Nonviral Delivery Systems of mRNA Vaccines for Cancer Gene Therapy
Abstract
1. Introduction
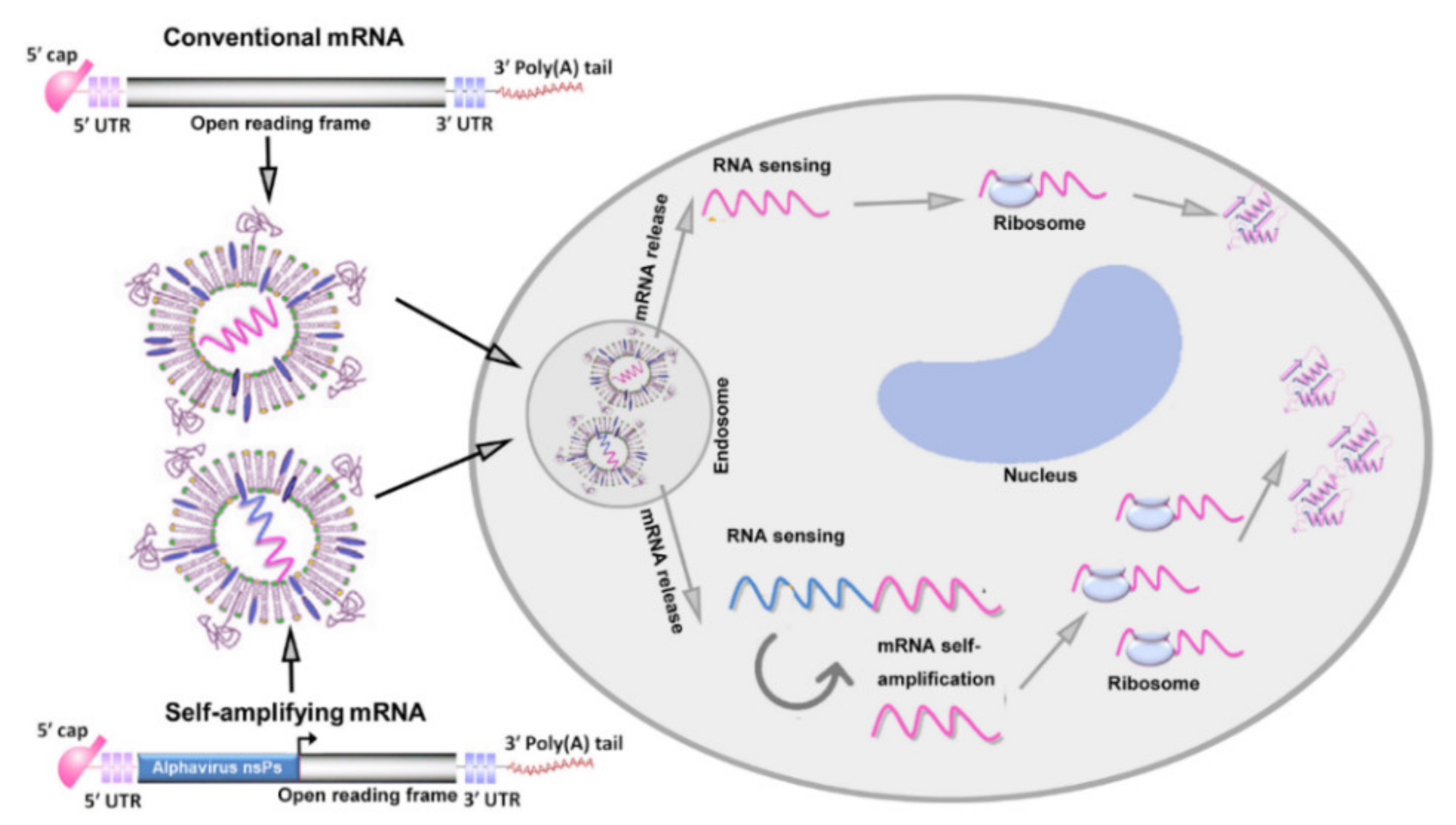
2. Major Types of mRNA Vaccine
2.1. Conventional mRNA Vaccine
2.2. Self-Amplifying mRNA Vaccine
3. The Core Mechanism of mRNA Vaccines for Cancer Therapy
4. The Principles of mRNA Vaccine Design and Modification
4.1. The 5′-Cap
4.2. The 5′-UTR
4.3. The ORF
4.4. The 3′-UTR
4.5. The 3′-poly(A) Tail
5. The Nonviral Nanodelivery Systems of mRNA Vaccines
5.1. Lipid Nanoparticles (LNPs)
5.2. Polymer Nanoparticles
5.3. Polypeptide Nanoparticles
5.4. Hybrid Nanoparticles
5.5. Gold Nanoparticle–DNA Oligonucleotide Conjugates
5.6. mRNA-Loaded Exosomes
6. Clinical mRNA Vaccines for Cancer Therapy
6.1. mRNA Cancer Vaccines Based on the Transfection of DCs In Vitro
6.2. mRNA Cancer Vaccine Based on Direct Injection In Vivo
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| mRNA | messenger ribonucleic acid |
| WHO | World Health Organization |
| DNA | deoxyribonucleic acid |
| FDA | Food and Drug Administration |
| DC | dendritic cell |
| TALENs | transcription activator-like effector nucleases |
| EP | electroporation |
| saRNA | self-amplifying mRNA |
| UTR | untranslated region |
| ORF | open reading frame |
| Poly (A) | polyadenylic acid |
| nsPs | nonstructural proteins |
| IVT mRNA | in vitro transcribed mRNA |
| TAA | tumor-associated antigen |
| TSA | tumor-specific antigen |
| APC | antigen-presenting cell |
| PAMPs | pathogen-associated molecular patterns |
| PRRs | pattern recognition receptors |
| TLRs | Toll-like receptors |
| RIG-I | retinoic acid-inducible gene-I |
| RLRs | retinoic acid-inducible gene-I-like receptors |
| hnRNPs | nuclear heterogeneous ribonucleoprotein |
| ZBP1 | Z-DNA-binding protein 1 |
| Th | helper T Cell |
| MHC | major histocompatibility complex |
| CTLs | cytotoxic T lymphocytes |
| CD | cluster of differentiation |
| AKP | alkaline phosphatase |
| eIF4E | eukaryotic initiation factor 4E |
| eIF4G | eukaryotic initiation factor 4G |
| PKR | double-stranded RNA-dependent protein kinase |
| Ψ | pseudouridine |
| m5C | 5-methyl cytosine |
| m6A | 6-methyl adenosine |
| m5U | 5-methyl uracil |
| s2U | 2-thiouracil |
| PABP | poly(A)-binding protein |
| LNPs | lipid nanoparticles |
| PEG | polyethylene glycol |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine |
| DSPC | 1,2-distearoyl-sn-glycero-3-phosphocholine |
| NHPs | nonhuman primates |
| PEI | polyethyleneimine |
| CP 2k | cationic cyclodextrin-PEI 2k conjugate |
| PAMAM | polyamide amine |
| PLGA | poly (lactic-co-glycolic acid) |
| GFP | green fluorescent protein |
| RNase | ribonuclease |
| LPNs | lipid-polymer hybrid nanoparticles |
| LLPs | lipid-like nanoparticles |
| DOTAP | 1,2-dioleoyl-3-trimethylammonium-propane |
| iPSCs | induced pluripotent stem cells |
| CPPs | cell-penetrating peptides |
| WNT | wingless integrated |
| SD | spray drying |
| SFD | spray freeze drying |
| SEND | selective endogenous encapsidation |
| AuNP | gold nanoparticle |
| i.v. | intravenous injection |
| i.m. | intramuscular injection |
| i.h. | hypodermic injection |
| i.d. | intradermal injection |
| i.p. | intraperitoneal injection |
| i.t. | intratumoral injection |
| i.n. | nasal administration |
| eGFP | enhanced green fluorescent protein |
| Luc | luciferase |
| Fluc | firefly luciferase |
| hGLuc | humanized Gaussia luciferase |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| IL-4 | interleukin-4 |
| DI-TMZ | dose-intensified temozolomide |
| CR | complete response |
| HCV | hepatitis C virus |
| HCC | hepatitis C cancer |
| GBM | glioblastoma multiforme |
| PD-1 | programmed cell death protein 1 |
| CTLA-4 | cytotoxic T-lymphocyte-associated protein 4 |
| NSCLC | non-small-cell lung cancer |
| HPV | human papillomavirus |
| PDL-1 | programmed cell death ligand-1 |
References
- Bray, F.; Soerjomataram, I. The Changing Global Burden of Cancer: Transitions in Human Development and Implications for Cancer Prevention and Control. In Cancer: Disease Control Priorities, 3rd ed.; Gelband, H., Jha, P., Sankaranarayanan, R., Horton, S., Eds.; The International Bank for Reconstruction and Development; The World Bank © 2015 International Bank for Reconstruction and Development; The World Bank: Washington, DC, USA, 2015; Volume 3. [Google Scholar]
- Morales, A.; Eidinger, D.; Bruce, A.W. Intracavitary Bacillus Calmette-Guerin in the treatment of superficial bladder tumors. J. Urol. 1976, 116, 180–183. [Google Scholar] [CrossRef]
- Cheever, M.A.; Higano, C.S. PROVENGE (Sipuleucel-T) in prostate cancer: The first FDA-approved therapeutic cancer vaccine. Clin. Cancer Res. 2011, 17, 3520–3526. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.; Luo, J.; Han, X.; Wei, Y.; Wei, X. mRNA vaccine: A potential therapeutic strategy. Mol. Cancer 2021, 20, 33. [Google Scholar] [CrossRef]
- Nichols, W.W.; Ledwith, B.J.; Manam, S.V.; Troilo, P.J. Potential DNA vaccine integration into host cell genome. Ann. N. Y. Acad. Sci. 1995, 772, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; O’Keeffe-Ahern, J.; Lyu, J.; Pierucci, L.; Zhou, D.; Wang, W. A new developing class of gene delivery: Messenger RNA-based therapeutics. Biomater. Sci. 2017, 5, 2381–2392. [Google Scholar] [CrossRef]
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug. Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Linares-Fernández, S.; Lacroix, C.; Exposito, J.Y.; Verrier, B. Tailoring mRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends. Mol. Med. 2020, 26, 311–323. [Google Scholar] [CrossRef]
- Kantoff, P.W.; Higano, C.S.; Shore, N.D.; Berger, E.R.; Small, E.J.; Penson, D.F.; Redfern, C.H.; Ferrari, A.C.; Dreicer, R.; Sims, R.B.; et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010, 363, 411–422. [Google Scholar] [CrossRef]
- Dimitriadis, G.J. Translation of rabbit globin mRNA introduced by liposomes into mouse lymphocytes. Nature 1978, 274, 923–924. [Google Scholar] [CrossRef]
- Ostro, M.J.; Giacomoni, D.; Lavelle, D.; Paxton, W.; Dray, S. Evidence for translation of rabbit globin mRNA after liposome-mediated insertion into a human cell line. Nature 1978, 274, 921–923. [Google Scholar] [CrossRef]
- Krieg, P.A.; Melton, D.A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984, 12, 7057–7070. [Google Scholar] [CrossRef] [PubMed]
- Malone, R.W.; Felgner, P.L.; Verma, I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA 1989, 86, 6077–6081. [Google Scholar] [CrossRef] [PubMed]
- Conry, R.M.; LoBuglio, A.F.; Wright, M.; Sumerel, L.; Pike, M.J.; Johanning, F.; Benjamin, R.; Lu, D.; Curiel, D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995, 55, 1397–1400. [Google Scholar] [PubMed]
- Yoon, S.H.; Lee, J.M.; Cho, H.I.; Kim, E.K.; Kim, H.S.; Park, M.Y.; Kim, T.G. Adoptive immunotherapy using human peripheral blood lymphocytes transferred with RNA encoding Her-2/neu-specific chimeric immune receptor in ovarian cancer xenograft model. Cancer Gene. Ther. 2009, 16, 489–497. [Google Scholar] [CrossRef]
- Wood, A.J.; Lo, T.W.; Zeitler, B.; Pickle, C.S.; Ralston, E.J.; Lee, A.H.; Amora, R.; Miller, J.C.; Leung, E.; Meng, X.; et al. Targeted genome editing across species using ZFNs and TALENs. Science 2011, 333, 307. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Mahvi, D.M.; Sheehy, M.J.; Yang, N.S. DNA cancer vaccines: A gene gun approach. Immunol. Cell. Biol. 1997, 75, 456–460. [Google Scholar] [CrossRef]
- Van Meirvenne, S.; Straetman, L.; Heirman, C.; Dullaers, M.; De Greef, C.; Van Tendeloo, V.; Thielemans, K. Efficient genetic modification of murine dendritic cells by electroporation with mRNA. Cancer Gene. Ther. 2002, 9, 787–797. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Minnaert, A.K.; Vanluchene, H.; Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Raemdonck, K.; Sanders, N.N.; Remaut, K. Strategies for controlling the innate immune activity of conventional and self-amplifying mRNA therapeutics: Getting the message across. Adv. Drug. Deliv. Rev. 2021, 176, 113900. [Google Scholar] [CrossRef]
- Liang, F.; Lindgren, G.; Lin, A.; Thompson, E.A.; Ols, S.; Röhss, J.; John, S.; Hassett, K.; Yuzhakov, O.; Bahl, K.; et al. Efficient Targeting and Activation of Antigen-Presenting Cells In Vivo after Modified mRNA Vaccine Administration in Rhesus Macaques. Mol. Ther. 2017, 25, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Bloom, K.; van den Berg, F.; Arbuthnot, P. Self-amplifying RNA vaccines for infectious diseases. Gene. Ther. 2021, 28, 117–129. [Google Scholar] [CrossRef]
- Kääriäinen, L.; Takkinen, K.; Keränen, S.; Söderlund, H. Replication of the genome of alphaviruses. J. Cell. Sci. Suppl. 1987, 7, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, K.; Liljeström, P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccines 2015, 14, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.A.; Kommareddy, S.; Maione, D.; Uematsu, Y.; Giovani, C.; Berlanda Scorza, F.; Otten, G.R.; Yu, D.; Mandl, C.W.; Mason, P.W.; et al. Self-amplifying mRNA vaccines. Adv. Genet. 2015, 89, 179–233. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.B.; Lambert, L.; Kinnear, E.; Busse, D.; Erbar, S.; Reuter, K.C.; Wicke, L.; Perkovic, M.; Beissert, T.; Haas, H.; et al. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018, 26, 446–455. [Google Scholar] [CrossRef]
- Guo, Y.; Lei, K.; Tang, L. Neoantigen Vaccine Delivery for Personalized Anticancer Immunotherapy. Front. Immunol. 2018, 9, 1499. [Google Scholar] [CrossRef] [PubMed]
- Linette, G.P.; Carreno, B.M. Neoantigen Vaccines Pass the Immunogenicity Test. Trends Mol. Med. 2017, 23, 869–871. [Google Scholar] [CrossRef]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- Sahay, G.; Querbes, W.; Alabi, C.; Eltoukhy, A.; Sarkar, S.; Zurenko, C.; Karagiannis, E.; Love, K.; Chen, D.; Zoncu, R.; et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013, 31, 653–658. [Google Scholar] [CrossRef]
- Guevara, M.L.; Persano, F.; Persano, S. Advances in Lipid Nanoparticles for mRNA-Based Cancer Immunotherapy. Front. Chem. 2020, 8, 589959. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lu, Y.; Thulasi Raman, S.N.; Xu, F.; Wu, Q.; Li, Z.; Brownlie, R.; Liu, Q.; Zhou, Y. Nuclear-resident RIG-I senses viral replication inducing antiviral immunity. Nat. Commun. 2018, 9, 3199. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Hattinger, E.; Scheiblhofer, S.; Roesler, E.; Thalhamer, T.; Thalhamer, J.; Weiss, R. Prophylactic mRNA Vaccination against Allergy Confers Long-Term Memory Responses and Persistent Protection in Mice. J. Immunol. Res. 2015, 2015, 797421. [Google Scholar] [CrossRef] [PubMed]
- Heine, A.; Juranek, S.; Brossart, P. Clinical and immunological effects of mRNA vaccines in malignant diseases. Mol. Cancer 2021, 20, 52. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef]
- Kurts, C.; Robinson, B.W.; Knolle, P.A. Cross-priming in health and disease. Nat. Rev. Immunol. 2010, 10, 403–414. [Google Scholar] [CrossRef]
- De Beuckelaer, A.; Pollard, C.; Van Lint, S.; Roose, K.; Van Hoecke, L.; Naessens, T.; Udhayakumar, V.K.; Smet, M.; Sanders, N.; Lienenklaus, S.; et al. Type I Interferons Interfere with the Capacity of mRNA Lipoplex Vaccines to Elicit Cytolytic T Cell Responses. Mol. Ther. 2016, 24, 2012–2020. [Google Scholar] [CrossRef]
- Andries, O.; De Filette, M.; De Smedt, S.C.; Demeester, J.; Van Poucke, M.; Peelman, L.; Sanders, N.N. Innate immune response and programmed cell death following carrier-mediated delivery of unmodified mRNA to respiratory cells. J. Control Release 2013, 167, 157–166. [Google Scholar] [CrossRef]
- Loomis, K.H.; Lindsay, K.E.; Zurla, C.; Bhosle, S.M.; Vanover, D.A.; Blanchard, E.L.; Kirschman, J.L.; Bellamkonda, R.V.; Santangelo, P.J. In Vitro Transcribed mRNA Vaccines with Programmable Stimulation of Innate Immunity. Bioconjug Chem. 2018, 29, 3072–3083. [Google Scholar] [CrossRef]
- Baptista, B.; Carapito, R.; Laroui, N.; Pichon, C.; Sousa, F. mRNA, a Revolution in Biomedicine. Pharmaceutics 2021, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liang, Y.; Huang, L. Development and Delivery Systems of mRNA Vaccines. Front. Bioeng. Biotechnol. 2021, 9, 718753. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Stevens, A. Yeast cells lacking 5′-->3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5’ cap structure. Mol. Cell. Biol. 1993, 13, 4826–4835. [Google Scholar] [CrossRef]
- Sonenberg, N.; Rupprecht, K.M.; Hecht, S.M.; Shatkin, A.J. Eukaryotic mRNA cap binding protein: Purification by affinity chromatography on sepharose-coupled m7GDP. Proc. Natl. Acad. Sci. USA 1979, 76, 4345–4349. [Google Scholar] [CrossRef] [PubMed]
- Nwokoye, E.C.; AlNaseem, E.; Crawford, R.A.; Castelli, L.M.; Jennings, M.D.; Kershaw, C.J.; Pavitt, G.D. Overlapping regions of Caf20 mediate its interactions with the mRNA-5’cap-binding protein eIF4E and with ribosomes. Sci. Rep. 2021, 11, 13467. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E.; Dahlberg, J.E.; Lund, E. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA 1995, 1, 957–967. [Google Scholar] [PubMed]
- van Dulmen, M.; Muthmann, N.; Rentmeister, A. Chemo-Enzymatic Modification of the 5’ Cap Maintains Translation and Increases Immunogenic Properties of mRNA. Angew. Chem. Int. Ed. Engl. 2021, 60, 13280–13286. [Google Scholar] [CrossRef]
- Leppek, K.; Das, R.; Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell. Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Insertion mutagenesis to increase secondary structure within the 5’ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 1985, 40, 515–526. [Google Scholar] [CrossRef]
- Kim, S.C.; Sekhon, S.S.; Shin, W.R.; Ahn, G.; Cho, B.K.; Ahn, J.Y.; Kim, Y.H. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol. Cell. Toxicol. 2021, 18, 1–8. [Google Scholar] [CrossRef]
- Jia, L.; Mao, Y.; Ji, Q.; Dersh, D.; Yewdell, J.W.; Qian, S.B. Decoding mRNA translatability and stability from the 5’ UTR. Nat. Struct. Mol. Biol. 2020, 27, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Gerstein, M. Analysis of the yeast transcriptome with structural and functional categories: Characterizing highly expressed proteins. Nucleic Acids. Res. 2000, 28, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Marín, A.; Gallardo, M.; Kato, Y.; Shirahige, K.; Gutiérrez, G.; Ohta, K.; Aguilera, A. Relationship between G+C content, ORF-length and mRNA concentration in Saccharomyces cerevisiae. Yeast 2003, 20, 703–711. [Google Scholar] [CrossRef]
- Kudla, G.; Lipinski, L.; Caffin, F.; Helwak, A.; Zylicz, M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006, 4, e180. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Cannarozzi, G.; Schraudolph, N.N.; Faty, M.; von Rohr, P.; Friberg, M.T.; Roth, A.C.; Gonnet, P.; Gonnet, G.; Barral, Y. A role for codon order in translation dynamics. Cell 2010, 141, 355–367. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Verbeke, R.; Lentacker, I.; Wayteck, L.; Breckpot, K.; Van Bockstal, M.; Descamps, B.; Vanhove, C.; De Smedt, S.C.; Dewitte, H. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: Restoring the immunogenicity of immunosilent mRNA. J. Control Release 2017, 266, 287–300. [Google Scholar] [CrossRef]
- Tanguay, R.L.; Gallie, D.R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996, 16, 146–156. [Google Scholar] [CrossRef]
- Murray, E.L.; Schoenberg, D.R. A+U-rich instability elements differentially activate 5′-3′ and 3′-5′ mRNA decay. Mol. Cell. Biol. 2007, 27, 2791–2799. [Google Scholar] [CrossRef]
- Rodgers, N.D.; Wang, Z.; Kiledjian, M. Regulated alpha-globin mRNA decay is a cytoplasmic event proceeding through 3’-to-5’ exosome-dependent decapping. RNA 2002, 8, 1526–1537. [Google Scholar] [PubMed]
- Adibzadeh, S.; Fardaei, M.; Takhshid, M.A.; Miri, M.R.; Rafiei Dehbidi, G.; Farhadi, A.; Ranjbaran, R.; Alavi, P.; Nikouyan, N.; Seyyedi, N.; et al. Enhancing Stability of Destabilized Green Fluorescent Protein Using Chimeric mRNA Containing Human Beta-Globin 5′ and 3′ Untranslated Regions. Avicenna J. Med. Biotechnol. 2019, 11, 112–117. [Google Scholar] [PubMed]
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Löwer, M.; Vallazza, B.; Beissert, T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3’ UTRs Identified by Cellular Library Screening. Mol Ther 2019, 27, 824–836. [Google Scholar] [CrossRef]
- Kelly, J.M.; Cox, R.A. Periodicity in the length of 3’-poly(A) tails from native globin mRNA of rabbit. Nucleic Acids Res. 1982, 10, 4173–4179. [Google Scholar] [CrossRef] [PubMed]
- Subtelny, A.O.; Eichhorn, S.W.; Chen, G.R.; Sive, H.; Bartel, D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014, 508, 66–71. [Google Scholar] [CrossRef]
- Silvestri, L.S.; Parilla, J.M.; Morasco, B.J.; Ogram, S.A.; Flanegan, J.B. Relationship between poliovirus negative-strand RNA synthesis and the length of the 3’ poly(A) tail. Virology 2006, 345, 509–519. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Türeci, O.; Sahin, U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.A.; Chipman, L.B.; Nicholson, A.L.; Chen, Y.H.; Yee, B.A.; Yeo, G.W.; Coller, J.; Pasquinelli, A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017, 24, 1057–1063. [Google Scholar] [CrossRef]
- Tenforde, M.W.; Patel, M.M.; Ginde, A.A.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; Mohr, N.M.; Zepeski, A.; Gaglani, M.; McNeal, T.; et al. Effectiveness of SARS-CoV-2 mRNA Vaccines for Preventing Covid-19 Hospitalizations in the United States. Clin. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Kuntsche, J.; Horst, J.C.; Bunjes, H. Cryogenic transmission electron microscopy (cryo-TEM) for studying the morphology of colloidal drug delivery systems. Int. J. Pharm. 2011, 417, 120–137. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Eygeris, Y.; Gupta, M.; Sahay, G. Self-assembled mRNA vaccines. Adv. Drug. Deliv. Rev. 2021, 170, 83–112. [Google Scholar] [CrossRef]
- Miao, L.; Li, L.; Huang, Y.; Delcassian, D.; Chahal, J.; Han, J.; Shi, Y.; Sadtler, K.; Gao, W.; Lin, J.; et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019, 37, 1174–1185. [Google Scholar] [CrossRef]
- Magarkar, A.; Dhawan, V.; Kallinteri, P.; Viitala, T.; Elmowafy, M.; Róg, T.; Bunker, A. Cholesterol level affects surface charge of lipid membranes in saline solution. Sci. Rep. 2014, 4, 5005. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Chonn, A.; Cullis, P.R. Influence of cholesterol on the association of plasma proteins with liposomes. Biochemistry 1996, 35, 2521–2525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; El-Mayta, R.; Murdoch, T.J.; Warzecha, C.C.; Billingsley, M.M.; Shepherd, S.J.; Gong, N.; Wang, L.; Wilson, J.M.; Lee, D.; et al. Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater. Sci. 2021, 9, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Tam, Y.Y.; Cullis, P.R. Lipid nanoparticles for short interfering RNA delivery. Adv. Genet. 2014, 88, 71–110. [Google Scholar] [CrossRef] [PubMed]
- Schlich, M.; Palomba, R.; Costabile, G.; Mizrahy, S.; Pannuzzo, M.; Peer, D.; Decuzzi, P. Cytosolic delivery of nucleic acids: The case of ionizable lipid nanoparticles. Bioeng. Transl. Med. 2021, 6, e10213. [Google Scholar] [CrossRef]
- Billingsley, M.M.; Hamilton, A.G.; Mai, D.; Patel, S.K.; Swingle, K.L.; Sheppard, N.C.; June, C.H.; Mitchell, M.J. Orthogonal Design of Experiments for Optimization of Lipid Nanoparticles for mRNA Engineering of CAR T Cells. Nano Lett. 2021. [Google Scholar] [CrossRef]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stöter, M.; et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Richner, J.M.; Himansu, S.; Dowd, K.A.; Butler, S.L.; Salazar, V.; Fox, J.M.; Julander, J.G.; Tang, W.W.; Shresta, S.; Pierson, T.C.; et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125. [Google Scholar] [CrossRef]
- Aldrich, C.; Leroux-Roels, I.; Huang, K.B.; Bica, M.A.; Loeliger, E.; Schoenborn-Kellenberger, O.; Walz, L.; Leroux-Roels, G.; von Sonnenburg, F.; Oostvogels, L. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: A phase 1 trial. Vaccine 2021, 39, 1310–1318. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.J.; Moreira, E.D., Jr.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Polack, F.P.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine through 6 Months. N. Engl. J. Med. 2021, 385, 1761–1773. [Google Scholar] [CrossRef] [PubMed]
- Sedic, M.; Senn, J.J.; Lynn, A.; Laska, M.; Smith, M.; Platz, S.J.; Bolen, J.; Hoge, S.; Bulychev, A.; Jacquinet, E.; et al. Safety Evaluation of Lipid Nanoparticle-Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2018, 55, 341–354. [Google Scholar] [CrossRef]
- Zhang, H.; You, X.; Wang, X.; Cui, L.; Wang, Z.; Xu, F.; Li, M.; Yang, Z.; Liu, J.; Huang, P.; et al. Delivery of mRNA vaccine with a lipid-like material potentiates antitumor efficacy through Toll-like receptor 4 signaling. Proc. Natl. Acad. Sci. USA 2021, 118, e2005191118. [Google Scholar] [CrossRef]
- Kelso, J.M. IgE-mediated allergy to polyethylene glycol (PEG) as a cause of anaphylaxis to mRNA COVID-19 vaccines. Clin. Exp. Allergy 2021. [Google Scholar] [CrossRef]
- Suzuki, T.; Suzuki, Y.; Hihara, T.; Kubara, K.; Kondo, K.; Hyodo, K.; Yamazaki, K.; Ishida, T.; Ishihara, H. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: Faster PEG shedding attenuates anti-PEG IgM production. Int. J. Pharm. 2020, 588, 119792. [Google Scholar] [CrossRef]
- Kedmi, R.; Ben-Arie, N.; Peer, D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials 2010, 31, 6867–6875. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K.J.; Dorkin, J.R.; Yang, J.H.; Heartlein, M.W.; DeRosa, F.; Mir, F.F.; Fenton, O.S.; Anderson, D.G. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015, 15, 7300–7306. [Google Scholar] [CrossRef] [PubMed]
- Ball, R.L.; Hajj, K.A.; Vizelman, J.; Bajaj, P.; Whitehead, K.A. Lipid Nanoparticle Formulations for Enhanced Co-delivery of siRNA and mRNA. Nano Lett. 2018, 18, 3814–3822. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; Kim, J.; Eygeris, Y.; Jozic, A.; Sahay, G. Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery. Biomater. Sci. 2021, 9, 4289–4300. [Google Scholar] [CrossRef]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Holtta, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019, 10, 4333. [Google Scholar] [CrossRef]
- Lee, S.M.; Cheng, Q.; Yu, X.; Liu, S.; Johnson, L.T.; Siegwart, D.J. A Systematic Study of Unsaturation in Lipid Nanoparticles Leads to Improved mRNA Transfection In Vivo. Angew. Chem. Int. Ed. Engl. 2021, 60, 5848–5853. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1–17. [Google Scholar] [CrossRef]
- Qiu, M.; Li, Y.; Bloomer, H.; Xu, Q. Developing Biodegradable Lipid Nanoparticles for Intracellular mRNA Delivery and Genome Editing. ACC Chem. Res. 2021, 54, 4001–4011. [Google Scholar] [CrossRef]
- Zukancic, D.; Suys, E.J.A.; Pilkington, E.H.; Algarni, A.; Al-Wassiti, H.; Truong, N.P. The Importance of Poly(ethylene glycol) and Lipid Structure in Targeted Gene Delivery to Lymph Nodes by Lipid Nanoparticles. Pharmaceutics 2020, 12, 1068. [Google Scholar] [CrossRef]
- Hassett, K.J.; Higgins, J.; Woods, A.; Levy, B.; Xia, Y.; Hsiao, C.J.; Acosta, E.; Almarsson, O.; Moore, M.J.; Brito, L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control Release 2021, 335, 237–246. [Google Scholar] [CrossRef]
- Barros, S.A.; Gollob, J.A. Safety profile of RNAi nanomedicines. Adv. Drug. Deliv. Rev. 2012, 64, 1730–1737. [Google Scholar] [CrossRef] [PubMed]
- Demeneix, B.; Behr, J.P. Polyethylenimine (PEI). Adv. Genet. 2005, 53, 215–230. [Google Scholar] [CrossRef]
- Wu, G.Y.; Wu, C.H. Receptor-mediated in vitro gene transformation by a soluble DNA carrier system. J. Biol. Chem. 1987, 262, 4429–4432. [Google Scholar] [CrossRef]
- Blakney, A.K.; Zhu, Y.; McKay, P.F.; Bouton, C.R.; Yeow, J.; Tang, J.; Hu, K.; Samnuan, K.; Grigsby, C.L.; Shattock, R.J.; et al. Big Is Beautiful: Enhanced saRNA Delivery and Immunogenicity by a Higher Molecular Weight, Bioreducible, Cationic Polymer. ACS Nano 2020, 14, 5711–5727. [Google Scholar] [CrossRef]
- Miyata, K.; Nishiyama, N.; Kataoka, K. Rational design of smart supramolecular assemblies for gene delivery: Chemical challenges in the creation of artificial viruses. Chem. Soc. Rev. 2012, 41, 2562–2574. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, T.; Carlisle, R.C.; Read, M.L.; Ogris, M.; Seymour, L.W. Peptide-mediated RNA delivery: A novel approach for enhanced transfection of primary and post-mitotic cells. Nucleic Acids Res. 2001, 29, 3882–3891. [Google Scholar] [CrossRef] [PubMed]
- Moghimi, S.M.; Symonds, P.; Murray, J.C.; Hunter, A.C.; Debska, G.; Szewczyk, A. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. [Google Scholar] [CrossRef]
- Brunot, C.; Ponsonnet, L.; Lagneau, C.; Farge, P.; Picart, C.; Grosgogeat, B. Cytotoxicity of polyethyleneimine (PEI), precursor base layer of polyelectrolyte multilayer films. Biomaterials 2007, 28, 632–640. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef]
- Zhao, M.; Li, M.; Zhang, Z.; Gong, T.; Sun, X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug. Deliv. 2016, 23, 2596–2607. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, M.; Fu, Y.; Li, Y.; Gong, T.; Zhang, Z.; Sun, X. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Control Release 2016, 228, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; MacKay, J.A.; Fréchet, J.M.; Szoka, F.C. Designing dendrimers for biological applications. Nat. Biotechnol. 2005, 23, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.M.; Sunoqrot, S.; Hsu, H.J.; Bae, J.W.; Hong, S. Dendritic nanoparticles: The next generation of nanocarriers? Ther. Deliv. 2012, 3, 941–959. [Google Scholar] [CrossRef] [PubMed]
- Pourianazar, N.T.; Mutlu, P.; Gunduz, U. Bioapplications of poly(amidoamine) (PAMAM) dendrimers in nanomedicine. J. Nanopart. Res. 2014, 16, 38. [Google Scholar] [CrossRef]
- Bugno, J.; Hsu, H.J.; Hong, S. Tweaking dendrimers and dendritic nanoparticles for controlled nano-bio interactions: Potential nanocarriers for improved cancer targeting. J. Drug. Target. 2015, 23, 642–650. [Google Scholar] [CrossRef]
- Jain, K.; Kesharwani, P.; Gupta, U.; Jain, N.K. Dendrimer toxicity: Let’s meet the challenge. Int. J. Pharm. 2010, 394, 122–142. [Google Scholar] [CrossRef]
- Zhang, D.; Atochina-Vasserman, E.N.; Maurya, D.S.; Huang, N.; Xiao, Q.; Ona, N.; Liu, M.; Shahnawaz, H.; Ni, H.; Kim, K.; et al. One-Component Multifunctional Sequence-Defined Ionizable Amphiphilic Janus Dendrimer Delivery Systems for mRNA. J. Am. Chem. Soc. 2021, 143, 12315–12327. [Google Scholar] [CrossRef]
- Yang, X.; Du, H.; Liu, J.; Zhai, G. Advanced nanocarriers based on heparin and its derivatives for cancer management. Biomacromolecules 2015, 16, 423–436. [Google Scholar] [CrossRef]
- Barclay, T.G.; Day, C.M.; Petrovsky, N.; Garg, S. Review of polysaccharide particle-based functional drug delivery. Carbohydr. Polym. 2019, 221, 94–112. [Google Scholar] [CrossRef]
- Steinle, H.; Ionescu, T.M.; Schenk, S.; Golombek, S.; Kunnakattu, S.J.; Özbek, M.T.; Schlensak, C.; Wendel, H.P.; Avci-Adali, M. Incorporation of Synthetic mRNA in Injectable Chitosan-Alginate Hybrid Hydrogels for Local and Sustained Expression of Exogenous Proteins in Cells. Int. J. Mol. Sci. 2018, 19, 1313. [Google Scholar] [CrossRef]
- Yan, J.; Chen, R.; Zhang, H.; Bryers, J.D. Injectable Biodegradable Chitosan-Alginate 3D Porous Gel Scaffold for mRNA Vaccine Delivery. Macromol. Biosci. 2019, 19, e1800242. [Google Scholar] [CrossRef]
- Soliman, O.Y.; Alameh, M.G.; De Cresenzo, G.; Buschmann, M.D.; Lavertu, M. Efficiency of Chitosan/Hyaluronan-Based mRNA Delivery Systems In Vitro: Influence of Composition and Structure. J. Pharm. Sci. 2020, 109, 1581–1593. [Google Scholar] [CrossRef] [PubMed]
- Jeandupeux, E.; Alameh, M.G.; Ghattas, M.; De Crescenzo, G.; Lavertu, M. Poly(2-Propylacrylic Acid) Increases In Vitro Bioactivity of Chitosan/mRNA Nanoparticles. J. Pharm. Sci. 2021, 110, 3439–3449. [Google Scholar] [CrossRef] [PubMed]
- Sharifnia, Z.; Bandehpour, M.; Hamishehkar, H.; Mosaffa, N.; Kazemi, B.; Zarghami, N. In-vitro Transcribed mRNA Delivery Using PLGA/PEI Nanoparticles into Human Monocyte-derived Dendritic Cells. Iran. J. Pharm. Res. 2019, 18, 1659–1675. [Google Scholar] [CrossRef]
- Yoshinaga, N.; Naito, M.; Tachihara, Y.; Boonstra, E.; Osada, K.; Cabral, H.; Uchida, S. PEGylation of mRNA by Hybridization of Complementary PEG-RNA Oligonucleotides Stabilizes mRNA without Using Cationic Materials. Pharmaceutics 2021, 13, 800. [Google Scholar] [CrossRef] [PubMed]
- Jarzebska, N.T.; Lauchli, S.; Iselin, C.; French, L.E.; Johansen, P.; Guenova, E.; Kündig, T.M.; Pascolo, S. Functional differences between protamine preparations for the transfection of mRNA. Drug. Deliv. 2020, 27, 1231–1235. [Google Scholar] [CrossRef]
- Amos, H. Protamine Enhancement of RNA Uptake by Cultured Chick Cells. Biochem. Biophys. Res. Commun. 1961, 5, 1–4. [Google Scholar] [CrossRef]
- Sebastian, M.; Schröder, A.; Scheel, B.; Hong, H.S.; Muth, A.; von Boehmer, L.; Zippelius, A.; Mayer, F.; Reck, M.; Atanackovic, D.; et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol. Immunother. 2019, 68, 799–812. [Google Scholar] [CrossRef]
- Morris, M.C.; Vidal, P.; Chaloin, L.; Heitz, F.; Divita, G. A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res. 1997, 25, 2730–2736. [Google Scholar] [CrossRef]
- Ramsey, J.D.; Flynn, N.H. Cell-penetrating peptides transport therapeutics into cells. Pharmacol. Ther. 2015, 154, 78–86. [Google Scholar] [CrossRef]
- Lehto, T.; Ezzat, K.; Wood, M.J.A.; El Andaloussi, S. Peptides for nucleic acid delivery. Adv. Drug. Deliv. Rev. 2016, 106, 172–182. [Google Scholar] [CrossRef]
- van den Brand, D.; Gorris, M.A.J.; van Asbeck, A.H.; Palmen, E.; Ebisch, I.; Dolstra, H.; Hällbrink, M.; Massuger, L.; Brock, R. Peptide-mediated delivery of therapeutic mRNA in ovarian cancer. Eur. J. Pharm. Biopharm. 2019, 141, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Udhayakumar, V.K.; De Beuckelaer, A.; McCaffrey, J.; McCrudden, C.M.; Kirschman, J.L.; Vanover, D.; Van Hoecke, L.; Roose, K.; Deswarte, K.; De Geest, B.G.; et al. Arginine-Rich Peptide-Based mRNA Nanocomplexes Efficiently Instigate Cytotoxic T Cell Immunity Dependent on the Amphipathic Organization of the Peptide. Adv. Healthc. Mater. 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Rosenecker, J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene. Ther. 2017, 24, 133–143. [Google Scholar] [CrossRef]
- Gómez-Aguado, I.; Rodríguez-Castejón, J.; Vicente-Pascual, M.; Rodríguez-Gascón, A.; Solinís, M.; Del Pozo-Rodríguez, A. Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials 2020, 10, 364. [Google Scholar] [CrossRef]
- Peer, D.; Karp, J.M.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef]
- Zhang, L.; Chan, J.M.; Gu, F.X.; Rhee, J.W.; Wang, A.Z.; Radovic-Moreno, A.F.; Alexis, F.; Langer, R.; Farokhzad, O.C. Self-assembled lipid--polymer hybrid nanoparticles: A robust drug delivery platform. ACS Nano 2008, 2, 1696–1702. [Google Scholar] [CrossRef]
- Siewert, C.D.; Haas, H.; Cornet, V.; Nogueira, S.S.; Nawroth, T.; Uebbing, L.; Ziller, A.; Al-Gousous, J.; Radulescu, A.; Schroer, M.A.; et al. Hybrid Biopolymer and Lipid Nanoparticles with Improved Transfection Efficacy for mRNA. Cells 2020, 9, 2034. [Google Scholar] [CrossRef]
- Yasar, H.; Biehl, A.; De Rossi, C.; Koch, M.; Murgia, X.; Loretz, B.; Lehr, C.M. Kinetics of mRNA delivery and protein translation in dendritic cells using lipid-coated PLGA nanoparticles. J. Nanobiotechnol. 2018, 16, 72. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, J.C.; Patel, A.K.; Rhym, L.H.; Palmiero, U.C.; Bhat, B.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Systemic delivery of mRNA and DNA to the lung using polymer-lipid nanoparticles. Biomaterials 2021, 275, 120966. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Arya, S.; Lung, P.; Lin, Q.; Huang, J.; Li, Q. Hybrid nanovaccine for the co-delivery of the mRNA antigen and adjuvant. Nanoscale 2019, 11, 21782–21789. [Google Scholar] [CrossRef]
- Zhang, R.; Tang, L.; Tian, Y.; Ji, X.; Hu, Q.; Zhou, B.; Ding, Z.; Xu, H.; Yang, L. DP7-C-modified liposomes enhance immune responses and the antitumor effect of a neoantigen-based mRNA vaccine. J. Control Release 2020, 328, 210–221. [Google Scholar] [CrossRef]
- Coolen, A.L.; Lacroix, C.; Mercier-Gouy, P.; Delaune, E.; Monge, C.; Exposito, J.Y.; Verrier, B. Poly(lactic acid) nanoparticles and cell-penetrating peptide potentiate mRNA-based vaccine expression in dendritic cells triggering their activation. Biomaterials 2019, 195, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Man, R.C.H.; Liao, Q.; Kung, K.L.K.; Chow, M.Y.T.; Lam, J.K.W. Effective mRNA pulmonary delivery by dry powder formulation of PEGylated synthetic KL4 peptide. J. Control Release 2019, 314, 102–115. [Google Scholar] [CrossRef]
- Gao, Y.; Men, K.; Pan, C.; Li, J.; Wu, J.; Chen, X.; Lei, S.; Gao, X.; Duan, X. Functionalized DMP-039 Hybrid Nanoparticle as a Novel mRNA Vector for Efficient Cancer Suicide Gene Therapy. Int. J. Nanomed. 2021, 16, 5211–5232. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.Y.; Lee, T.J.; Yang, G.M.; Oh, J.; Won, J.; Han, J.; Jeong, G.J.; Kim, J.; Kim, J.H.; Kim, B.S.; et al. Efficient mRNA delivery with graphene oxide-polyethylenimine for generation of footprint-free human induced pluripotent stem cells. J. Control Release 2016, 235, 222–235. [Google Scholar] [CrossRef]
- Taton, T.A. Preparation of gold nanoparticle-DNA conjugates. Cur.r Protoc. Nucleic Acid. Chem. 2002, 12. Unit 12.12. [Google Scholar] [CrossRef]
- Zhang, X. Gold Nanoparticles: Recent Advances in the Biomedical Applications. Cell. Biochem. Biophys. 2015, 72, 771–775. [Google Scholar] [CrossRef]
- Cho, E.C.; Glaus, C.; Chen, J.; Welch, M.J.; Xia, Y. Inorganic nanoparticle-based contrast agents for molecular imaging. Trends Mol. Med. 2010, 16, 561–573. [Google Scholar] [CrossRef]
- Jeong, E.H.; Jung, G.; Hong, C.A.; Lee, H. Gold nanoparticle (AuNP)-based drug delivery and molecular imaging for biomedical applications. Arch. Pharm. Res. 2014, 37, 53–59. [Google Scholar] [CrossRef]
- Alivisatos, A.P.; Johnsson, K.P.; Peng, X.; Wilson, T.E.; Loweth, C.J.; Bruchez, M.P., Jr.; Schultz, P.G. Organization of ‘nanocrystal molecules’ using DNA. Nature 1996, 382, 609–611. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J. Am. Chem. Soc. 2012, 134, 7266–7269. [Google Scholar] [CrossRef] [PubMed]
- Talamantez-Lyburn, S.; Brown, P.; Hondrogiannis, N.; Ratliff, J.; Wicks, S.L.; Nana, N.; Zheng, Z.; Rosenzweig, Z.; Hondrogiannis, E.; Devadas, M.S.; et al. Gold nanoparticles loaded with cullin-5 DNA increase sensitivity to 17-AAG in cullin-5 deficient breast cancer cells. Int. J. Pharm. 2019, 564, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Glancy, D.; Chan, W.C. DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 2016, 351, 841–845. [Google Scholar] [CrossRef]
- Ye, W.; Li, H.; Li, X.; Fan, X.; Jin, Q.; Ji, J. mRNA Guided Intracellular Self-Assembly of DNA-Gold Nanoparticle Conjugates as a Precise Trigger to Up-Regulate Cell Apoptosis and Activate Photothermal Therapy. Bioconjug Chem. 2019, 30, 1763–1772. [Google Scholar] [CrossRef]
- Yeom, J.H.; Ryou, S.M.; Won, M.; Park, M.; Bae, J.; Lee, K. Inhibition of Xenograft tumor growth by gold nanoparticle-DNA oligonucleotide conjugates-assisted delivery of BAX mRNA. PLoS ONE 2013, 8, e75369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, F.; Han, M.; Wang, X.; Du, J.; Zhang, H.; Li, W. Co-delivery of 5-fluorodeoxyuridine and doxorubicin via gold nanoparticle equipped with affibody-DNA hybrid strands for targeted synergistic chemotherapy of HER2 overexpressing breast cancer. Sci. Rep. 2020, 10, 22015. [Google Scholar] [CrossRef]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.G. A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Kim, H.; Jang, H.; Cho, H.; Choi, J.; Hwang, K.Y.; Choi, Y.; Kim, S.H.; Yang, Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers 2021, 13, 4435. [Google Scholar] [CrossRef]
- Rosas, L.E.; Elgamal, O.A.; Mo, X.; Phelps, M.A.; Schmittgen, T.D.; Papenfuss, T.L. In vitro immunotoxicity assessment of culture-derived extracellular vesicles in human monocytes. J. Immunotoxicol. 2016, 13, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Aslan, C.; Kiaie, S.H.; Zolbanin, N.M.; Lotfinejad, P.; Ramezani, R.; Kashanchi, F.; Jafari, R. Exosomes for mRNA delivery: A novel biotherapeutic strategy with hurdles and hope. BMC Biotechnol. 2021, 21, 20. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes). Biochem. Soc. Trans. 2013, 41, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.J.; Atai, N.A.; Cacciottolo, M.; Nice, J.; Salehi, A.; Guo, C.; Sedgwick, A.; Kanagavelu, S.; Gould, S.J. Exosome-mediated mRNA Delivery in vivo is safe and can be used to induce SARS-CoV-2 immunity. J. Biol. Chem. 2021, 297, 101266. [Google Scholar] [CrossRef] [PubMed]
- Forterre, A.V.; Wang, J.H.; Delcayre, A.; Kim, K.; Green, C.; Pegram, M.D.; Jeffrey, S.S.; Matin, A.C. Extracellular Vesicle-Mediated In Vitro Transcribed mRNA Delivery for Treatment of HER2(+) Breast Cancer Xenografts in Mice by Prodrug CB1954 without General Toxicity. Mol. Cancer Ther. 2020, 19, 858–867. [Google Scholar] [CrossRef]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta. Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef]
- Kojima, R.; Bojar, D.; Rizzi, G.; Hamri, G.C.; El-Baba, M.D.; Saxena, P.; Ausländer, S.; Tan, K.R.; Fussenegger, M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018, 9, 1305. [Google Scholar] [CrossRef]
- Yang, Z.; Shi, J.; Xie, J.; Wang, Y.; Sun, J.; Liu, T.; Zhao, Y.; Zhao, X.; Wang, X.; Ma, Y.; et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020, 4, 69–83. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Palucka, K.; Banchereau, J. Dendritic-cell-based therapeutic cancer vaccines. Immunity 2013, 39, 38–48. [Google Scholar] [CrossRef]
- Diken, M.; Kreiter, S.; Selmi, A.; Britten, C.M.; Huber, C.; Türeci, Ö.; Sahin, U. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011, 18, 702–708. [Google Scholar] [CrossRef]
- Boczkowski, D.; Nair, S.K.; Snyder, D.; Gilboa, E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J. Exp. Med. 1996, 184, 465–472. [Google Scholar] [CrossRef]
- Zhang, R.; Yuan, F.; Shu, Y.; Tian, Y.; Zhou, B.; Yi, L.; Zhang, X.; Ding, Z.; Xu, H.; Yang, L. Personalized neoantigen-pulsed dendritic cell vaccines show superior immunogenicity to neoantigen-adjuvant vaccines in mouse tumor models. Cancer Immunol. Immunother. 2020, 69, 135–145. [Google Scholar] [CrossRef]
- Batich, K.A.; Reap, E.A.; Archer, G.E.; Sanchez-Perez, L.; Nair, S.K.; Schmittling, R.J.; Norberg, P.; Xie, W.; Herndon, J.E., 2nd; Healy, P.; et al. Long-term Survival in Glioblastoma with Cytomegalovirus pp65-Targeted Vaccination. Clin. Cancer Res. 2017, 23, 1898–1909. [Google Scholar] [CrossRef]
- Wang, Q.T.; Nie, Y.; Sun, S.N.; Lin, T.; Han, R.J.; Jiang, J.; Li, Z.; Li, J.Q.; Xiao, Y.P.; Fan, Y.Y.; et al. Tumor-associated antigen-based personalized dendritic cell vaccine in solid tumor patients. Cancer Immunol. Immunother. 2020, 69, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Vik-Mo, E.O.; Nyakas, M.; Mikkelsen, B.V.; Moe, M.C.; Due-Tønnesen, P.; Suso, E.M.; Sæbøe-Larssen, S.; Sandberg, C.; Brinchmann, J.E.; Helseth, E.; et al. Therapeutic vaccination against autologous cancer stem cells with mRNA-transfected dendritic cells in patients with glioblastoma. Cancer Immunol. Immunother. 2013, 62, 1499–1509. [Google Scholar] [CrossRef]
- Sahin, U.; Oehm, P.; Derhovanessian, E.; Jabulowsky, R.A.; Vormehr, M.; Gold, M.; Maurus, D.; Schwarck-Kokarakis, D.; Kuhn, A.N.; Omokoko, T.; et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 2020, 585, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Komdeur, F.L.; Singh, A.; van de Wall, S.; Meulenberg, J.J.M.; Boerma, A.; Hoogeboom, B.N.; Paijens, S.T.; Oyarce, C.; de Bruyn, M.; Schuuring, E.; et al. First-in-Human Phase I Clinical Trial of an SFV-Based RNA Replicon Cancer Vaccine against HPV-Induced Cancers. Mol. Ther. 2021, 29, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Reichmuth, A.M.; Oberli, M.A.; Jaklenec, A.; Langer, R.; Blankschtein, D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016, 7, 319–334. [Google Scholar] [CrossRef]
- Ndeupen, S.; Qin, Z.; Jacobsen, S.; Bouteau, A.; Estanbouli, H.; Igyártó, B.Z. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 2021, 24, 103479. [Google Scholar] [CrossRef] [PubMed]
- Packer, M.; Gyawali, D.; Yerabolu, R.; Schariter, J.; White, P. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat. Commun. 2021, 12, 6777. [Google Scholar] [CrossRef]
- Shepherd, S.J.; Warzecha, C.C.; Yadavali, S.; El-Mayta, R.; Alameh, M.G.; Wang, L.; Weissman, D.; Wilson, J.M.; Issadore, D.; Mitchell, M.J. Scalable mRNA and siRNA Lipid Nanoparticle Production Using a Parallelized Microfluidic Device. Nano Lett. 2021, 21, 5671–5680. [Google Scholar] [CrossRef]
- Blakney, A.K.; Deletic, P.; McKay, P.F.; Bouton, C.R.; Ashford, M.; Shattock, R.J.; Sabirsh, A. Effect of complexing lipids on cellular uptake and expression of messenger RNA in human skin explants. J. Control Release 2021, 330, 1250–1261. [Google Scholar] [CrossRef]
- Kaczmarek, J.C.; Kauffman, K.J.; Fenton, O.S.; Sadtler, K.; Patel, A.K.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Optimization of a Degradable Polymer-Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells. Nano Lett. 2018, 18, 6449–6454. [Google Scholar] [CrossRef]
- Zhang, R.; Men, K.; Zhang, X.; Huang, R.; Tian, Y.; Zhou, B.; Yu, C.; Wang, Y.; Ji, X.; Hu, Q.; et al. Delivery of a Modified mRNA Encoding IL-22 Binding Protein (IL-22BP) for Colon Cancer Gene Therapy. J. Biomed. Nanotechnol. 2018, 14, 1239–1251. [Google Scholar] [CrossRef]
- Apavaloaei, A.; Hardy, M.P.; Thibault, P.; Perreault, C. The Origin and Immune Recognition of Tumor-Specific Antigens. Cancers 2020, 12, 2607. [Google Scholar] [CrossRef]
- Lancaster, E.M.; Jablons, D.; Kratz, J.R. Applications of Next-Generation Sequencing in Neoantigen Prediction and Cancer Vaccine Development. Genet. Test. Mol. Biomarkers 2020, 24, 59–66. [Google Scholar] [CrossRef]
- Vormehr, M.; Türeci, Ö.; Sahin, U. Harnessing Tumor Mutations for Truly Individualized Cancer Vaccines. Annu. Rev. Med. 2019, 70, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L.; Demaria, S.; Rodriguez-Ruiz, M.E.; Zarour, H.M.; Melero, I. Emerging Opportunities and Challenges in Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
- Butts, C.; Socinski, M.A.; Mitchell, P.L.; Thatcher, N.; Havel, L.; Krzakowski, M.; Nawrocki, S.; Ciuleanu, T.E.; Bosquée, L.; Trigo, J.M.; et al. Tecemotide (L-BLP25) versus placebo after chemoradiotherapy for stage III non-small-cell lung cancer (START): A randomised, double-blind, phase 3 trial. Lancet. Oncol. 2014, 15, 59–68. [Google Scholar] [CrossRef]




| Vectors | Advantages | Disadvantages |
|---|---|---|
| Viral | High transfection efficiency, sustained expression of vector, the complex assembly process is completed by the cells | Elicit immune response, risk of insertion, carcinogenesis, broad tropism, limited DNA packaging capacity, difficulty of vector production |
| Nonviral | Low immunogenic response, high loading capacity, chemical design flexibility, safe and stable, high transfection efficiency, biocompatible, easier to synthesize | Increased cytotoxicity for cationic lipids short half-lives, nonspecific binding to serum proteins for cationic carrier |
| Category | Key Component | Core Technology | Size (nm) | Delivery Route | Gene | Ref |
| LNPs | Ionizable lipids, DOPE, C14-PEG2000 | Heterocyclic lipids formed by one-step three-component reaction (3-CR) | 100 | In vitro i.m. (mouse) | Fluc | [75] |
| LNPs | DOPE, DSPC, PEG ionizable lipids | Helper lipid structure | 170.5/167.2 | i.v. (mouse) | Fluc | [78] |
| LNPs | C14–4, DOPE, Chol PEG | Optimized ratios in LNPs | 57–151 | In vitro | Luc | [81] |
| LNPs | Ionizable lipid, cholesterol, DSPC, DMPE-PEG | Optimized molar ratio between ionizable lipids and mRNA | 82–90 | In vitro | Human erythropoietin | [96] |
| LNPs | Synthesized lipid, DOPE, cholesterol, DMG-PEG2k | Lipid with unsaturated tail | 143 | In vitro i.v. (mouse) | Fluc | [97] |
| LNPs | DSPC, cholesterol, MC3, PEG2k-DMG | Generated via stepwise ethanol dilution | 61 | i.v. (rat) i.v. (monkey) | hEPO | [88] |
| LNPs | Cationic lipids, DSPE-PEG 2000 | Optimized cationic lipid-like materials | 140–160 | DC-mediated (mouse) | OVA | [89] |
| Polymers | Ionizable lipids, phospholipids, PEG, cholesterol | Ionizable amphiphilic Janus dendrimer (IAJD) | 75/92 | In vitro i.p. (mouse) | Luc | [119] |
| Polymers | Alginate, chitosan | Hydrogels | Not mentioned | In vitro | hGLuc | [122] |
| Polymers | Chitosan, hyaluronic acid, trehalose | Hydrogels | 80–180 | In vitro | Luc | [124] |
| Polymers | PLGA, PEI | Nontoxic PLGA | 428.9 ± 12 | DC-based | GFP | [126] |
| Polymers | PEG | Cation-free delivery strategy with PEG | 10–90 | In vitro | GLuc | [127] |
| Polypeptide | Protamine | Natural cationic peptide | 90–180 | In vitro | Luc | [128] |
| Polypeptide | Protamine | Natural cationic peptide | 30–110 | i.d. (human) | TAA for NSCLC | [130] |
| Polypeptide | Pepfect14 | Cationic CPP | 70–110 | i.p. (3D model) | eGFP mCherry | [134] |
| Polypeptide | RALA CPP | Arginine-rich peptide | 89–144 | In vitro i.v. (mouse) | eGFP OVA | [135] |
| Hybrid | Protamine, DOTAP | Heterogenous internal organization | 146–234 | In vitro; i.m. (mouse) | Luc | [141] |
| Hybrid | PLGA, DOTMA | Lipid-coated PLGA | 231 ± 7.0 | DC-based | m-cherry | [142] |
| Hybrid | PBAE, PEG-lipid | PBAE terpolymers formulated with PEG-lipid | 200–370 | i.v. (mouse) | Luc | [143] |
| Hybrid | Lipid, PLGA | adjuvant-loaded hybrid | 300 | DC-based i.v. (mouse) | EGFP | [144] |
| Hybrid | DP-7, DOTAP, cholesterol | DP7-C with double functions | 100.23 ± 7.5 | In vitro i.v./s.c. (mouse) | eGFP neoantigen | [145] |
| Polypeptide | PLA-NP, LAH4-L1 | Cationic CPP | 220.1 ± 22.9 | In vitro | eGFP | [146] |
| Polypeptide | PEG, KL4 peptide | Monodisperse linear PEG with peptide | 467.9 ± 24.9 | In vitro i.n. (mouse) | eGFP | [147] |
| AuNP–DNA conjugates | AuNP, DNA | Targeted DNA | Not mentioned | In vitro s.c. (mouse) | BAX GFP | [159] |
| Exosome | Exosome | HEK-293F derived Exo | 120 ± 50 | i.m. (mouse) | Antares2 | [167] |
| Exosome | Exosome, EVHB | HER2-targeted peptide | Not mentioned | i.p. (mouse) | HChrR6 | [168] |
| Exosome | Exosomes | Exosome derived from RBC devoid of DNA | 100–250 | In vitro i.p., i.t. (mouse) | 125b ASO | [173] |
| Company or Institution | Delivery Vehicle | Tumor Types | Antigens | Phase | Status | NCT Number |
|---|---|---|---|---|---|---|
| Oslo University Hospital | Not mentioned | Prostate cancer | - | Phase I/II | Completed | NCT01278914 |
| Inge Marie Svane | Not mentioned | Breast cancer Malignant melanoma | Survivin, hTERT, p53 | Phase I | Completed | NCT00978913 |
| Oslo University Hospital | Not mentioned | Glioblastoma Brain tumor | Tumor stem cell | Phase I/II | Completed | NCT00846456 |
| Oslo University Hospital | Not mentioned | Prostate cancer | hTERT, Survivin | Phase I/II | Active, not recruiting | NCT01197625 |
| Radboud University | Electroporated | Uveal melanoma | Tyrosinase, gp100 | Phase I/II | Terminated | NCT00929019 |
| National Cancer Institute | Not mentioned | Leukemia | - | Phase I | Terminated | NCT00514189 |
| Steinar Aamdal | Not mentioned | Recurrent epithelial ovarian cancer | hTERT, survivin | Phase I/II | Terminated | NCT01334047 |
| Steinar Aamdal | Not mentioned | Metastatic malignant melanoma | hTERT, survivin | Phase I/II | Terminated | NCT00961844 |
| Trinomab Biotech Co., Ltd. | Not mentioned | Brain cancer, neoplasm metastases | - | Phase I | Unknown | NCT02808416 |
| University of Florida | Not mentioned | Metastatic prostate cancer | hTERT | Phase I/II | Withdrawn | NCT01153113 |
| Inge Marie Svane | Electroporated | Prostatic neoplasms | PSA, PAP, survivin, hTERT | Phase II | Completed | NCT01446731 |
| Radboud University | Electroporated | Colorectal cancer, liver Metastases | Carcinoembryonic antigen | Phase I/II | Completed | NCT00228189 |
| National Cancer Institute | Not mentioned | Recurrent central nervous, system neoplasm | Brain tumor stem cell-specific mRNA | Phase I | Completed | NCT00890032 |
| University Hospital, Antwerp | Not mentioned | Glioblastoma, renal cell carcinoma, sarcomas, breast cancers, malignant mesothelioma, colorectal tumors | WT1 protein | Phase I/II | Unknown | NCT01291420 |
| Memorial Sloan Kettering Cancer Center | Electroporated | Melanoma | Tumor-associated antigen | Phase I | Active, not recruiting | NCT01456104 |
| Affiliated Hospital to Academy of Military Medical Sciences | Not mentioned | Esophagus cancer | MUC1, survivin | Phase I/II | Unknown | NCT02693236 |
| National Cancer Institute | Not mentioned | Malignant neoplasms of brain | pp65-LAMP | Phase I | Active, not recruiting | NCT00639639 |
| University Hospital, Antwerp | Electroporated | Acute myeloid leukemia | Wilms’ tumor antigen 1 | Phase I | Completed | NCT00834002 |
| Life Research Technologies GmbH | Not mentioned | Ovarian epithelial cancer | TERT- | Phase I | Unknown | NCT01456065 |
| Memorial Sloan Kettering Cancer Center | Electroporated | Multiple myeloma | CT7, MAGE-A3, WT1 | Phase I | Active, not recruiting | NCT01995708 |
| Radboud University | Not mentioned | Melanoma stage III or IV | gp100 and tyrosinase | Phase I/II | Completed | NCT00243529 |
| Radboud University | Electroporated | Hematological malignancies | Minor histocompatibility antigens | Phase I/II | Completed | NCT02528682 |
| National Cancer Institute | Not mentioned | Malignant neoplasms brain | Cytomegalovirus (CMV) pp65-lysosome-associated membrane protein (LAMP) | Phase I | Completed | NCT00626483 |
| Zwi Berneman | Electroporated | Acute myeloid leukemia | Wilms’ tumor antigen (WT1) | Phase II | Recruiting | NCT01686334 |
| CureVac AG | Not mentioned | Non-small-cell lung cancer | - | Phase I/II | Completed | NCT00923312 |
| Radboud University | Protamine | Prostatic neoplasms | - | Phase II | Completed | NCT02692976 |
| Oslo University Hospital | Not mentioned | Glioblastoma | Survivin and hTERT | Phase II/III | Recruiting | NCT03548571 |
| Ludwig Maximilian University of Munich | Electroporated | Acute myeloid leukemia | WT1, PRAME, CMVpp65 | Phase I/II | Completed | NCT01734304 |
| University Hospital, Antwerp | Electroporated | Malignant pleural mesothelioma | Wilms’ tumor protein 1 (WT1) | Phase I/II | Recruiting | NCT02649829 |
| Asterias Biotherapeutics, Inc. | Not mentioned | Acute myelogenous leukemia | hTERT and a portion of the lysosome-associated membrane protein (LAMP-1) | Phase II | Completed | NCT00510133 |
| Oslo University Hospital | Not mentioned | Malignant melanoma | - | Phase I/II | Completed | NCT01278940 |
| Radboud University | Electroporated | Melanoma | gp100 and tyrosinase | Phase I/II | Completed | NCT01530698 |
| University Hospital, Antwerp | Electroporated | High-grade glioma, diffuse intrinsic pontine glioma | WT1 | Phase I/II | Recruiting | NCT04911621 |
| Radboud University | Electroporated | Melanoma | gp100 and tyrosinase | Phase I/II | Completed | NCT00940004 |
| Bart Neyns | Electroporated | Melanoma | - | Phase I | Completed | NCT01066390 |
| Radboud University | Electroporated | Melanoma | gp100 and tyrosinase | Phase II | Completed | NCT02285413 |
| Immunomic Therapeutics, Inc. | Electroporated | Glioblastoma multiforme, glioblastoma, malignant glioma, astrocytoma, grade IVGBM | pp65-shLAMP | Phase II | Recruiting | NCT02465268 |
| University Hospital, Antwerp | Electroporated | Glioblastoma multiforme of brain | WT1 | Phase I/II | Recruiting | NCT02649582 |
| Gary Archer, Ph.D. | Not mentioned | Glioblastoma | Human CMV pp65-LAMP | Phase II | Recruiting | NCT03688178 |
| University Hospital, Antwerp | Electroporated | Acute myeloid leukemia | WT1 | Phase I | Completed | NCT00834002 |
| Gary Archer, Ph.D. | Electroporated | Glioblastoma | Human CMV pp65-LAMP | Phase II | Suspended | NCT03927222 |
| Universitair Ziekenhuis Brussel | Electroporated | Malignant melanoma | - | Phase II | Completed | NCT01676779 |
| Guangdong 999 Brain Hospital | Not mentioned | Glioblastoma | Personalized TAAs | Phase I | Active, Not recruiting | NCT02808364 |
| Company or Institution | Delivery Vehicle | Tumor Types | Antigens | Phase | Status | NCT Number |
|---|---|---|---|---|---|---|
| CureVac AG | Protamine | Non-small-cell lung cancer | MUC1, survivin, NY-ESO-1, 5T4, MAGE-C2, MAGE-C1 | Phase I/II | Completed | NCT03164772 |
| University Hospital Tuebingen | Not mentioned | Malignant melanoma | Melan-A, Mage-A1, Mage-A3, Survivin, GP100, tyrosinase | Phase I/II | Completed | NCT00204516 |
| University Hospital Tuebingen | Protamine | Malignant melanoma | Melan-A, Mage-A1, Mage-A3, Survivin, GP100, and tyrosinase | Phase I/II | Completed | NCT00204607 |
| Merck | Not mentioned | Non-small-cell lung cancer, pancreatic neoplasms, colorectal neoplasms | mRNA-5671/V941 | Phase I | Recruiting | NCT03948763 |
| BioNTech | Lipid-based vector | Ovarian cancer | Ova | Phase I | Recruiting | NCT04163094 |
| Moderna | Lipid-based vector | Melanoma | Personalized mRNA-4157 | Phase II | Recruiting | NCT03897881 |
| Moderna | Lipid-based vector | Solid tumors | Personalized mRNA-4157 | Phase I | Recruiting | NCT03313778 |
| BioNTech | Lipid-based vector | Prostate cancer | W_pro1 | Phase I/II | Recruiting | NCT04382898 |
| Universitair Ziekenhuis Brussel | Naked mRNA | Early-stage breast cancer | - | Phase I | Recruiting | NCT03788083 |
| National Cancer Institute | Lipid-based vector | Melanoma, colon cancer, gastrointestinal cancer, genitourinary cancer, hepatocellular cancer | Personalized mRNA | Phase I/II | Terminated | NCT03480152 |
| University Hospital Tuebingen | Protamine | Recurrent prostate cancer | - | Phase I/II | Unknown | NCT02452307 |
| University of Florida | Lipid-based vector | Adult glioblastoma | Autologous total tumor and pp65 full-length (fl) lysosome-associated membrane protein (LAMP) | Phase I | Recruiting | NCT04573140 |
| Moderna | Lipid-based vector | Relapsed/refractory solid tumor malignancies or lymphoma | human OX40L | Phase I/II | Active, not recruiting | NCT03323398 |
| BioNTech | Lipid-based vector | Melanoma | NY-ESO-1, tyrosinase, MAGE-A3, TPTE | Phase I | Active, not recruiting | NCT02410733 |
| BioNTech SE | Lipid-based vector | Breast cancer | Tumor relevant and immunogenic RNA | Phase I | Active, not recruiting | NCT02316457 |
| CureVac AG | Peptide-based vector | Non-small-cell lung cancer | CV9201 | Phase I/II | Completed | NCT00923312 |
| CureVac AG | Peptide-based vector | Non-small-cell lung cancer | CV9202 | Phase I | Terminated | NCT01915524 |
| CureVac AG | Peptide-based vector | Prostate cancer | CV9103 | Phase I/II | Completed | NCT00831467 |
| CureVac AG | Peptide-based vector | Prostate cancer | CV9103 | Phase I/II | Terminated | NCT00906243 |
| CureVac AG | Peptide-based vector | Prostate cancer | CV9104 | Phase I/II | Terminated | NCT01817738 |
| CureVac AG | Peptide-based vector | Prostate cancer | CV9104 | Phase II | Terminated | NCT02140138 |
| Moderna | Lipid-based vector | Solid tumor malignancies or lymphoma | mRNA-2752 | Phase I | Recruiting | NCT03739931 |
| Stemirna Therapeutics | Not mentioned | Esophageal cancer, non-small-cell lung cancer | Personalized mRNA | Not applicable | Not yet recruiting | NCT03908671 |
| Changhai Hospital | Not mentioned | Advanced Esophageal Squamous; Carcinoma; Gastric Adenocarcinoma, pancreatic adenocarcinoma, colorectal adenocarcinoma | Personalized mRNA | Not applicable | Recruiting | NCT03468244 |
| Laura Esserman | Not mentioned | Carcinoma, intraductal, noninfiltrating | mRNA 2752 | Phase I | Recruiting | NCT02872025 |
| University Medical Center Groningen | Not mentioned | Cervical cancer | HPV-derived tumor antigens | Phase I | Completed | NCT03141463 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, R.; Tang, L.; Yang, L. Nonviral Delivery Systems of mRNA Vaccines for Cancer Gene Therapy. Pharmaceutics 2022, 14, 512. https://doi.org/10.3390/pharmaceutics14030512
Wang Y, Zhang R, Tang L, Yang L. Nonviral Delivery Systems of mRNA Vaccines for Cancer Gene Therapy. Pharmaceutics. 2022; 14(3):512. https://doi.org/10.3390/pharmaceutics14030512
Chicago/Turabian StyleWang, Yusi, Rui Zhang, Lin Tang, and Li Yang. 2022. "Nonviral Delivery Systems of mRNA Vaccines for Cancer Gene Therapy" Pharmaceutics 14, no. 3: 512. https://doi.org/10.3390/pharmaceutics14030512
APA StyleWang, Y., Zhang, R., Tang, L., & Yang, L. (2022). Nonviral Delivery Systems of mRNA Vaccines for Cancer Gene Therapy. Pharmaceutics, 14(3), 512. https://doi.org/10.3390/pharmaceutics14030512





