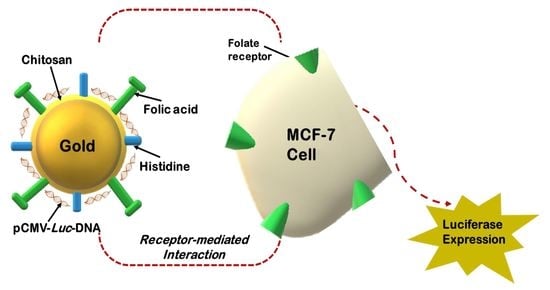
Histidine-Tagged Folate-Targeted Gold Nanoparticles for Enhanced Transgene Expression in Breast Cancer Cells In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Gold Nanoparticle Synthesis
2.3. Synthesis of Chitosan Functionalized Gold Nanoparticles
2.4. Synthesis of Chitosan and Histidine Functionalized Gold Nanoparticles
2.5. Synthesis of Chitosan, Histidine and Folate Functionalized Gold Nanoparticles
2.6. Preparation of Nanocomplexes
2.7. Nanoparticle and Nanocomplex Characterization
2.8. Band Shift Assay
2.9. Ethidium Bromide Intercalation Assay
2.10. Nuclease Protection Assay
2.11. MTT Cell Viability Assay
2.12. Apoptosis Assay
2.13. Luciferase Assay
2.14. Competition Binding Assay
2.15. Statistical Analysis
3. Results
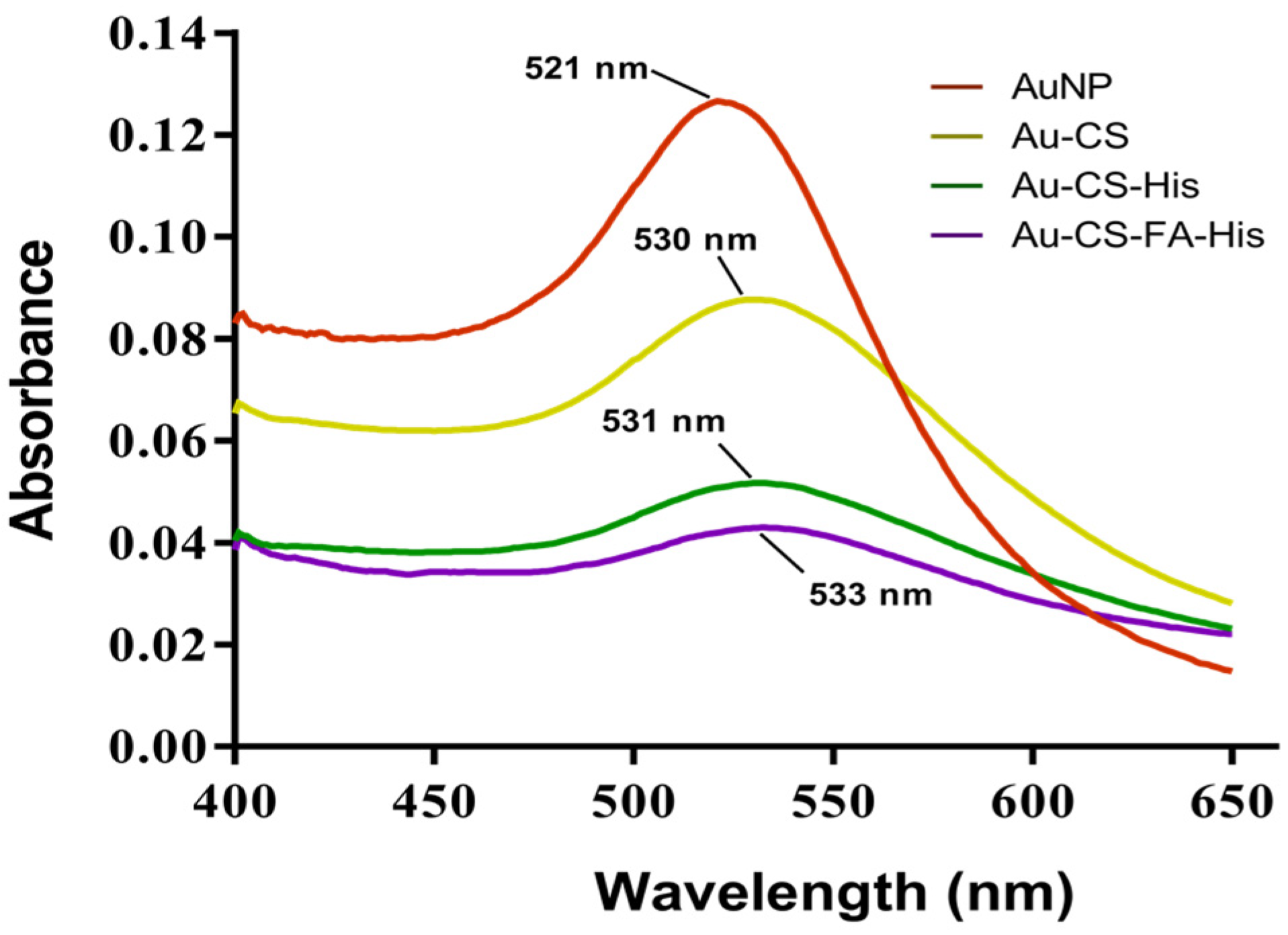
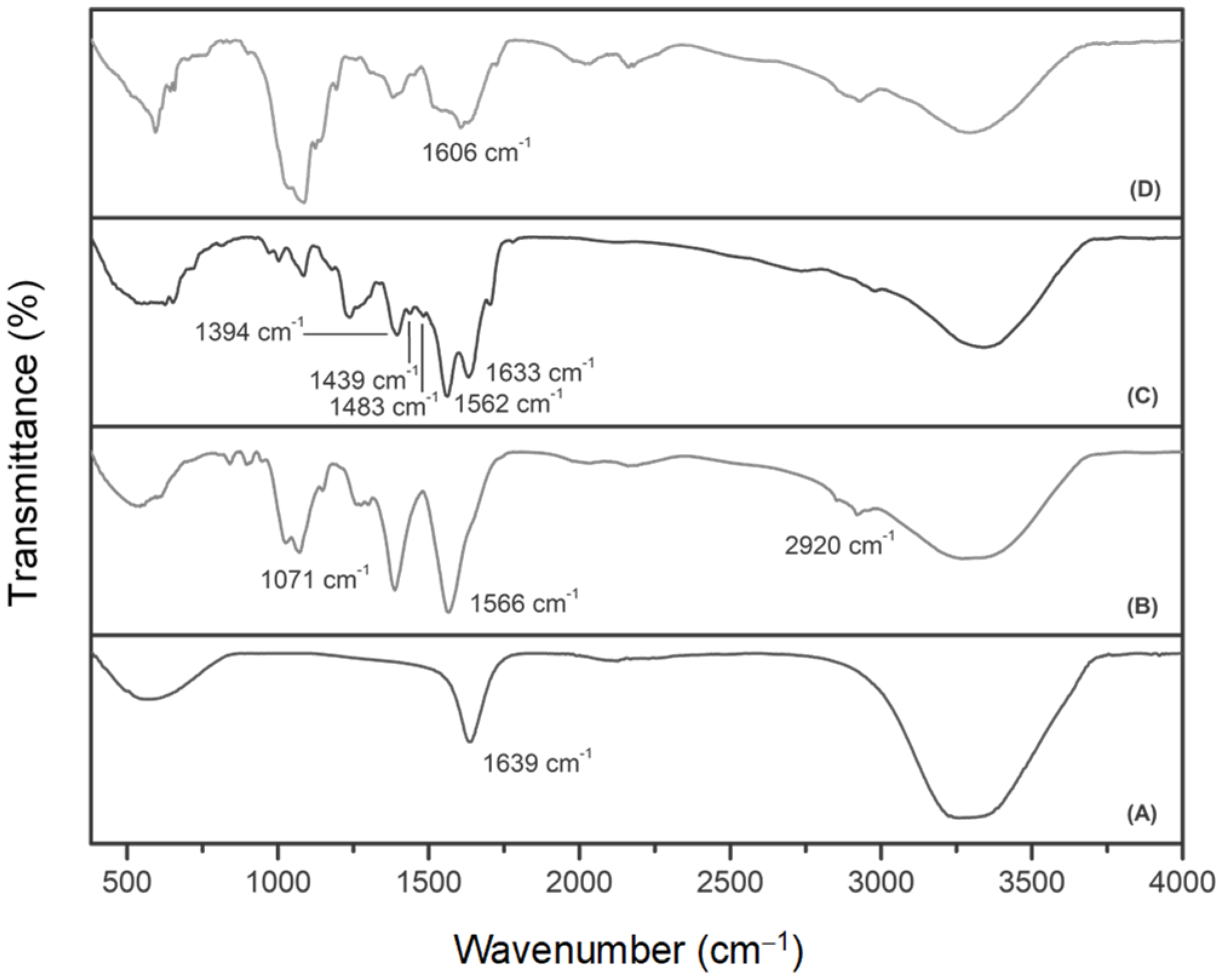
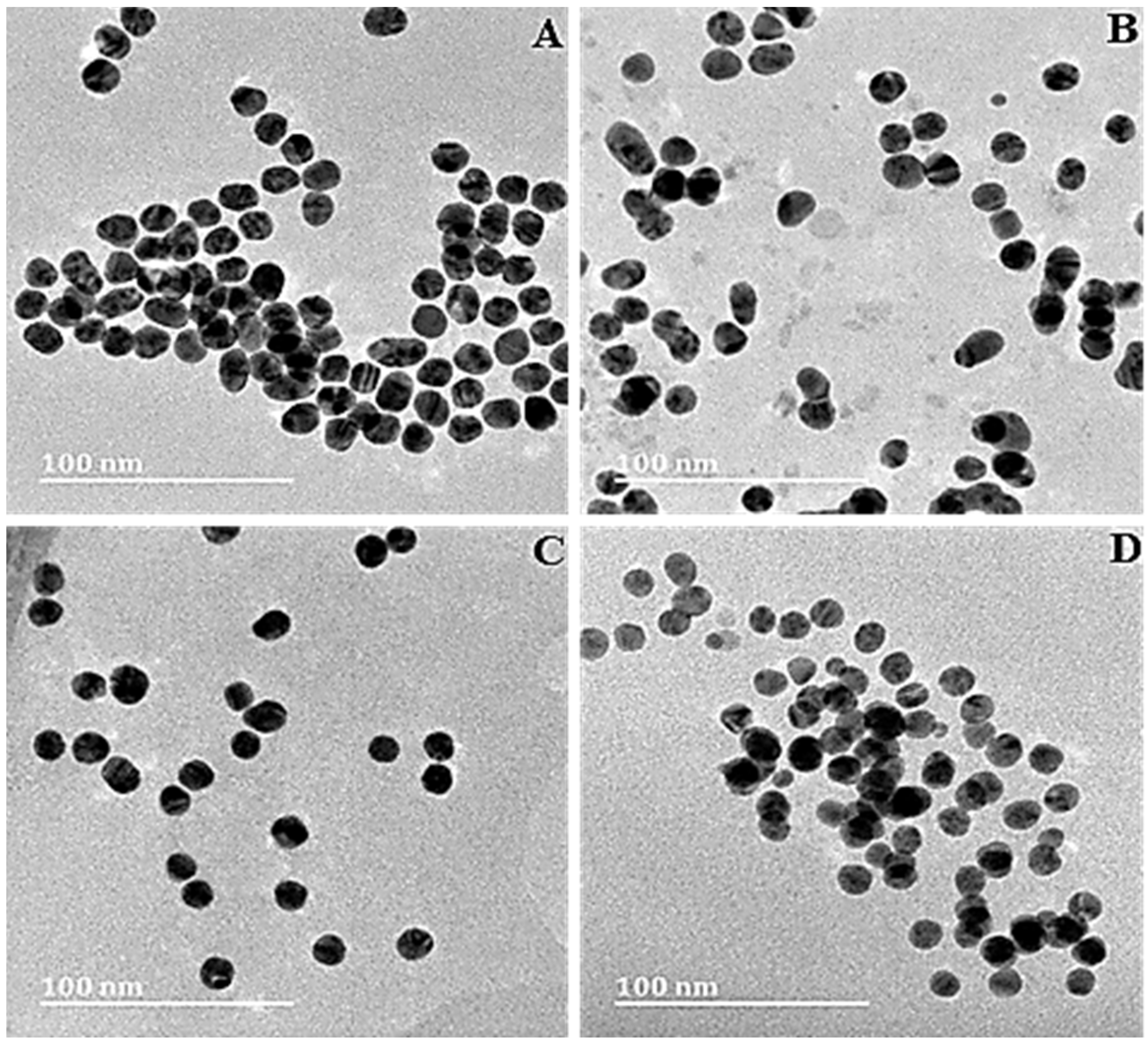
3.1. Nanoparticle Synthesis and Characterization
3.2. Band Shift Assay
3.3. Dye Displacement Assay
3.4. Nuclease Protection Assay
3.5. MTT Cytotoxicity Assay
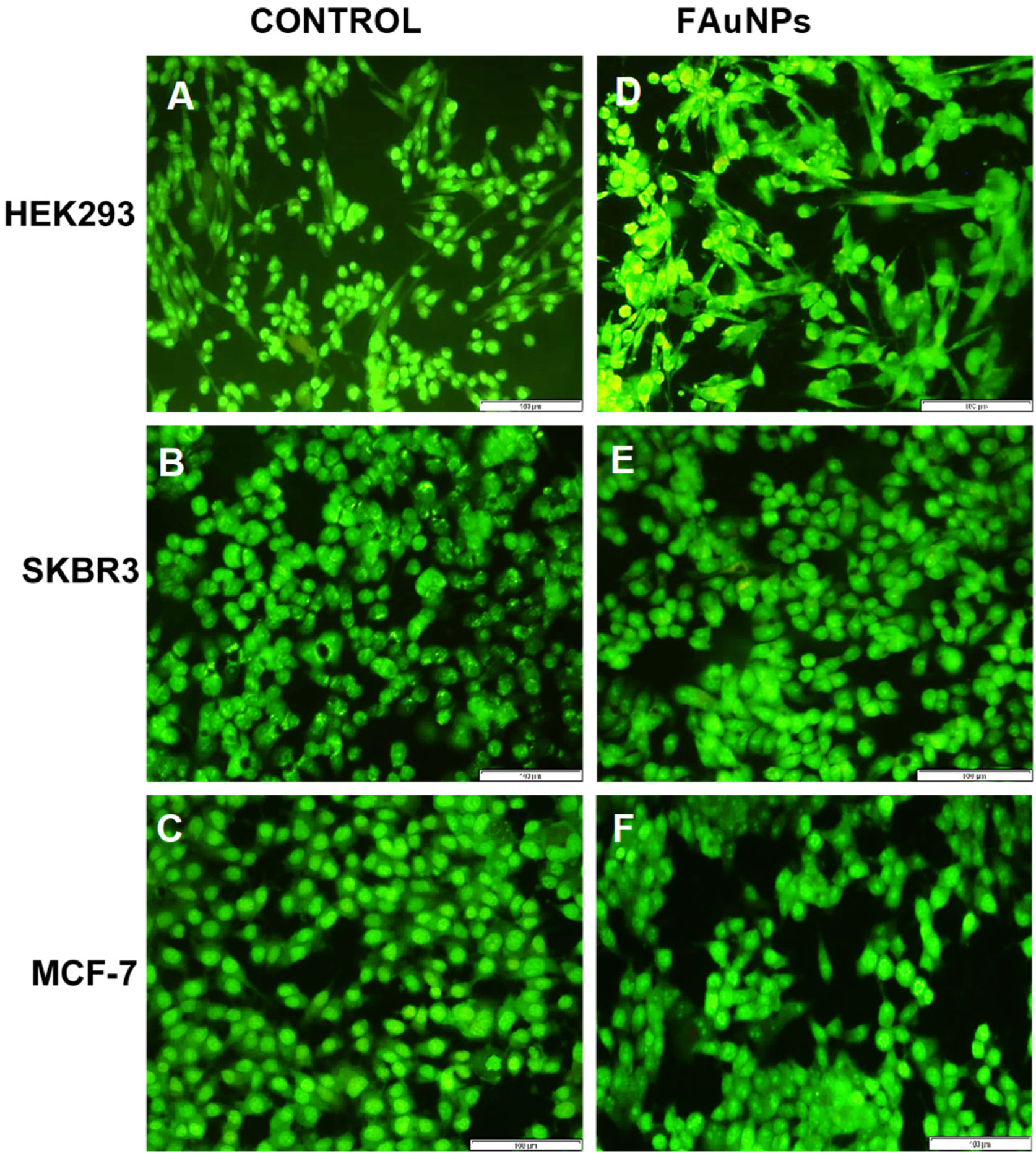
3.6. Apoptosis Assay
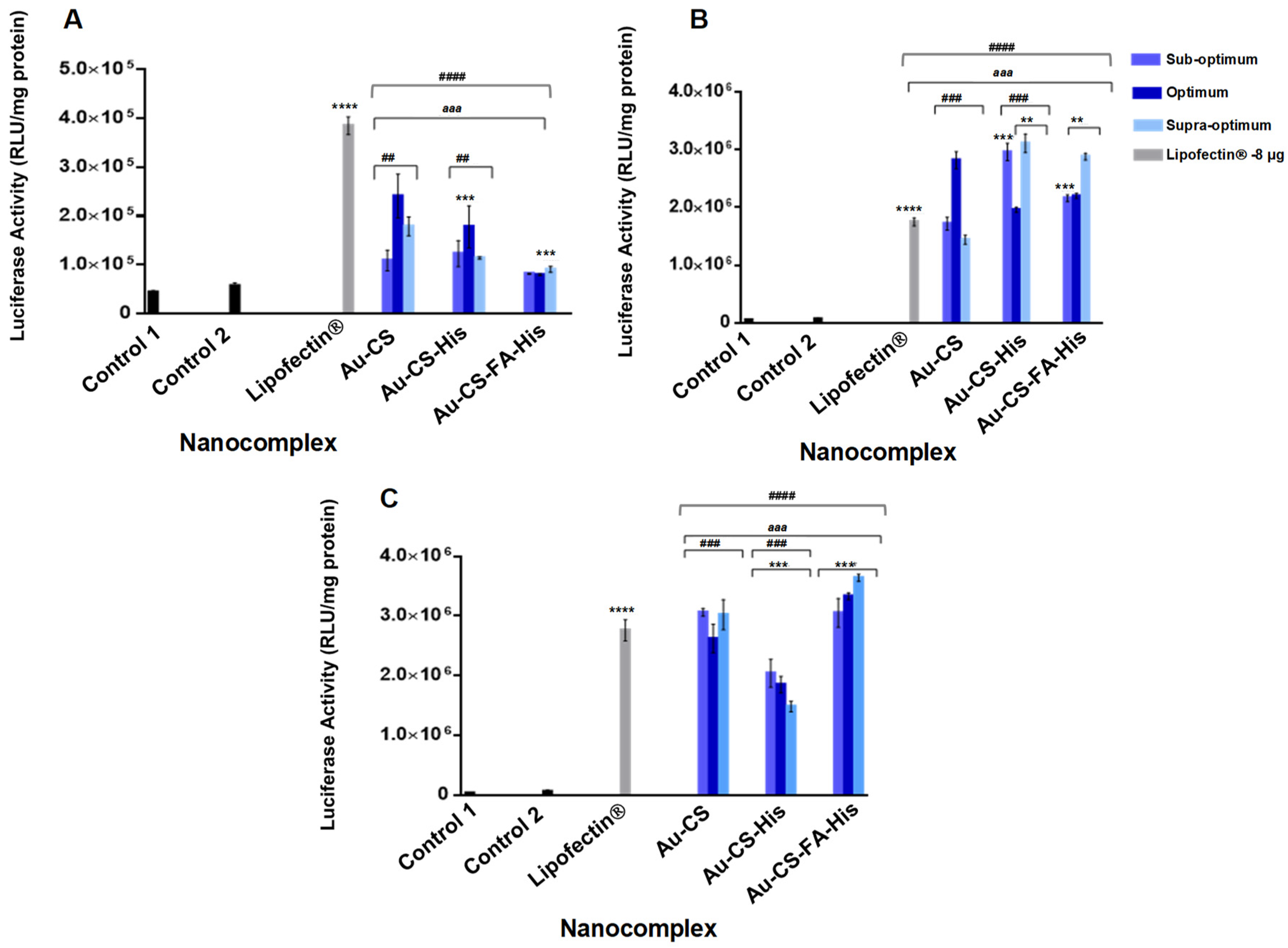
3.7. Transfection Studies
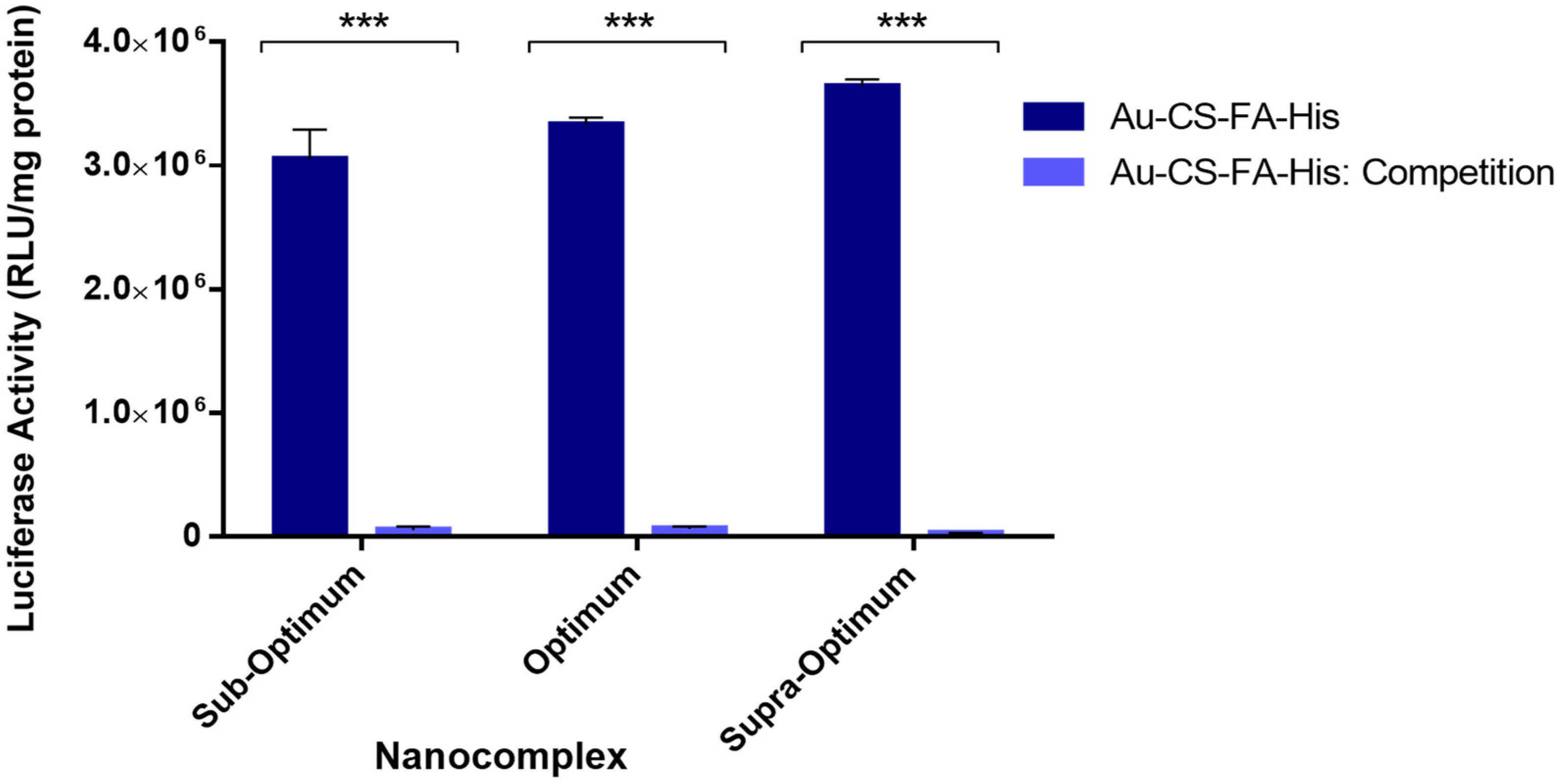
3.8. Receptor Competition Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Islami, F.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global cancer in women: Burden and trends. Cancer Epidemiol. Biomark. Prev. 2017, 26, 444–457. [Google Scholar] [CrossRef] [Green Version]
- Joko-Fru, W.Y.; Miranda-Filho, A.; Soerjomataram, I.; Egue, M.; Akele-Akpo, M.T.; N’da, G.; Assefa, M.; Buziba, N.; Korir, A.; Kamate, B. Breast cancer survival in sub-Saharan Africa by age, stage at diagnosis and human development index: A population-based registry study. Int. J. Cancer 2020, 146, 1208–1218. [Google Scholar] [CrossRef] [Green Version]
- Cross, D.; Burmester, J.K. Gene therapy for cancer treatment: Past, present and future. Clin. Med. Res. 2006, 4, 218–227. [Google Scholar] [CrossRef] [Green Version]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [Green Version]
- El-Tamer, M.B.; Ward, B.M.; Schifftner, T.; Neumayer, L.; Khuri, S.; Henderson, W. Morbidity and mortality following breast cancer surgery in women: National benchmarks for standards of care. Ann. Surg. 2007, 245, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Lind, P.A.; Wennberg, B.; Gagliardi, G.; Fornander, T. Pulmonary complications following different radiotherapy techniques for breast cancer, and the association to irradiated lung volume and dose. Breast Cancer Res. Treat. 2001, 68, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Langlands, F.; Horgan, K.; Dodwell, D.; Smith, L. Breast cancer subtypes: Response to radiotherapy and potential radiosensitization. Br. J. Radiol. 2013, 86, 20120601. [Google Scholar] [CrossRef]
- Gadi, V.K.; Davidson, N.E. Practical approach to triple-negative breast cancer. J. Oncol. Pract. 2017, 13, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Ariatti, M. A cationic cytofectin with long spacer mediates favourable transfection in transformed human epithelial cells. Int. J. Pharm. 2006, 309, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Dykman, L.; Khlebtsov, N. Gold nanoparticles in biomedical applications: Recent advances and perspectives. Chem. Soc. Rev. 2012, 41, 2256–2282. [Google Scholar] [CrossRef]
- Fernandes Queiroz, M.; Melo, K.R.T.; Sabry, D.A.; Sassaki, G.L.; Rocha, H.A.O. Does the use of chitosan contribute to oxalate kidney stone formation? Mar. Drugs 2015, 13, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, A.; Pawlowska, R.; Jedrzejczyk, D.; Chworos, A. Gold nanoparticles in conjunction with nucleic acids as a modern molecular system for cellular delivery. Molecules 2020, 25, 204. [Google Scholar] [CrossRef] [Green Version]
- Padayachee, J.; Daniels, A.; Balgobind, A.; Ariatti, M.; Singh, M. Her2/neu and MYC gene silencing in breast cancer: Therapeutic potential and advancement in nonviral nanocarrier systems. Nanomedicine 2020, 15, 1437–1452. [Google Scholar] [CrossRef]
- Kalaivani, R.; Maruthupandy, M.; Muneeswaran, T.; Beevi, A.H.; Anand, M.; Ramakritinan, C.; Kumaraguru, A. Synthesis of chitosan mediated silver nanoparticles (Ag NPs) for potential antimicrobial applications. Front. Lab. Med. 2018, 2, 30–35. [Google Scholar] [CrossRef]
- Leedy, M.R.; Martin, H.J.; Norowski, P.A.; Jennings, J.A.; Haggard, W.O.; Bumgardner, J.D. Use of chitosan as a bioactive implant coating for bone-implant applications. In Chitosan for Biomaterials II; Jayakumar, R., Prabaharan, M., Muzzarelli, R., Eds.; Springer: Cham, Switzerland, 2011; Volume 244, pp. 129–165. [Google Scholar]
- Mansouri, S.; Lavigne, P.; Corsi, K.; Benderdour, M.; Beaumont, E.; Fernandes, J.C. Chitosan-DNA nanoparticles as non-viral vectors in gene therapy: Strategies to improve transfection efficacy. Eur. J. Pharm. Biopharm. 2004, 57, 1–8. [Google Scholar] [CrossRef]
- Nimesh, S.; Thibault, M.M.; Lavertu, M.; Buschmann, M.D. Enhanced gene delivery mediated by low molecular weight chitosan/DNA complexes: Effect of pH and serum. Mol. Biotechnol. 2010, 46, 182–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, S.-M.; Du, Q.-S.; Meng, J.-Z.; Pang, Z.-W.; Huang, R.-B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 2013, 7, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mrozek, A.; Karolak-Wojciechowska, J.; Kieć-Kononowicz, K. Five-membered heterocycles. Part III. Aromaticity of 1, 3-imidazole in 5+n hetero-bicyclic molecules. J. Mol. Struct. 2003, 655, 397–403. [Google Scholar] [CrossRef]
- Marti, E.M.; Methivier, C.; Dubot, P.; Pradier, C. Adsorption of (S)-histidine on Cu (110) and oxygen-covered Cu (110), a combined Fourier transform reflection absorption infrared spectroscopy and force field calculation study. J. Phys. Chem. B 2003, 107, 10785–10792. [Google Scholar] [CrossRef]
- Sreevalsa, V.; Jeeju, P.; Augustine, M.S.; Anilkumar, K.; Jayalekshmi, S. L-Histidine-modified biocompatible zinc oxide nanocrystals. J. Exp. Nanosci. 2013, 8, 937–946. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Li, S.; Su, Y.; Yi, X.; Zheng, A.; Deng, F. Interaction between histidine and Zn (II) metal ions over a wide pH as revealed by solid-state NMR spectroscopy and DFT calculations. J. Phys. Chem. B 2013, 117, 8954–8965. [Google Scholar] [CrossRef]
- Habib, S.; Singh, M.; Ariatti, M. Glycosylated liposomes with proton sponge capacity: Novel hepatocyte-specific gene carriers. Curr. Drug Deliv. 2013, 10, 685–695. [Google Scholar] [CrossRef]
- Nundkumar, N.; Singh, S.; Singh, M. Amino acid functionalized hydrotalcites for gene silencing. J. Nanosci. Nanotechnol. 2020, 20, 3387–3397. [Google Scholar] [CrossRef] [PubMed]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Hirsjarvi, S.; Passirani, C.; Benoit, J.-P. Passive and active tumour targeting with nanocarriers. Curr. Drug Discov. Technol. 2011, 8, 188–196. [Google Scholar] [CrossRef]
- Choi, C.H.J.; Alabi, C.A.; Webster, P.; Davis, M.E. Mechanism of active targeting in solid tumors with transferrin-containing gold nanoparticles. Proc. Natl. Acad. Sci. USA 2010, 107, 1235–1240. [Google Scholar] [CrossRef] [Green Version]
- Shmeeda, H.; Mak, L.; Tzemach, D.; Astrahan, P.; Tarshish, M.; Gabizon, A. Intracellular uptake and intracavitary targeting of folate-conjugated liposomes in a mouse lymphoma model with up-regulated folate receptors. Mol. Cancer Ther. 2006, 5, 818–824. [Google Scholar] [CrossRef] [Green Version]
- Mahapatro, A.; Singh, D.K. Biodegradable nanoparticles are excellent vehicle for site directed in-vivo delivery of drugs and vaccines. J. Nanobiotechnol. 2011, 9, 55. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Tai, H.C.; Xue, W.; Lee, L.J.; Lee, R.J. Receptor-targeted nanocarriers for therapeutic delivery to cancer. Mol. Membr. Biol. 2010, 27, 286–298. [Google Scholar] [CrossRef] [Green Version]
- Samadian, H.; Hosseini-Nami, S.; Kamrava, S.K.; Ghaznavi, H.; Shakeri-Zadeh, A. Folate-conjugated gold nano-particle as a new nanoplatform for targeted cancer therapy. J. Cancer Res. Clin. Oncol. 2016, 142, 2217–2229. [Google Scholar] [CrossRef] [PubMed]
- Turkevich, J.; Stevenson, P.; Hiller, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Akinyelu, J.; Singh, M. Folate-tagged chitosan-functionalized gold nanoparticles for enhanced delivery of 5-fluorouracil to cancer cells. Appl. Nanosci. 2019, 9, 7–17. [Google Scholar] [CrossRef]
- Guan, J.; Jiang, L.; Li, J.; Yang, W. pH-dependent aggregation of histidine-functionalized Au nanoparticles induced by Fe3+ ions. J. Phys. Chem. C 2008, 112, 3267–3271. [Google Scholar] [CrossRef]
- Singh, M. Assessing nucleic acid: Cationic nanoparticle interaction for gene delivery. In Bio-Carrier Vectors; Narayanan, K., Ed.; Springer: New York, NY, USA, 2021; Volume 2211, pp. 43–55. [Google Scholar]
- Huang, X.; Jain, P.; El-Sayed, I.; El-Sayed, M. Gold nanoparticles: Interesting optical properties and recent applications in cancer diagnostics and therapy. Nanomedicine 2007, 2, 681–693. [Google Scholar] [CrossRef] [Green Version]
- Amendola, V.; Meneghetti, M. Size evaluation of gold nanoparticles by UV−vis spectroscopy. J. Phys. Chem. C 2009, 113, 4277–4285. [Google Scholar] [CrossRef]
- Chen, X.; Jensen, L. Understanding the shape effect on the plasmonic response of small ligand coated nanoparticles. J. Opt. 2016, 18, 074009. [Google Scholar] [CrossRef]
- Wulandari, P.; Li, X.; Tamada, K.; Hara, M. Conformational study of citrates adsorbed on gold nanoparticles using Fourier transform infrared spectroscopy. J. Nonlinear Opt. Phys. Mater. 2008, 17, 185–192. [Google Scholar] [CrossRef]
- Wulandari, P.; Nagahiro, T.; Fukada, N.; Kimura, Y.; Niwano, M.; Tamada, K. Characterization of citrates on gold and silver nanoparticles. J. Colloid Interf. Sci. 2015, 438, 244–248. [Google Scholar] [CrossRef]
- Rajam, M.; Pulavendran, S.; Rose, C.; Mandal, A. Chitosan nanoparticles as a dual growth factor delivery system for tissue engineering applications. Int. J. Pharm. 2011, 410, 145–152. [Google Scholar] [CrossRef]
- Nivethaa, E.; Dhanavel, S.; Narayanan, V.; Vasu, C.A.; Stephen, A. An in vitro cytotoxicity study of 5-fluorouracil encapsulated chitosan/gold nanocomposites towards MCF-7 cells. RSC Adv. 2015, 5, 1024–1032. [Google Scholar] [CrossRef]
- Iswarya, V.; Manivannan, J.; De, A.; Paul, S.; Roy, R.; Johnson, J.; Kundu, R.; Chandrasekaran, N.; Mukherjee, A.; Mukherjee, A. Surface capping and size-dependent toxicity of gold nanoparticles on different trophic levels. Environ. Sci. Pollut. Res. 2016, 23, 4844–4858. [Google Scholar] [CrossRef] [PubMed]
- George, D.; Maheswari, P.U.; Begum, K.M.S. Chitosan-cellulose hydrogel conjugated with L-histidine and Zinc oxide nanoparticles for sustained drug delivery: Kinetics and in-vitro biological studies. Carbohydr. Polym. 2020, 236, 116101. [Google Scholar] [CrossRef]
- Morris, V.B.; Sharma, C.P. Folate mediated histidine derivative of quaternised chitosan as a gene delivery vector. Int. J. Pharm. 2010, 389, 176–185. [Google Scholar] [CrossRef]
- Wang, W.; Tong, C.-Y.; Liu, X.-Y.; Li, T.; Liu, B.; Xiong, W. Preparation and functional characterization of tumour-targeted folic acid-chitosan conjugated nanoparticles loaded with mitoxantrone. J. Cent. South Univ. 2015, 22, 3311–3317. [Google Scholar] [CrossRef]
- Wilson, D.R.; Green, J.J. Nanoparticle tracking analysis for determination of hydrodynamic diameter, concentration, and zeta-potential of polyplex nanoparticles. In Biomedical Nanotechnology: Methods and Protocols; Petrosko, S.H., Day, E.S., Eds.; Springer: New York, NY, USA, 2017; pp. 31–46. [Google Scholar] [CrossRef]
- Ghosh, P.S. Tailoring the Surface-Coating of Gold Nanoparticles for Bio-Applications. Open Access Diss.; University of Massachusetts Amherst: Amherst, MA, USA, 2009; p. 111. [Google Scholar] [CrossRef]
- Alex, S.; Tiwari, A. Functionalized gold nanoparticles: Synthesis, properties and applications—A review. J. Nanosci. Nanotechnol. 2015, 15, 1869–1894. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.R.; Agrawal, Y. Nanosuspension: An approach to enhance solubility of drugs. J. Adv. Pharm. Technol. Res. 2011, 2, 81. [Google Scholar] [CrossRef] [PubMed]
- Forest, V.; Pourchez, J. Preferential binding of positive nanoparticles on cell membranes is due to electrostatic interactions: A too simplistic explanation that does not take into account the nanoparticle protein corona. Mater. Sci. Eng. C 2017, 70, 889–896. [Google Scholar] [CrossRef] [Green Version]
- Cebrián, V.; Martín-Saavedra, F.; Yagüe, C.; Arruebo, M.; Santamaría, J.; Vilaboa, N. Size-dependent transfection efficiency of PEI-coated gold nanoparticles. Acta Biomater. 2011, 7, 3645–3655. [Google Scholar] [CrossRef]
- Clayton, K.N.; Salameh, J.W.; Wereley, S.T.; Kinzer-Ursem, T.L. Physical characterization of nanoparticle size and surface modification using particle scattering diffusometry. Biomicrofluidics 2016, 10, 054107. [Google Scholar] [CrossRef] [Green Version]
- Nafisi, S.; Saboury, A.A.; Keramat, N.; Neault, J.-F.; Tajmir-Riahi, H.-A. Stability and structural features of DNA intercalation with ethidium bromide, acridine orange and methylene blue. J. Mol. Struct. 2007, 827, 35–43. [Google Scholar] [CrossRef]
- Roy, S.; Sadhukhan, R.; Ghosh, U.; Das, T.K. Interaction studies between biosynthesized silver nanoparticle with calf thymus DNA and cytotoxicity of silver nanoparticles. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 141, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Prazeres, D.M.; Ferreira, G.N.; Monteiro, G.A.; Cooney, C.L.; Cabral, J.M. Large-scale production of pharmaceutical grade plasmid DNA for gene therapy: Problems and bottlenecks. Trends Biotechnol. 1999, 17, 169–174. [Google Scholar] [CrossRef]
- Senthilraja, P.; Kathiresan, K. In vitro cytotoxicity MTT assay in Vero, HepG2 and MCF-7 cell lines study of Marine Yeast. J. Appl. Pharm. Sci. 2015, 5, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Fotakis, G.; Timbrell, J.A. In vitro cytotoxicity assays: Comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 2006, 160, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Low, P.S. Folate-targeted therapies for cancer. J. Med. Chem. 2010, 53, 6811–6824. [Google Scholar] [CrossRef] [PubMed]
- Akinyelu, J.; Singh, M. Chitosan stabilized gold-folate-poly (lactide-co-glycolide) nanoplexes facilitate efficient gene delivery in hepatic and breast cancer cells. J. Nanosci. Nanotechnol. 2018, 18, 4478–4486. [Google Scholar] [CrossRef]
- Mbatha, L.S.; Maiyo, F.; Daniels, A.; Singh, M. Dendrimer-coated gold nanoparticles for efficient folate-targeted mRNA delivery in vitro. Pharmaceutics 2021, 13, 900. [Google Scholar] [CrossRef]
- Kimling, J.; Maier, M.; Okenve, B.; Kotaidis, V.; Ballot, H.; Plech, A. Turkevich method for gold nanoparticle synthesis revisited. J. Phys. Chem. B 2006, 110, 15700–15707. [Google Scholar] [CrossRef]
- Alexandridis, P. Gold nanoparticle synthesis, morphology control, and stabilization facilitated by functional polymers. Chem. Eng. Technol. 2011, 34, 15–28. [Google Scholar] [CrossRef]
- Ramnandan, D.; Mokhosi, S.; Daniels, A.; Singh, M. Chitosan, Polyethylene glycol and Polyvinyl alcohol modified MgFe2O4 ferrite magnetic nanoparticles in Doxorubicin delivery: A comparative study in vitro. Molecules 2021, 26, 3893. [Google Scholar] [CrossRef]
- Singh, D.; Singh, M. Hepatocellular-targeted mRNA delivery using functionalized selenium nanoparticles in vitro. Pharmaceutics 2021, 13, 298. [Google Scholar] [CrossRef] [PubMed]
- Petros, R.A.; Desimone, J.M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Al-Johani, H.; Abou-Hamad, E.; Jedidi, A.; Widdifield, C.M.; Viger-Gravel, J.; Sangaru, S.S.; Gajan, D.; Anjum, D.H.; Ould-Chikh, S.; Hedhili, M.N. The structure and binding mode of citrate in the stabilization of gold nanoparticles. Nat. Chem. 2017, 9, 890. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Anaya, L.M.; Soltero, J.A.; Rinaudo, M. DNA/chitosan electrostatic complex. Int. J. Biol. Macromol. 2016, 88, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Haddad, Y.; Xhaxhiu, K.; Kopel, P.; Hynek, D.; Zitka, O.; Adam, V. The isolation of DNA by polycharged magnetic particles: An analysis of the interaction by zeta potential and particle size. Int. J. Mol. Sci. 2016, 17, 550. [Google Scholar] [CrossRef] [Green Version]
- John, A.A.; Jaganathan, S.K.; Ayyar, M.; Krishnasamy, N.P.; Rajasekar, R.; Supriyanto, E. Folic acid decorated chitosan nanoparticles and its derivatives for the delivery of drugs and genes to cancer cells. Curr. Sci. 2017, 113, 1530. [Google Scholar] [CrossRef]
- Okuda, T.; Sugiyama, A.; Niidome, T.; Aoyagi, H. Characters of dendritic poly (L-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro. Biomaterials 2004, 25, 537–544. [Google Scholar] [CrossRef]
- Midoux, P.; Pichon, C.; Yaouanc, J.J.; Jaffrès, P.A. Chemical vectors for gene delivery: A current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 2009, 157, 166–178. [Google Scholar] [CrossRef] [Green Version]
- Liao, R.; Yi, S.; Liu, M.; Jin, W.; Yang, B. Folic-acid-targeted self-assembling supramolecular carrier for gene delivery. ChemBioChem 2015, 16, 1622–1628. [Google Scholar] [CrossRef]
- Akinyelu, A.; Oladimeji, O.; Singh, M. Lactobionic acid-chitosan functionalized gold coated poly(lactide-co-glycolide) nanoparticles for hepatocyte targeted gene delivery. Adv. Nat. Sci. Nanosci. Nanotechnol. 2020, 11, 045017. [Google Scholar] [CrossRef]
- Xue, S.-F.; Lu, L.-F.; Wang, Q.-X.; Zhang, S.; Zhang, M.; Shi, G. An integrated logic system for time-resolved fluorescent “turn-on” detection of cysteine and histidine base on terbium (III) coordination polymer–copper (II) ensemble. Talanta 2016, 158, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Janaszewska, A.; Lazniewska, J.; Trzepiński, P.; Marcinkowska, M.; Klajnert-Maculewicz, B. Cytotoxicity of dendrimers. Biomolecules 2019, 9, 330. [Google Scholar] [CrossRef] [Green Version]
- Talekar, M.; Kendall, J.; Denny, W.; Garg, S. Targeting of nanoparticles in cancer: Drug delivery and diagnostics. Anti-Cancer Drugs 2011, 22, 949–962. [Google Scholar] [CrossRef]
- Ziegler, U.; Groscurth, P. Morphological features of cell death. Physiology 2004, 19, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Amini, N.; Majid, F.A.A.; Marvibaigi, M.; Supriyanto, E.; Jaganathan, S.K.; Soon, W.T.; Nasiri, R.; Hamzehalipour, J. Cervicare™ induces apoptosis in HeLa and CaSki cells through ROS production and loss of mitochondrial membrane potential. RSC Adv. 2016, 6, 24391–24417. [Google Scholar] [CrossRef]
- Zhu, J.; Qiao, M.; Wang, Q.; Ye, Y.; Ba, S.; Ma, J.; Hu, H.; Zhao, X.; Chen, D. Dual-responsive polyplexes with enhanced disassembly and endosomal escape for efficient delivery of siRNA. Biomaterials 2018, 162, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Pillay, N.S.; Daniels, A.; Singh, M. Folate-targeted transgenic activity of dendrimer functionalized selenium nanoparticles in vitro. Int. J. Mol. Sci. 2020, 21, 7177. [Google Scholar] [CrossRef]
- Jang, Y.; Kim, S.; Oh, W.-K.; Kim, C.; Lee, I.; Jang, J. A folic acid conjugated silica-titania porous hollow nanosphere for im-proved topical photodynamic therapy. Chem. Comm. 2014, 50, 15345–15347. [Google Scholar] [CrossRef]
- Gorle, S.; Sewbalas, A.; Ariatti, M.; Singh, M. Ligand-tagged cationic liposome facilitates efficient gene delivery to folate receptors. Curr. Sci. 2016, 111, 662–670. [Google Scholar] [CrossRef]
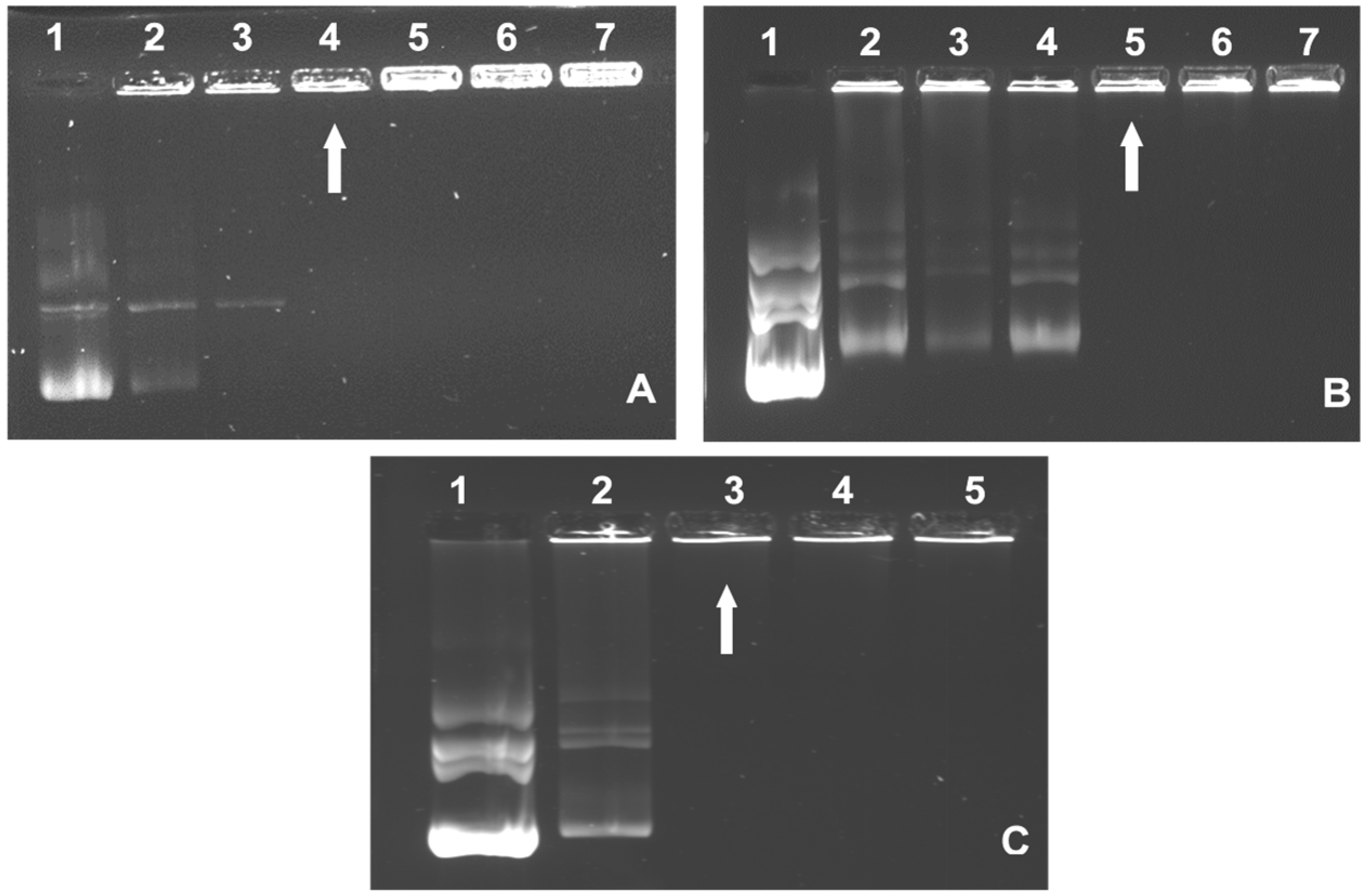
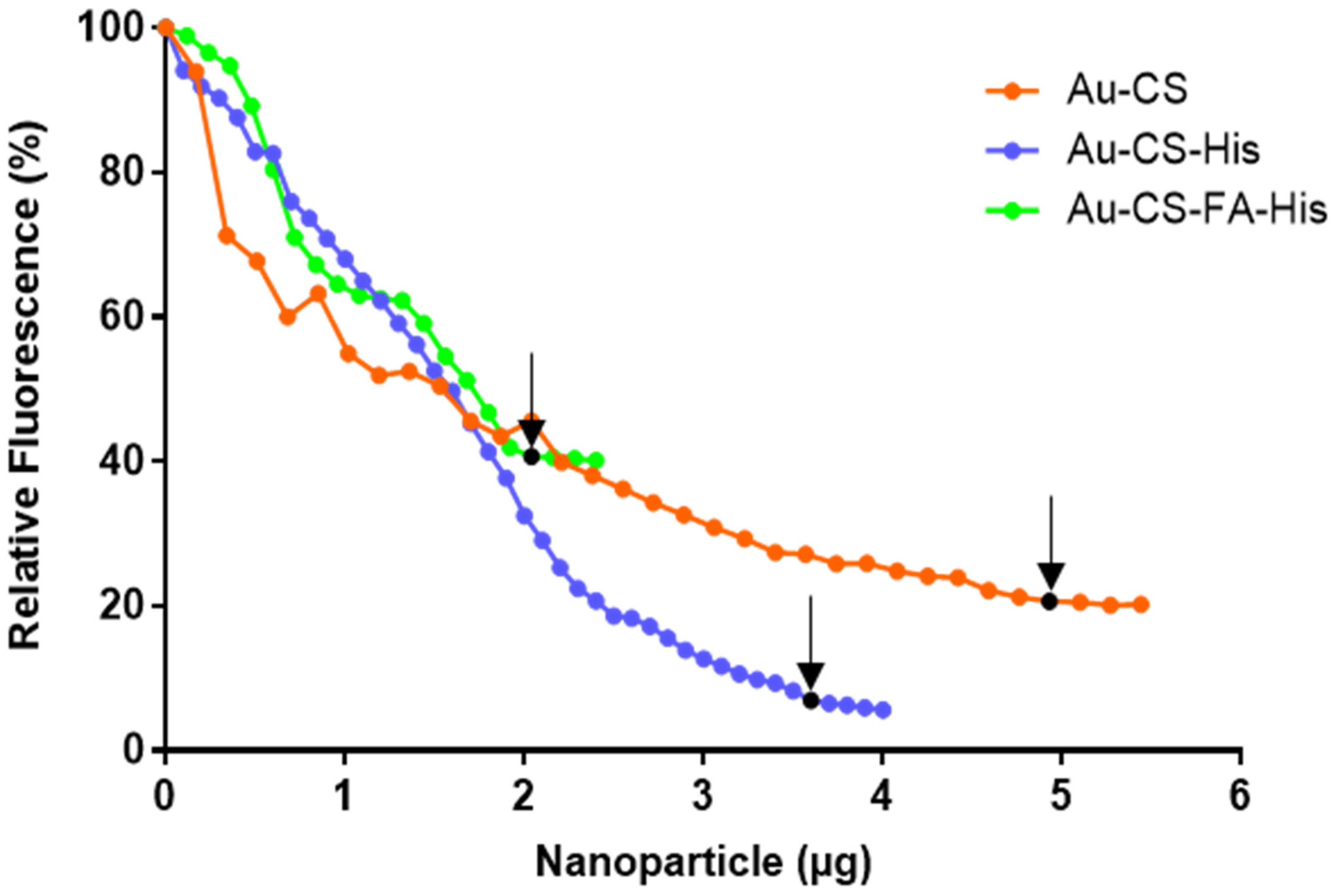
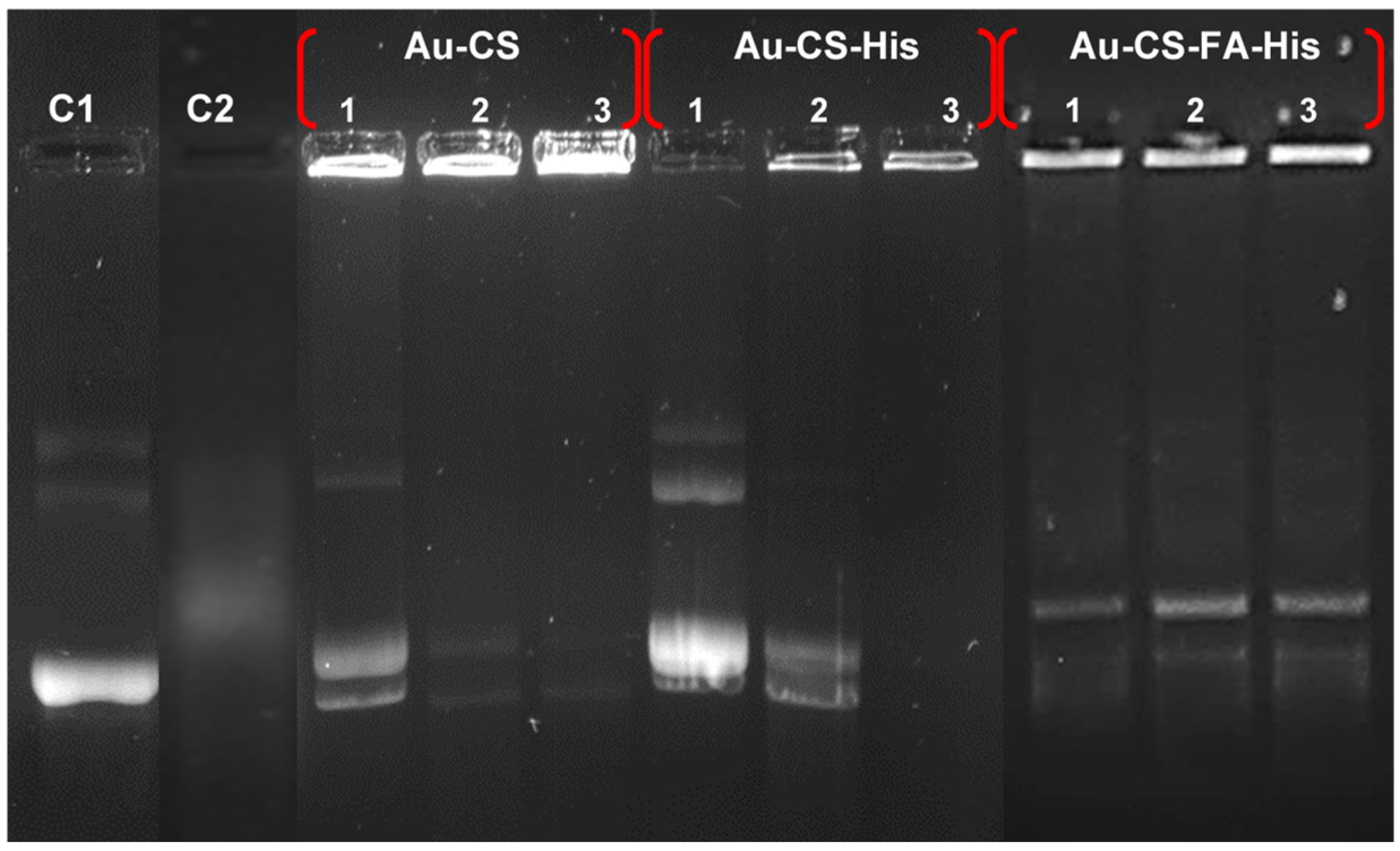
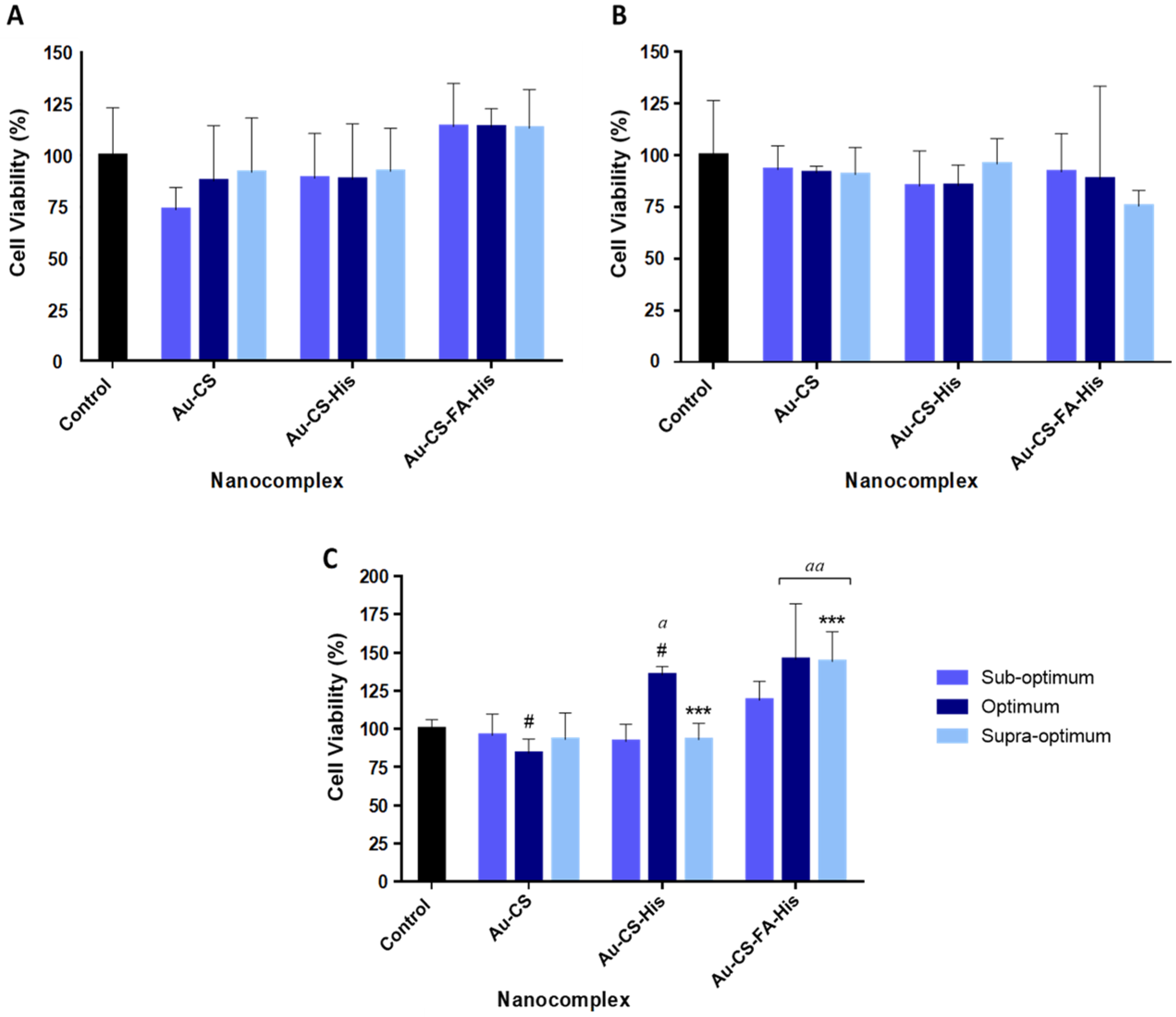
| Nanocomplexes | Suboptimum | Optimum | Supraoptimum |
|---|---|---|---|
| Au-CS: pDNA | 1.5:1 | 2:1 | 2.5:1 |
| Au-CS-His: pDNA | 2.8:1 | 3.2:1 | 3.6:1 |
| Au-CS-FA-His: pDNA | 4.5:1 | 5:1 | 5.5:1 |
| Nanoparticles | Nanocomplexes | ||||||
|---|---|---|---|---|---|---|---|
| Size (nm ± SD) | ζ Potential (mV ± SD) | PDI | pDNA:NP Ratio (w/w) | Size (nm ± SD) | ζ Potential (mV ± SD) | PDI | |
| AuNP | 79.9 ± 12.0 | −24.9 ± 0.2 | 0.023 | - | - | - | - |
| Au-CS | 116.5 ± 11.9 | 35.7 ± 0.2 | 0.011 | 1:2 | 129.7 ± 44.6 | −21.5 ± 2.1 | 0.118 |
| Au-CS-His | 121.5 ± 4.0 | 38.0 ± 1.6 | 0.0011 | 1:3.2 | 133.1 ± 13.5 | −23.4 ± 0.7 | 0.01 |
| Au-CS-FA-His | 122.2 ± 21.1 | 28.5 ± 3.3 | 0.029 | 1:5 | 126.3 ± 5.0 | −34.2 ± 0.3 | 0.0016 |
| Cell Lines | Apoptotic Index | ||
|---|---|---|---|
| Au-CS Suboptimum | Au-CS-FA-His Supraoptimum | Au-CS Optimum | |
| HEK293 | 0.032 | - | - |
| SKBR3 | - | 0.036 | - |
| MCF-7 | - | - | 0.026 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joseph, C.; Daniels, A.; Singh, S.; Singh, M. Histidine-Tagged Folate-Targeted Gold Nanoparticles for Enhanced Transgene Expression in Breast Cancer Cells In Vitro. Pharmaceutics 2022, 14, 53. https://doi.org/10.3390/pharmaceutics14010053
Joseph C, Daniels A, Singh S, Singh M. Histidine-Tagged Folate-Targeted Gold Nanoparticles for Enhanced Transgene Expression in Breast Cancer Cells In Vitro. Pharmaceutics. 2022; 14(1):53. https://doi.org/10.3390/pharmaceutics14010053
Chicago/Turabian StyleJoseph, Calrin, Aliscia Daniels, Sooboo Singh, and Moganavelli Singh. 2022. "Histidine-Tagged Folate-Targeted Gold Nanoparticles for Enhanced Transgene Expression in Breast Cancer Cells In Vitro" Pharmaceutics 14, no. 1: 53. https://doi.org/10.3390/pharmaceutics14010053
APA StyleJoseph, C., Daniels, A., Singh, S., & Singh, M. (2022). Histidine-Tagged Folate-Targeted Gold Nanoparticles for Enhanced Transgene Expression in Breast Cancer Cells In Vitro. Pharmaceutics, 14(1), 53. https://doi.org/10.3390/pharmaceutics14010053