucuxi Estimate | p-Value | Determination Coefficient (R2) a |
|---|---|---|---|---|
| Predicted concentrations (mg/L) | 46.6 ± 46.7 | 44.3 ± 44.5 | 0.29 | 0.93 |
| AUC24h (mg.h/L) | 975 ± 395 | 893 ± 345 | 0.19 | 0.66 |
| Dmin (mg/kg) | 6.4 ± 2.7 | 6.9 ± 2.8 | 0.14 | 0.81 |
| Dmax (mg/kg) | 12.0 ± 8.6 | 14.7 ± 10.7 | 0.046 | 0.77 |
| V1 (L) | 7.9 ± 3.5 | 5.9 ± 2.4 | <0.001 | 0.20 |
| T1/2 (h) b | 16.8 ± 10.6 | 13.1 ± 4.7 | 0.015 | 0.11 |
| CLdap (L/h) c | 0.74 ± 0.42 | 0.73 ± 0.29 | 0.50 | 0.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heitzmann, J.; Thoma, Y.; Bricca, R.; Gagnieu, M.-C.; Leclerc, V.; Roux, S.; Conrad, A.; Ferry, T.; Goutelle, S. Implementation and Comparison of Two Pharmacometric Tools for Model-Based Therapeutic Drug Monitoring and Precision Dosing of Daptomycin. Pharmaceutics 2022, 14, 114. https://doi.org/10.3390/pharmaceutics14010114
Heitzmann J, Thoma Y, Bricca R, Gagnieu M-C, Leclerc V, Roux S, Conrad A, Ferry T, Goutelle S. Implementation and Comparison of Two Pharmacometric Tools for Model-Based Therapeutic Drug Monitoring and Precision Dosing of Daptomycin. Pharmaceutics. 2022; 14(1):114. https://doi.org/10.3390/pharmaceutics14010114
Chicago/Turabian StyleHeitzmann, Justine, Yann Thoma, Romain Bricca, Marie-Claude Gagnieu, Vincent Leclerc, Sandrine Roux, Anne Conrad, Tristan Ferry, and Sylvain Goutelle. 2022. "Implementation and Comparison of Two Pharmacometric Tools for Model-Based Therapeutic Drug Monitoring and Precision Dosing of Daptomycin" Pharmaceutics 14, no. 1: 114. https://doi.org/10.3390/pharmaceutics14010114
APA StyleHeitzmann, J., Thoma, Y., Bricca, R., Gagnieu, M.-C., Leclerc, V., Roux, S., Conrad, A., Ferry, T., & Goutelle, S. (2022). Implementation and Comparison of Two Pharmacometric Tools for Model-Based Therapeutic Drug Monitoring and Precision Dosing of Daptomycin. Pharmaceutics, 14(1), 114. https://doi.org/10.3390/pharmaceutics14010114






