Enzyme-Responsive Amphiphilic Peptide Nanoparticles for Biocompatible and Efficient Drug Delivery
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Peptide Nanoparticles
2.3. Physical Characterization
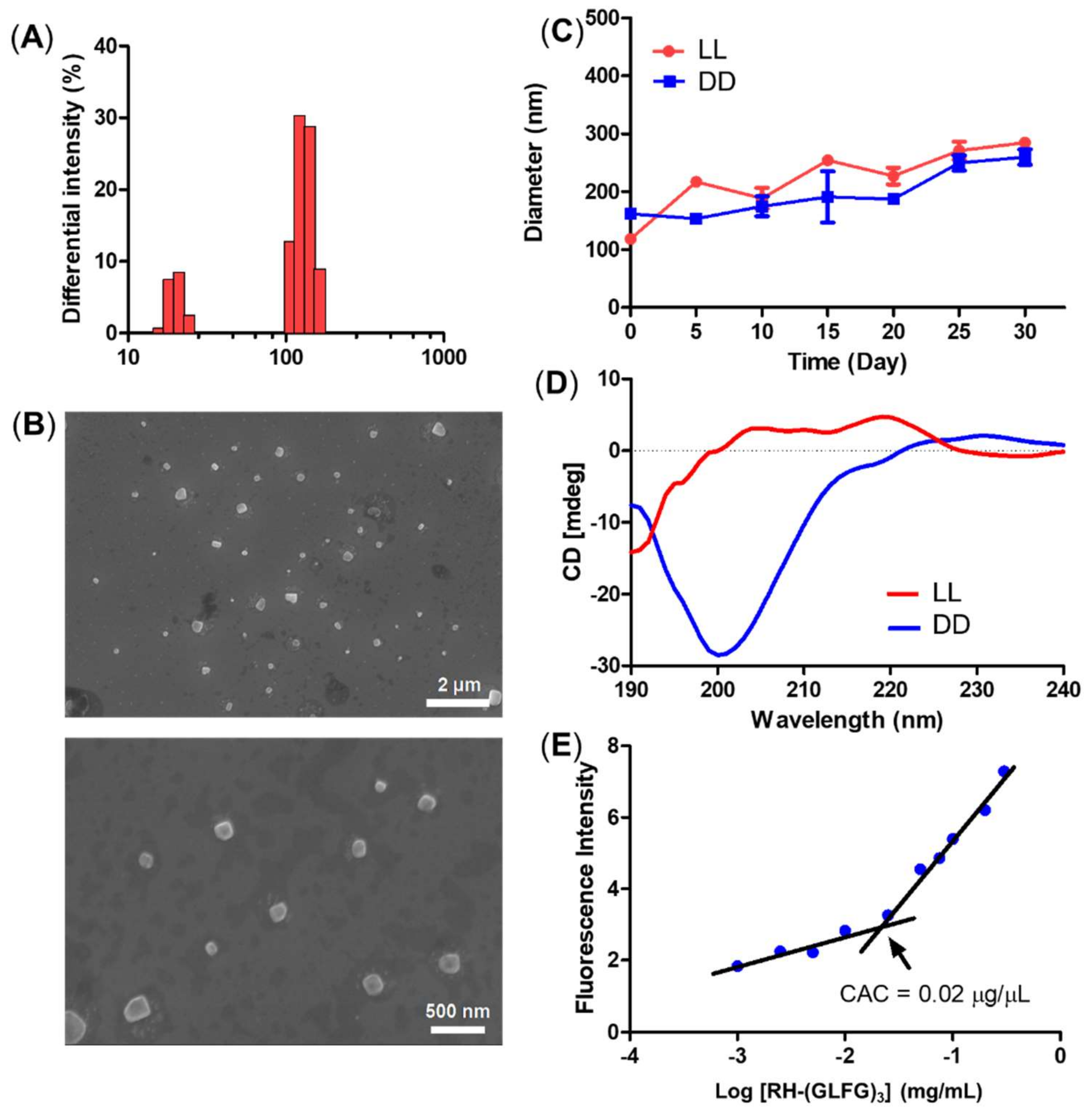
2.4. Determination of Critical Aggregation Concentration
2.5. Cell Culture
2.6. Cytotoxicity of Peptide Nanoparticles
2.7. Quantification of Hemolysis
2.8. Cellular Uptake Assay
2.9. Preparation of Doxorubicin-Loaded RH-(GFLG)3 Nanoparticles
2.10. Quantitation of Doxorubicin in Nanoparticles
2.11. Enzyme-Responsiveness Test
2.12. In Vitro Drug Release
2.13. Cellular Uptake of Dox-Loaded RH-(GFLG)3 Nanoparticles
2.14. Anticancer Effect of Doxorubicin-Loaded RH-(GFLG)3 Peptide Nanoparticles
2.15. In vivo Anticancer Capability in a Zebrafish Model
3. Results and Discussion
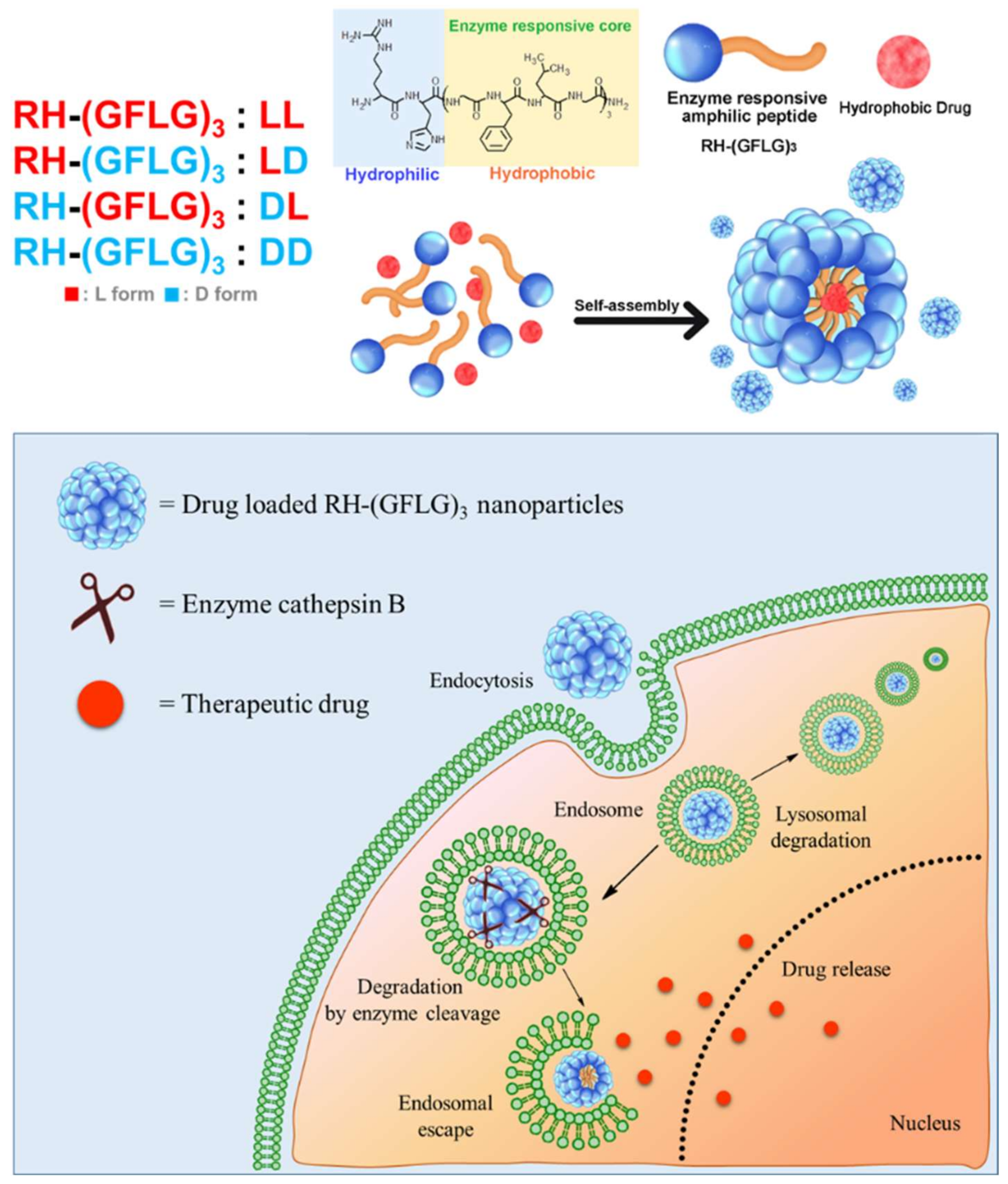
3.1. Preparation and Characterization of Peptide Nanoparticles
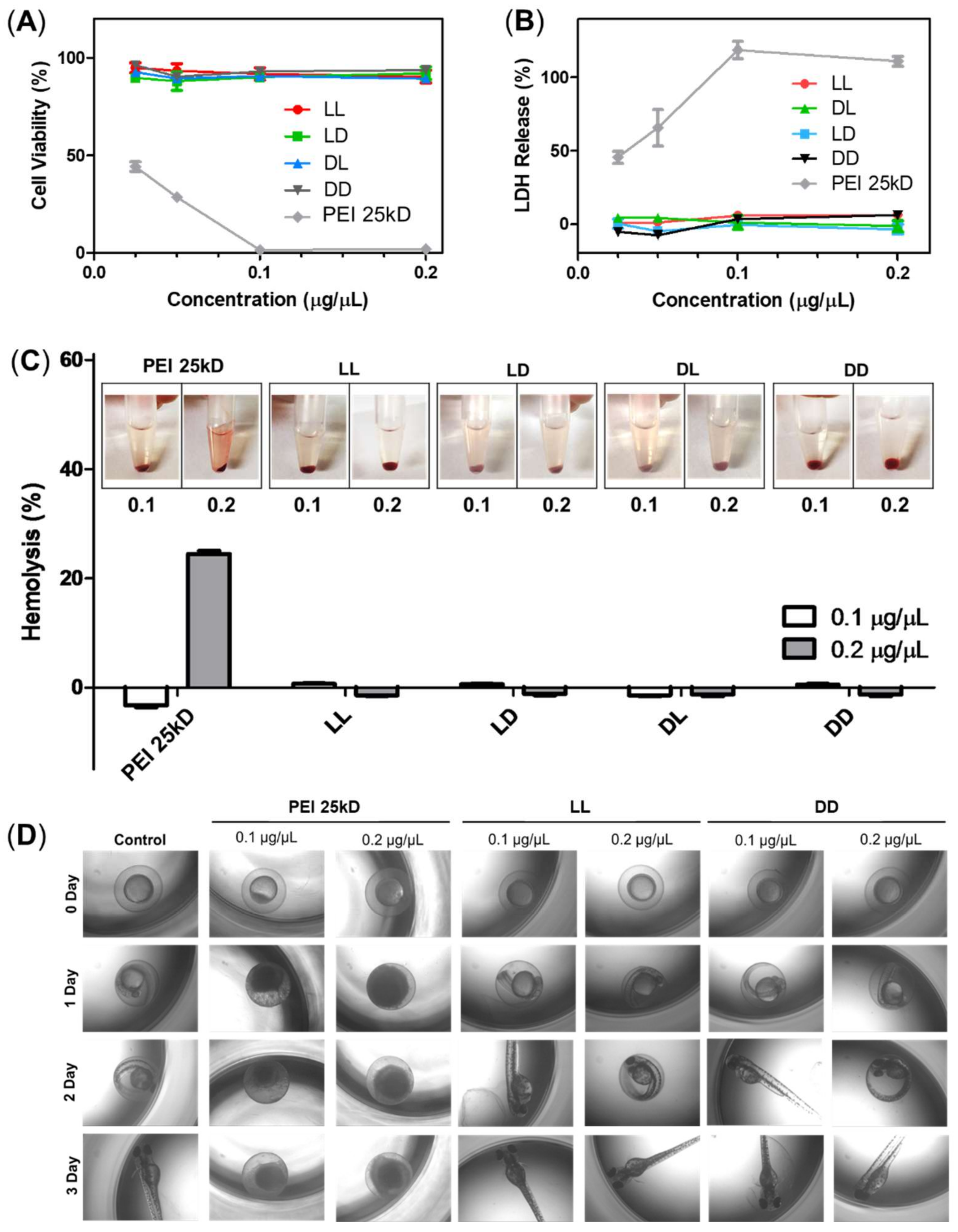
3.2. Cytotoxicity of RH-(GFLG)3 Peptide Nanoparticles
3.3. Preparation of Doxorubicin-Loaded RH-(GFLG)3 Nanoparticles
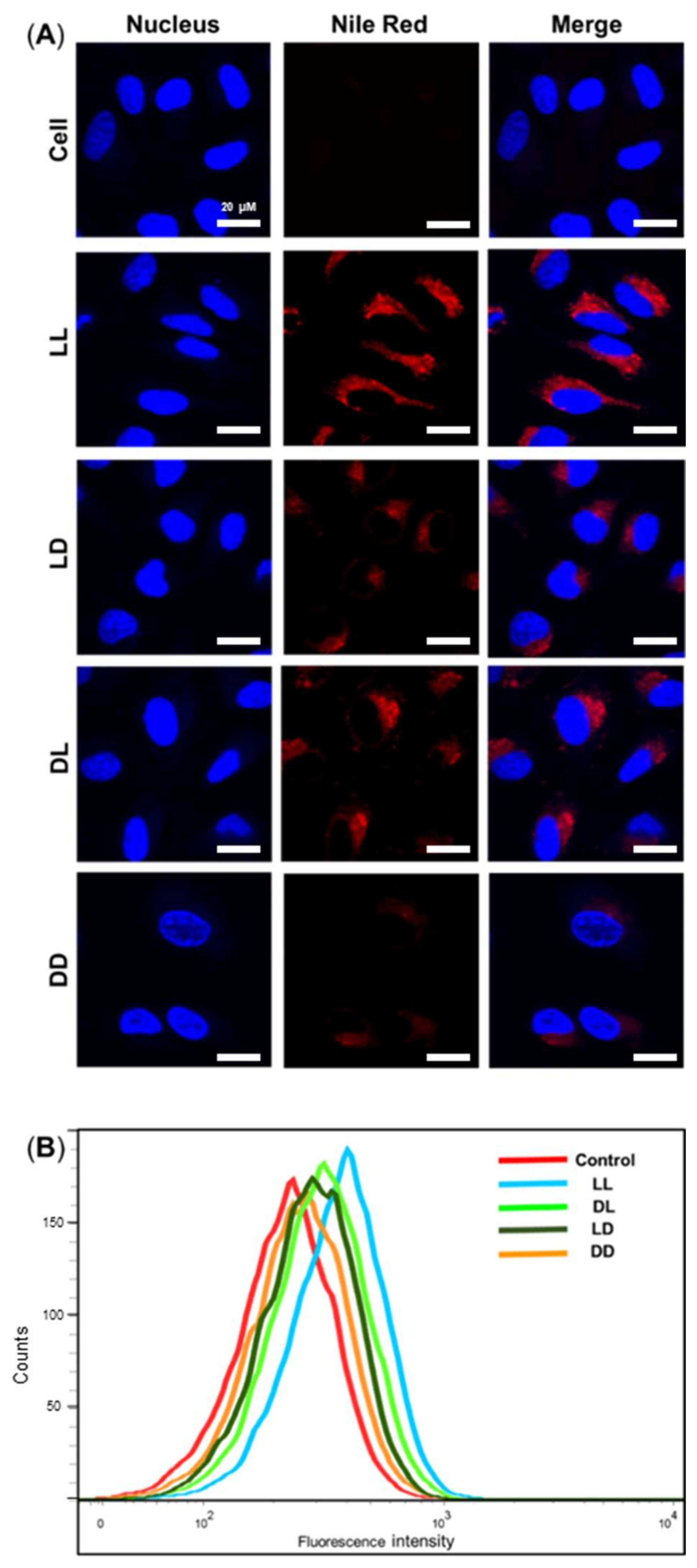
3.4. Cellular Uptake Assay
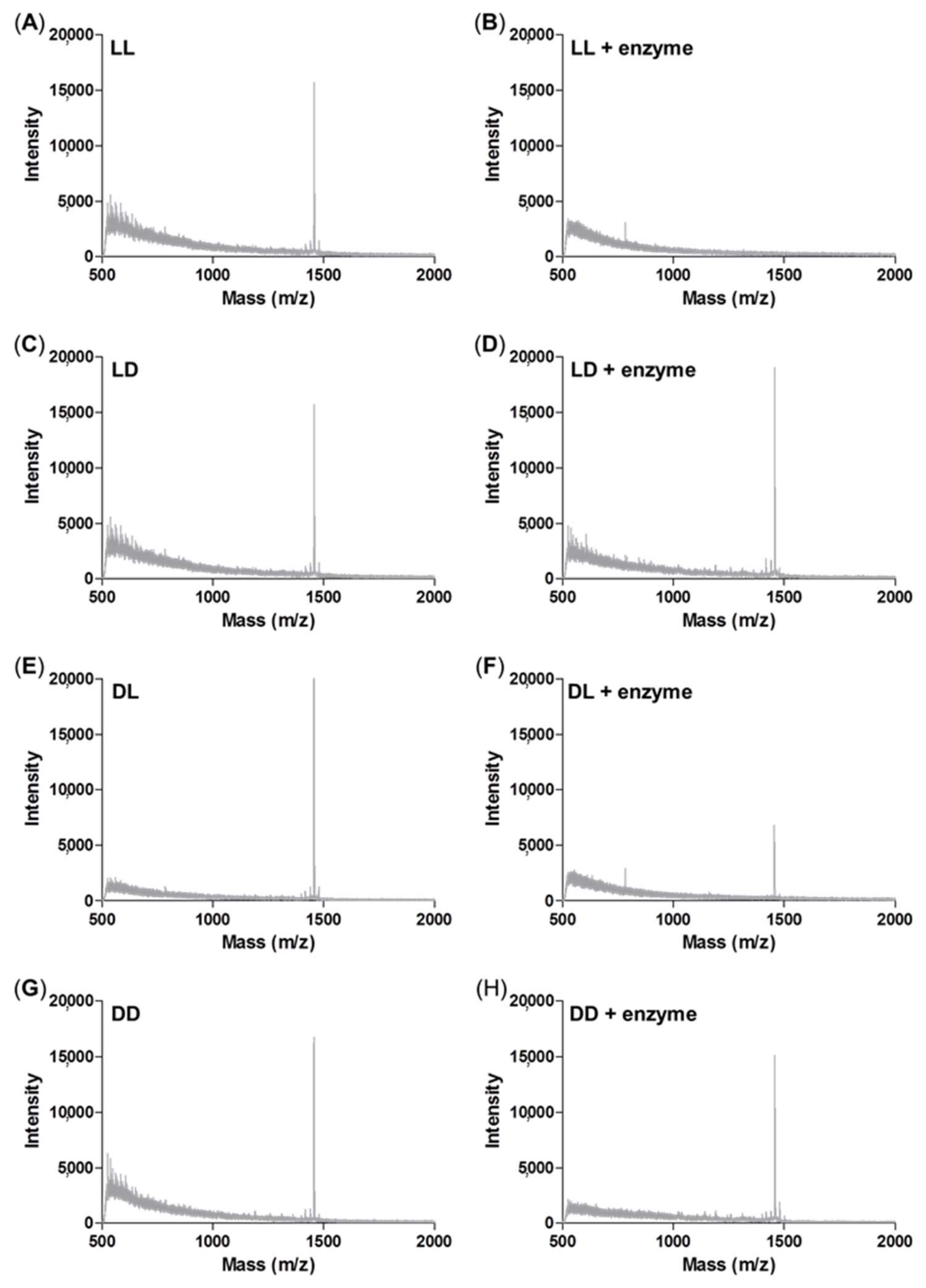
3.5. Enzyme Sensitivity Test
3.6. In Vitro Drug Release
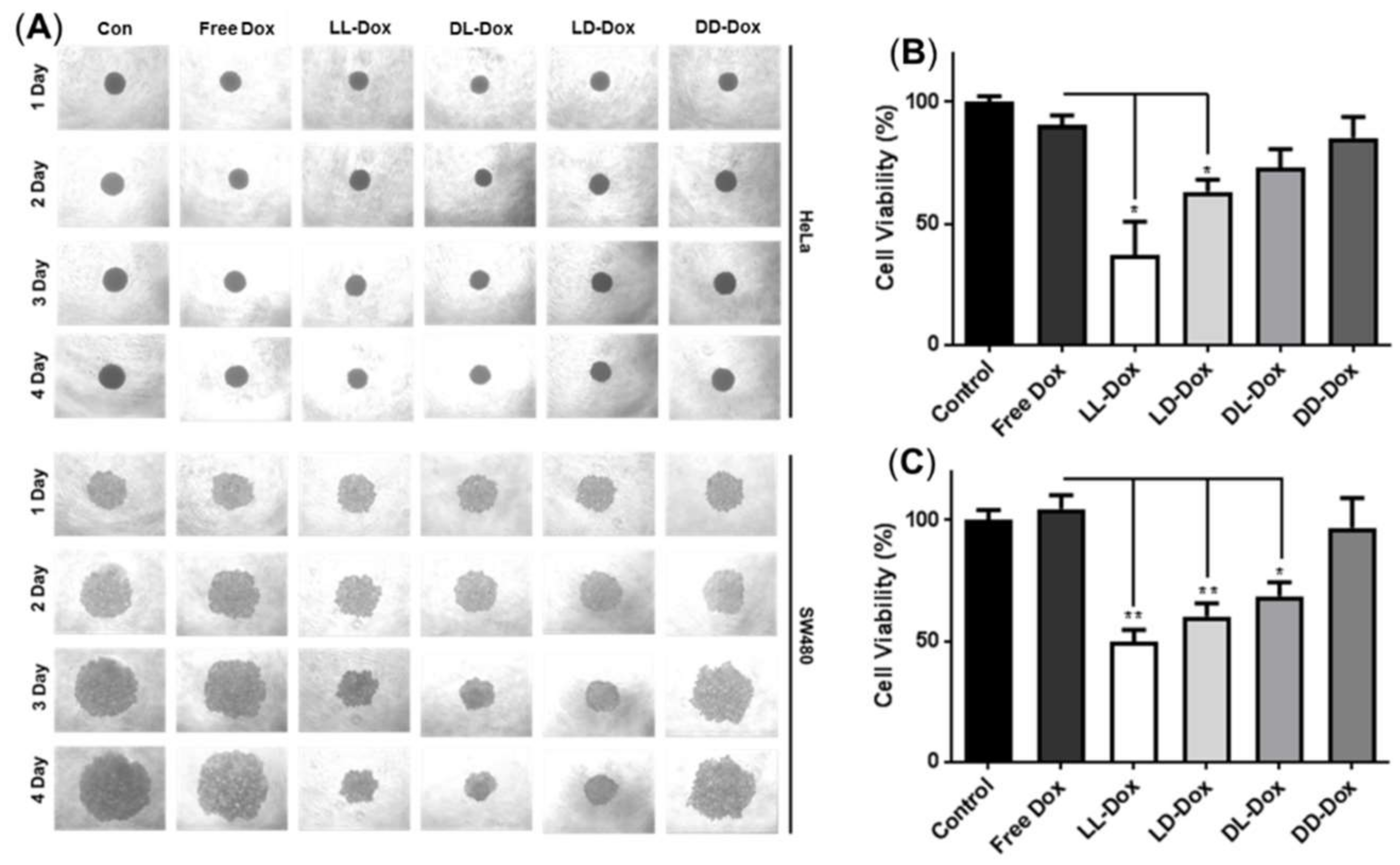
3.7. Cellular Uptake of Dox-Loaded RH-(GFLG)3 Nanoparticles in 3D Spheroids
3.8. Anticancer Effect of Doxorubicin-Loaded RH-(GFLG)3
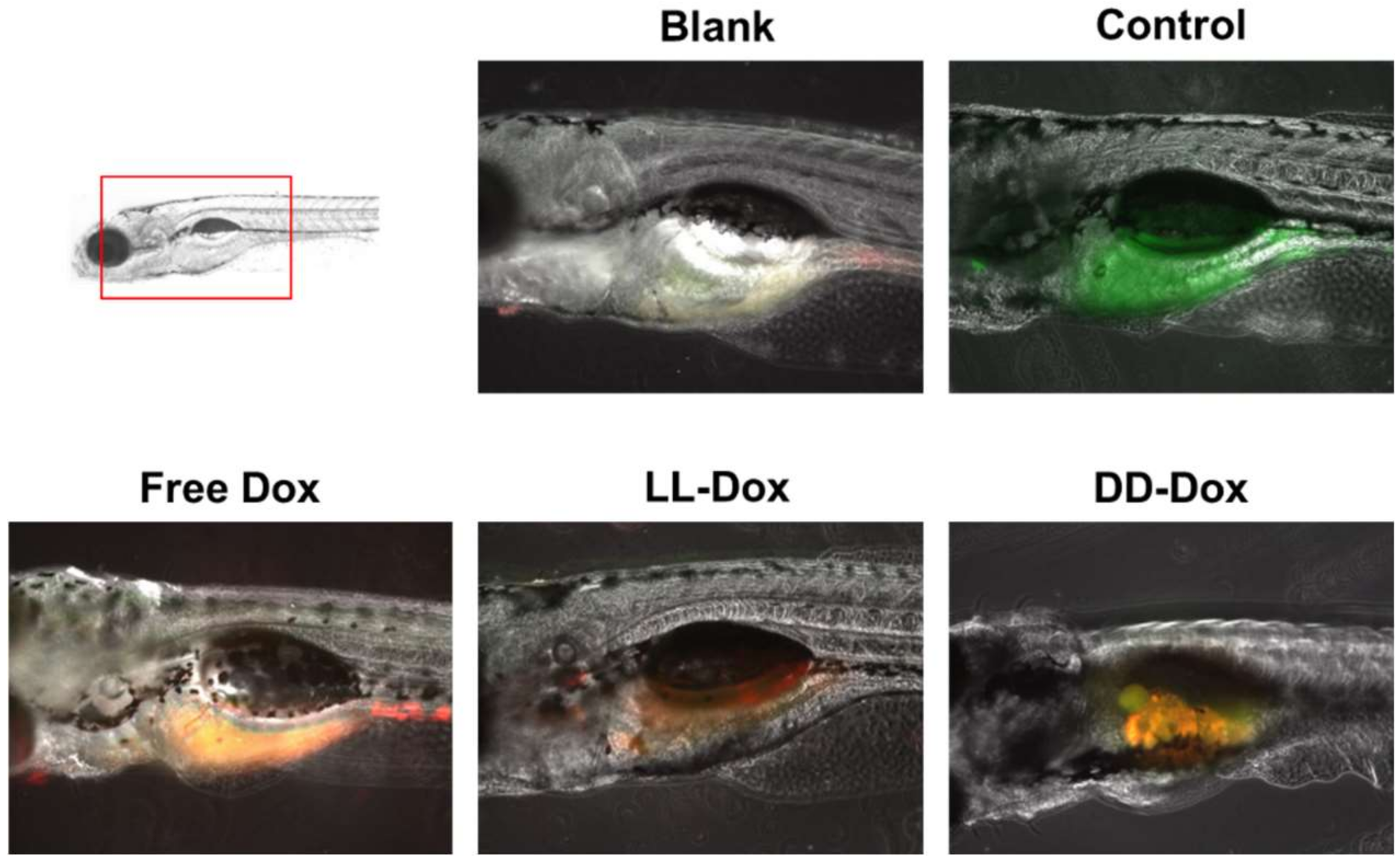
3.9. In Vivo Anticancer Capability in a Zebrafish Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Bruno, B.J.; Miller, G.D.; Lim, C.S. Basics and recent advances in peptide and protein drug delivery. Ther. Deliv. 2013, 4, 1443–1467. [Google Scholar] [CrossRef]
- Patel, A.; Patel, M.; Yang, X.; Mitra, A.K. Recent advances in protein and Peptide drug delivery: A special emphasis on polymeric nanoparticles. Protein Pept. Lett. 2014, 21, 1102–1120. [Google Scholar] [CrossRef] [PubMed]
- Rad-Malekshahi, M.; Lempsink, L.; Amidi, M.; Hennink, W.E.; Mastrobattista, E. Biomedical Applications of Self-Assembling Peptides. Bioconjug. Chem. 2016, 27, 3–18. [Google Scholar] [CrossRef]
- de la Rica, R.; Matsui, H. Applications of peptide and protein-based materials in bionanotechnology. Chem. Soc. Rev. 2010, 39, 3499–3509. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, R.; Malmsten, M. Delivery systems for antimicrobial peptides. Adv. Colloid Interface Sci. 2017, 242, 17–34. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef]
- Copolovici, D.M.; Langel, K.; Eriste, E.; Langel, U. Cell-penetrating peptides: Design, synthesis, and applications. ACS Nano 2014, 8, 1972–1994. [Google Scholar] [CrossRef]
- Giraud, G.; Stadhouders, R.; Conidi, A.; Dekkers, D.H.; Huylebroeck, D.; Demmers, J.A.; Soler, E.; Grosveld, F.G. NLS-tagging: An alternative strategy to tag nuclear proteins. Nucleic Acids Res. 2014, 42, e163. [Google Scholar] [CrossRef]
- van Gaal, E.V.; Oosting, R.S.; van Eijk, R.; Bakowska, M.; Feyen, D.; Kok, R.J.; Hennink, W.E.; Crommelin, D.J.; Mastrobattista, E. DNA nuclear targeting sequences for non-viral gene delivery. Pharm. Res. 2011, 28, 1707–1722. [Google Scholar] [CrossRef] [PubMed]
- Farsinejad, S.; Gheisary, Z.; Ebrahimi Samani, S.; Alizadeh, A.M. Mitochondrial targeted peptides for cancer therapy. Tumour Biol. 2015, 36, 5715–5725. [Google Scholar] [CrossRef]
- Chuah, J.A.; Matsugami, A.; Hayashi, F.; Numata, K. Self-Assembled Peptide-Based System for Mitochondrial-Targeted Gene Delivery: Functional and Structural Insights. Biomacromolecules 2016, 17, 3547–3557. [Google Scholar] [CrossRef]
- Cui, H.; Webber, M.J.; Stupp, S.I. Self-assembly of peptide amphiphiles: From molecules to nanostructures to biomaterials. Biopolymers 2010, 94, 1–18. [Google Scholar] [CrossRef]
- Liao, H.S.; Lin, J.; Liu, Y.; Huang, P.; Jin, A.; Chen, X. Self-assembly mechanisms of nanofibers from peptide amphiphiles in solution and on substrate surfaces. Nanoscale 2016, 8, 14814–14820. [Google Scholar] [CrossRef] [PubMed]
- Dehsorkhi, A.; Castelletto, V.; Hamley, I.W. Self-assembling amphiphilic peptides. J. Pept. Sci. 2014, 20, 453–467. [Google Scholar] [CrossRef]
- Habibi, N.; Kamaly, N.; Memic, A.; Shafiee, H. Self-assembled peptide-based nanostructures: Smart nanomaterials toward targeted drug delivery. Nano Today 2016, 11, 41–60. [Google Scholar] [CrossRef]
- Sun, L.; Zheng, C.; Webster, T.J. Self-assembled peptide nanomaterials for biomedical applications: Promises and pitfalls. Int. J. Nanomed. 2017, 12, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.Y.; Huang, Y.J.; Tan, C.; Presley, A.D.; Chang, J.; Xu, T. Amphiphilic peptide-polymer conjugates based on the coiled-coil helix bundle. Biomacromolecules 2010, 11, 1443–1452. [Google Scholar] [CrossRef]
- Gudlur, S.; Sukthankar, P.; Gao, J.; Avila, L.A.; Hiromasa, Y.; Chen, J.; Iwamoto, T.; Tomich, J.M. Peptide nanovesicles formed by the self-assembly of branched amphiphilic peptides. PLoS ONE 2012, 7, e45374. [Google Scholar] [CrossRef] [PubMed]
- Ryu, D.W.; Kim, H.A.; Ryu, J.H.; Lee, D.Y.; Lee, M. Amphiphilic peptides with arginine and valine residues as siRNA carriers. J. Cell. Biochem. 2012, 113, 619–628. [Google Scholar] [CrossRef]
- Trent, A.; Ulery, B.D.; Black, M.J.; Barrett, J.C.; Liang, S.; Kostenko, Y.; David, N.A.; Tirrell, M.V. Peptide amphiphile micelles self-adjuvant group A streptococcal vaccination. AAPS J. 2015, 17, 380–388. [Google Scholar] [CrossRef]
- Zarghami Dehaghani, M.; Yousefi, F.; Bagheri, B.; Seidi, F.; Hamed Mashhadzadeh, A.; Rabiee, N.; Zarrintaj, P.; Mostafavi, E.; Saeb, M.R.; Kim, Y.C. alpha-Helical Antimicrobial Peptide Encapsulation and Release from Boron Nitride Nanotubes: A Computational Study. Int. J. Nanomed. 2021, 16, 4277–4288. [Google Scholar] [CrossRef] [PubMed]
- Kalepu, S.; Nekkanti, V. Insoluble drug delivery strategies: Review of recent advances and business prospects. Acta Pharm. Sin. B 2015, 5, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Wais, U.; Jackson, A.W.; He, T.; Zhang, H. Nanoformulation and encapsulation approaches for poorly water-soluble drug nanoparticles. Nanoscale 2016, 8, 1746–1769. [Google Scholar] [CrossRef]
- Guo, S.; Huang, L. Nanoparticles containing insoluble drug for cancer therapy. Biotechnol. Adv. 2014, 32, 778–788. [Google Scholar] [CrossRef]
- Leite, D.M.; Barbu, E.; Pilkington, G.J.; Lalatsa, A. Peptide Self-Assemblies for Drug Delivery. Curr. Top. Med. Chem. 2015, 15, 2277–2289. [Google Scholar] [CrossRef][Green Version]
- Song, Z.; Chen, X.; You, X.; Huang, K.; Dhinakar, A.; Gu, Z.; Wu, J. Self-assembly of peptide amphiphiles for drug delivery: The role of peptide primary and secondary structures. Biomater. Sci. 2017, 5, 2369–2380. [Google Scholar] [CrossRef]
- Chow, D.; Nunalee, M.L.; Lim, D.W.; Simnick, A.J.; Chilkoti, A. Peptide-based Biopolymers in Biomedicine and Biotechnology. Mater. Sci. Eng. R Rep. 2008, 62, 125–155. [Google Scholar] [CrossRef]
- Song, S.J.; Lee, S.; Ryu, K.S.; Choi, J.S. Amphiphilic Peptide Nanorods Based on Oligo-Phenylalanine as a Biocompatible Drug Carrier. Bioconjug. Chem. 2017, 28, 2266–2276. [Google Scholar] [CrossRef]
- Hajebi, S.; Rabiee, N.; Bagherzadeh, M.; Ahmadi, S.; Rabiee, M.; Roghani-Mamaqani, H.; Tahriri, M.; Tayebi, L.; Hamblin, M.R. Stimulus-responsive polymeric nanogels as smart drug delivery systems. Acta Biomater. 2019, 92, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rabiee, N.; Ahmadi, S.; Afshari, R.; Khalaji, S.; Rabiee, M.; Bagherzadeh, M.; Fatahi, Y.; Dinarvand, R.; Tahriri, M.; Tayebi, L.; et al. Polymeric Nanoparticles for Nasal Drug Delivery to the Brain: Relevance to Alzheimer’s Disease. Adv. Ther. 2021, 4, 2000076. [Google Scholar] [CrossRef]
- Mahato, R.; Tai, W.; Cheng, K. Prodrugs for improving tumor targetability and efficiency. Adv. Drug Deliv. Rev. 2011, 63, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Cheetham, A.G.; Lock, L.L.; Cui, H. Cellular uptake and cytotoxicity of drug-peptide conjugates regulated by conjugation site. Bioconjug. Chem. 2013, 24, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824, 68–88. [Google Scholar] [CrossRef]
- Mitrovic, A.; Mirkovic, B.; Sosic, I.; Gobec, S.; Kos, J. Inhibition of endopeptidase and exopeptidase activity of cathepsin B impairs extracellular matrix degradation and tumour invasion. Biol. Chem. 2016, 397, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Sloane, B.F. Cathepsin B: Multiple roles in cancer. Proteom. Clin. Appl. 2014, 8, 427–437. [Google Scholar] [CrossRef]
- Durzynska, J.; Przysiecka, L.; Nawrot, R.; Barylski, J.; Nowicki, G.; Warowicka, A.; Musidlak, O.; Gozdzicka-Jozefiak, A. Viral and other cell-penetrating peptides as vectors of therapeutic agents in medicine. J. Pharmacol. Exp. Ther. 2015, 354, 32–42. [Google Scholar] [CrossRef]
- Chang, K.L.; Higuchi, Y.; Kawakami, S.; Yamashita, F.; Hashida, M. Efficient gene transfection by histidine-modified chitosan through enhancement of endosomal escape. Bioconjug. Chem. 2010, 21, 1087–1095. [Google Scholar] [CrossRef]
- Ashwanikumar, N.; Kumar, N.A.; Saneesh Babu, P.S.; Sivakumar, K.C.; Vadakkan, M.V.; Nair, P.; Hema Saranya, I.; Asha Nair, S.; Vinod Kumar, G.S. Self-assembling peptide nanofibers containing phenylalanine for the controlled release of 5-fluorouracil. Int. J. Nanomed. 2016, 11, 5583–5594. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, C.Y.; Gao, X.S.; You, X.Y.; Yao, C. Hg2+-selective ratiometric and “off-on” chemosensor based on the azadiene-pyrene derivative. Org. Lett. 2010, 12, 2566–2569. [Google Scholar] [CrossRef]
- Yu, G.S.; Bae, Y.M.; Choi, H.; Kong, B.; Choi, I.S.; Choi, J.S. Synthesis of PAMAM dendrimer derivatives with enhanced buffering capacity and remarkable gene transfection efficiency. Bioconjug. Chem. 2011, 22, 1046–1055. [Google Scholar] [CrossRef]
- Deng, X.; Xu, X.; Lai, Y.; He, B.; Gu, Z. Novel nanoparticles generated by polymeric amphiphiles with pi-pi conjugated small molecules for anti-tumor drug delivery. J. Biomed. Nanotechnol. 2013, 9, 1336–1344. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Lee, S.; Lee, Y.; Choi, J.S. Enzyme-responsive destabilization of stabilized plasmid-lipid nanoparticles as an efficient gene delivery. Eur. J. Pharm. Sci. 2016, 91, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Pan, D.; Luo, K.; She, W.; Guo, C.; Yang, Y.; Gu, Z. Peptide dendrimer-Doxorubicin conjugate-based nanoparticles as an enzyme-responsive drug delivery system for cancer therapy. Adv. Healthc. Mater. 2014, 3, 1299–1308. [Google Scholar] [CrossRef]
- Li, F.; Mahato, R.I. Bioconjugate Therapeutics: Current Progress and Future Perspective. Mol. Pharm. 2017, 14, 1321–1324. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.J.; Shao, L.H.; Li, Y. Cathepsin B-cleavable doxorubicin prodrugs for targeted cancer therapy (Review). Int. J. Oncol. 2013, 42, 373–383. [Google Scholar] [CrossRef]
- Luo, Z.; Zhao, X.; Zhang, S. Structural dynamic of a self-assembling peptide d-EAK16 made of only D-amino acids. PLoS ONE 2008, 3, e2364. [Google Scholar] [CrossRef] [PubMed]
- Veselinovic, J.B.; Kocic, G.M.; Pavic, A.; Nikodinovic-Runic, J.; Senerovic, L.; Nikolic, G.M.; Veselinovic, A.M. Selected 4-phenyl hydroxycoumarins: In vitro cytotoxicity, teratogenic effect on zebrafish (Danio rerio) embryos and molecular docking study. Chem. Biol. Interact. 2015, 231, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.R.; Guo, H. Evaluation of cytotoxicity, genotoxicity and embryotoxicity of insecticide propoxur using flounder gill (FG) cells and zebrafish embryos. Toxicol. In Vitro 2014, 28, 340–353. [Google Scholar] [CrossRef]
- Bian, B.; Mongrain, S.; Cagnol, S.; Langlois, M.J.; Boulanger, J.; Bernatchez, G.; Carrier, J.C.; Boudreau, F.; Rivard, N. Cathepsin B promotes colorectal tumorigenesis, cell invasion, and metastasis. Mol. Carcinog. 2016, 55, 671–687. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.J.; Choi, J.S. Enzyme-Responsive Amphiphilic Peptide Nanoparticles for Biocompatible and Efficient Drug Delivery. Pharmaceutics 2022, 14, 143. https://doi.org/10.3390/pharmaceutics14010143
Song SJ, Choi JS. Enzyme-Responsive Amphiphilic Peptide Nanoparticles for Biocompatible and Efficient Drug Delivery. Pharmaceutics. 2022; 14(1):143. https://doi.org/10.3390/pharmaceutics14010143
Chicago/Turabian StyleSong, Su Jeong, and Joon Sig Choi. 2022. "Enzyme-Responsive Amphiphilic Peptide Nanoparticles for Biocompatible and Efficient Drug Delivery" Pharmaceutics 14, no. 1: 143. https://doi.org/10.3390/pharmaceutics14010143
APA StyleSong, S. J., & Choi, J. S. (2022). Enzyme-Responsive Amphiphilic Peptide Nanoparticles for Biocompatible and Efficient Drug Delivery. Pharmaceutics, 14(1), 143. https://doi.org/10.3390/pharmaceutics14010143





