Isolation and Molecular Characterization of a Novel Lytic Bacteriophage That Inactivates MDR Klebsiella pneumoniae Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Antibiotic Susceptibility Assays
2.3. Phage Enrichment, Isolation and Enumeration
2.4. PEG Precipitation of Phage Suspensions
2.5. Transmission Electron Microscopy (TEM)
2.6. Purification of Phage DNA and Whole Genome Sequencing
2.7. Phage Genome Assembly and Annotation
2.8. Proteome-Based Clustering and Phylogenetic Analysis
2.9. Evaluation of the Host Range
2.10. One-Step Growth Curve (OSGC)
2.11. Adsorption Rate
2.12. Bacterial Kill Assay
2.13. UV-Vis Spectral Scans
2.14. X-ray Diffraction Analysis
2.15. Statistical Analyses
3. Results
3.1. Phage Isolation and Purification
3.2. Virion Morphology
3.3. Genomic Characterization of Phage Kpn31
3.4. Host Range of Phage Kpn31
3.5. Determination of Burst Size and Phage Adsorption Rate
3.6. In Vitro Inactivation of K. Pneumoniae CCCD-K001
3.7. Physicochemical Characterization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Ayobami, O.; Brinkwirth, S.; Eckmanns, T.; Markwart, R. Antibiotic resistance in hospital-acquired eskape-e infections in low- and lower-middle-income countries: A systematic review and meta-analysis. Emerg. Microbes Infect. 2022, 11, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Rolain, J.-M. Investigation of antibiotic resistance in the genomic era of multidrug-resistant Gram-negative bacilli, especially Enterobacteriaceae, Pseudomonas and Acinetobacter. Expert Rev. Anti-Infect. Ther. 2013, 11, 277–296. [Google Scholar] [CrossRef] [PubMed]
- Ashurst, J.V.; Dawson, A. Klebsiella Pneumonia. Available online: https://www.ncbi.nlm.nih.gov/books/NBK519004/ (accessed on 5 June 2022).
- Chang, D.; Sharma, L.; Cruz, C.S.D.; Zhang, D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front. Microbiol. 2021, 12, 750662. [Google Scholar] [CrossRef]
- Bengoechea, J.A.; Pessoa, J.S. Klebsiella pneumoniaeinfection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2018, 43, 123–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, P.Y. The emerging problems of Klebsiella pneumoniae infections: Carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 2016, 363, fnw219. [Google Scholar] [CrossRef] [Green Version]
- Wyres, K.; Holt, K. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Giuliano, S.; Ceccarelli, G.; Alessandri, F.; Giordano, A.; Brunetti, G.; Venditti, M. Comparison of Septic Shock Due to Multidrug-Resistant Acinetobacter baumannii or Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae in Intensive Care Unit Patients. Antimicrob. Agents Chemother. 2018, 62, e02562-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agodi, A.; Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Aldisio, E.; Marchese, A.E.; Mattaliano, A.R.; Tsakris, A. Antibiotic trends of Klebsiella pneumoniae and Acinetobacter baumannii resistance indicators in an intensive care unit of Southern Italy, 2008–2013. Antimicrob. Resist. Infect. Control 2015, 4, 43. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat Eskape Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; García, P.; Drulis-Kawa, Z. Targeting biofilms using phages and their enzymes. Curr. Opin. Biotechnol. 2021, 68, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 2022, 13, 302. [Google Scholar] [CrossRef]
- Fang, Q.; Feng, Y.; McNally, A.; Zong, Z. Characterization of phage resistance and phages capable of intestinal decolonization of carbapenem-resistant Klebsiella pneumoniae in mice. Commun. Biol. 2022, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Chadha, P.; Katare, O.P.; Chhibber, S. Liposome loaded phage cocktail: Enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections. Burns 2017, 43, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-H.; Kuo, C.-F.; Wang, C.-H.; Wu, C.-M.; Tsao, N. Experimental Phage Therapy in Treating Klebsiella pneumoniae -Mediated Liver Abscesses and Bacteremia in Mice. Antimicrob. Agents Chemother. 2011, 55, 1358–1365. [Google Scholar] [CrossRef] [Green Version]
- Nir-Paz, R.; Gelman, D.; Khouri, A.; Sisson, B.M.; Fackler, J.; Alkalay-Oren, S.; Khalifa, L.; Rimon, A.; Yerushalmy, O.; Bader, R.; et al. Successful Treatment of Antibiotic-resistant, Poly-microbial Bone Infection with Bacteriophages and Antibiotics Combination. Clin. Infect. Dis. 2019, 69, 2015–2018. [Google Scholar] [CrossRef]
- Singh, A.; Singh, A.N.; Rathor, N.; Chaudhry, R.; Singh, S.K.; Nath, G. Evaluation of Bacteriophage Cocktail on Septicemia Caused by Colistin-Resistant Klebsiella pneumoniae in Mice Model. Front. Pharmacol. 2022, 13, 778676. [Google Scholar] [CrossRef]
- Fayez, M.S.; Hakim, T.A.; Agwa, M.M.; Abdelmoteleb, M.; Aly, R.G.; Montaser, N.N.; Abdelsattar, A.S.; Rezk, N.; El-Shibiny, A. Topically Applied Bacteriophage to Control Multi-Drug Resistant Klebsiella pneumoniae Infected Wound in a Rat Model. Antibiotics 2021, 10, 1048. [Google Scholar] [CrossRef]
- Cano, E.J.; Caflisch, K.M.; Bollyky, P.L.; Van Belleghem, J.D.; Patel, R.; Fackler, J.; Brownstein, M.J.; Horne, B.; Biswas, B.; Henry, M.; et al. Phage Therapy for Limb-threatening Prosthetic Knee Klebsiella pneumoniae Infection: Case Report and In Vitro Characterization of Anti-biofilm Activity. Clin. Infect. Dis. 2020, 73, e144–e151. [Google Scholar] [CrossRef]
- Hesse, S.; Malachowa, N.; Porter, A.R.; Freedman, B.; Kobayashi, S.D.; Gardner, D.J.; Scott, D.P.; Adhya, S.; DeLeo, F.R. Bacteriophage Treatment Rescues Mice Infected with Multidrug-Resistant Klebsiella pneumoniae ST258. MBio 2021, 12, e00034-21. [Google Scholar] [CrossRef]
- Herridge, W.P.; Shibu, P.; O’Shea, J.; Brook, T.C.; Hoyles, L. Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J. Med. Microbiol. 2020, 69, 176–194. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, L.K.; Silva, E.C.; Rossi, F.P.; Cieza, B.; Oliveira, T.J.; Pereira, C.; Tomazetto, G.; Silva, B.B.; Squina, F.M.; Vila, M.M.; et al. Characterization and in vitro testing of newly isolated lytic bacteriophages for the biocontrol of Pseudomonas aeruginosa. Future Microbiol. 2022, 17, 111–141. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.C.; Oliveira, T.J.; Moreli, F.C.; Harada, L.K.; Vila, M.M.; Balcão, V.M. Newly isolated lytic bacteriophages for Staphylococcus intermedius, structurally and functionally stabilized in a hydroxyethylcellulose gel containing choline geranate: Potential for transdermal permeation in veterinary phage therapy. Res. Vet. Sci. 2020, 135, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers: London, UK, 1959. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Liang, C. Generic Repeat Finder: A High-Sensitivity Tool for Genome-Wide De Novo Repeat Detection. Plant Physiol. 2019, 180, 1803–1815. [Google Scholar] [CrossRef]
- Grazziotin, A.L.; Koonin, E.V.; Kristensen, D.M. Prokaryotic Virus Orthologous Groups (pVOGs): A resource for comparative genomics and protein family annotation. Nucleic Acids Res. 2016, 45, D491–D498. [Google Scholar] [CrossRef]
- Terzian, P.; Olo Ndela, E.; Galiez, C.; Lossouarn, J.; Pérez Bucio, R.E.; Mom, R.; Toussaint, A.; Petit, M.-A.; Enault, F. PHROG: Families of prokaryotic virus proteins clustered using remote homology. NAR Genom. Bioinform. 2021, 3, lqab067. [Google Scholar] [CrossRef] [PubMed]
- Abid, D.; Zhang, L. DeepCapTail: A Deep Learning Framework to Predict Capsid and Tail Proteins of Phage Ge-nomes. bioRxiv 2018, 477885. [Google Scholar] [CrossRef] [Green Version]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Cook, R.; Brown, N.; Redgwell, T.; Rihtman, B.; Barnes, M.; Clokie, M.; Stekel, D.J.; Hobman, J.; Jones, M.A.; Millard, A. INfrastructure for a PHAge REference Database: Identification of Large-Scale Biases in the Current Collection of Cultured Phage Genomes. Phage 2021, 2, 214–223. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [Green Version]
- Bin Jang, H.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R.; et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Takesue, S.; Ishibashi, K.; Nakahara, S. A Computer Simulation of the Adsorption ofLactobacillusPhage PL-1 to Host Cells: Some Factors Affecting the Process. Agric. Biol. Chem. 1982, 46, 697–702. [Google Scholar] [CrossRef]
- Moldovan, R.; Chapman-McQuiston, E.; Wu, X. On Kinetics of Phage Adsorption. Biophys. J. 2007, 93, 303–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storms, Z.J.; Sauvageau, D. Modeling tailed bacteriophage adsorption: Insight into mechanisms. Virology 2015, 485, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Rios, A.C.; Vila, M.M.; Lima, R.; Del Fiol, F.S.; Tubino, M.; Teixeira, J.A.; Balcão, V.M. Structural and functional stabilization of bacteriophage particles within the aqueous core of a W/O/W multiple emulsion: A potential biotherapeutic system for the inhalational treatment of bacterial pneumonia. Process Biochem. 2018, 64, 177–192. [Google Scholar] [CrossRef] [Green Version]
- Scherrer, P. Bestimmung der Größe und der inneren Struktur von Kolloidteilchen mittels Röntgenstrahlen. Nachr. Von Der Ges. Der Wiss. Zu Göttingen 1918, 1918, 98–100. [Google Scholar]
- Zinke, M.; Schröder, G.F.; Lange, A. Major tail proteins of bacteriophages of the order Caudovirales. J. Biol. Chem. 2021, 298, 101472. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Li, P.; Jiang, X.; Bi, D.; Xie, Y.; Tai, C.; Deng, Z.; Rajakumar, K.; Ou, H.-Y. Complete Genome Sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a Multidrug-Resistant Strain Isolated from Human Sputum. J. Bacteriol. 2012, 194, 1841–1842. [Google Scholar] [CrossRef] [Green Version]
- Xing, S.; Pan, X.; Sun, Q.; Pei, G.; An, X.; Mi, Z.; Huang, Y.; Zhao, B.; Tong, Y. Complete Genome Sequence of a Novel Multidrug-Resistant Klebsiella pneumoniae Phage, vB_Kpn_IME260. Genome Announc. 2017, 5, e00055-17. [Google Scholar] [CrossRef] [Green Version]
- Erickson, S.G.; Lessor, L.; O’Leary, C.; Gill, J.J.; Liu, M. Complete Genome Sequence of Klebsiella pneumoniae Siphophage Sugarland. Microbiol. Resour. Announc. 2018, 7, e01014–e01018. [Google Scholar] [CrossRef] [Green Version]
- Zurabov, F.; Zhilenkov, E. Complete Genome Sequences of Lytic Polysaccharide-Degrading Klebsiella pneumoniae Bacteriophages vB_KpnS_FZ10, vB_KpnP_FZ12, vB_KpnM_FZ14, and vB_KpnS_FZ41. Microbiol. Resour. Announc. 2019, 8, e00914–e00919. [Google Scholar] [CrossRef] [Green Version]
- Yerushalmy, O.; Coppenhagen-Glazer, S.; Nir-Paz, R.; Tuomala, H.; Skurnik, M.; Kiljunen, S.; Hazan, R. Complete Genome Sequences of Two Klebsiella pneumoniae Phages Isolated as Part of an International Effort. Microbiol. Resour. Announc. 2019, 8, e00843-19. [Google Scholar] [CrossRef] [Green Version]
- Venturini, C.; Ben Zakour, N.L.; Bowring, B.; Morales, S.; Cole, R.; Kovach, Z.; Branston, S.; Kettle, E.; Thomson, N.; Iredell, J.R. Fine capsule variation affects bacteriophage susceptibility in Klebsiella pneumoniae ST258. FASEB J. 2020, 34, 10801–10817. [Google Scholar] [CrossRef]
- Luo, Z.; Geng, S.; Lu, B.; Han, G.; Wang, Y.; Luo, Y.; Yang, Z.; Cao, S.; Yao, X. Isolation, Genomic Analysis, and Preliminary Application of a Bovine Klebsiella pneumoniae Bacteriophage vB_Kpn_B01. Front. Vet. Sci. 2021, 8, 622049. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of Phage φ6 for Biocontrol of Pseudomonas syringae pv. syringae: An in Vitro Preliminary Study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbonyiryivuze, A.; Omollo, I.; Ngom, B.D.; Mwakikunga, B.; Dhlamini, S.M.; Park, E.; Maaza, M. Natural Dye Sensitizer for Grӓtzel Cells: Sepia Melanin. Phys. Mater. Chem. 2015, 3, 1–6. [Google Scholar] [CrossRef]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and molecular perspectives. J. Intern. Med. 2019, 287, 283–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.; Alam, M.; Hasan, G.M.; Hassan, I. Potential therapeutic targets of Klebsiella pneumoniae: A multi-omics review perspective. Brief. Funct. Genom. 2021, 21, 63–77. [Google Scholar] [CrossRef]
- Kęsik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dąbrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Łusiak-Szelachowska, M.; Żaczek, M.; Górski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, 100. [Google Scholar] [CrossRef] [Green Version]
- Townsend, E.M.; Kelly, L.; Gannon, L.; Muscatt, G.; Dunstan, R.; Michniewski, S.; Sapkota, H.; Kiljunen, S.J.; Kolsi, A.; Skurnik, M.; et al. Isolation and Characterization of Klebsiella Phages for Phage Therapy. Phage 2021, 2, 26–42. [Google Scholar] [CrossRef]
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2018, 53, 16–21. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Hughes, K.; Sutherland, I.; Clark, J.; Jones, M. Bacteriophage and associated polysaccharide depolymerases—novel tools for study of bacterial biofilms. J. Appl. Microbiol. 1998, 85, 583–590. [Google Scholar] [CrossRef]
- Rios, A.C.; Moutinho, C.G.; Pinto, F.C.; Del Fiol, F.S.; Jozala, A.; Chaud, M.V.; Vila, M.M.; Teixeira, J.A.; Balcão, V.M. Alternatives to overcoming bacterial resistances: State-of-the-art. Microbiol. Res. 2016, 191, 51–80. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, C.; Harper, D.; Burch, D.; Änggård, E.; Soothill, J. Topical treatment of Pseudomonas aeruginosa otitis of dogs with a bacteriophage mixture: A before/after clinical trial. Vet. Microbiol. 2010, 146, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Characterization and in vitro evaluation of new bacteriophages for the biocontrol of Escherichia coli. Virus Res. 2017, 227, 171–182. [Google Scholar] [CrossRef]
- Kutter, E. Phage Host Range and Efficiency of Plating. In Bacteriophages; Humana Press: New York, NY, USA, 2009; Volume 501, pp. 141–149. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Zurabov, F.; Zhilenkov, E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol. J. 2021, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Li, Z.; Wang, S.; Wang, S.; Huang, D.; Li, Y.; Ma, Y.; Wang, J.; Liu, F.; Chen, X.; et al. Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 2012, 12, 156. [Google Scholar] [CrossRef] [Green Version]
- Ceyssens, P.-J. Isolation and Characterization of Lytic Bacteriophages Infecting Pseudomonas Aeruginosa; Katholieke Inuversiteit Leuven: Leuven, Belgium, 2009. [Google Scholar]
- Hyman, P.; Abedon, S.T. Practical methods for determining phage growth parameters. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; Volume 501, pp. 175–202. ISBN 9781603271646. [Google Scholar]
- Shao, Y.; Wang, I.-N. Bacteriophage Adsorption Rate and Optimal Lysis Time. Genetics 2008, 180, 471–482. [Google Scholar] [CrossRef] [Green Version]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological applications of bacteriophages: State of the art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef]
- Rakhuba, D.V.; Kolomiets, E.I.; Szwajcer Dey, E.; Novik, G.I. Bacteriophage receptors, mechanisms of phage ad-sorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Bull, J.J.; Gill, J.J. The habits of highly effective phages: Population dynamics as a framework for identifying therapeutic phages. Front. Microbiol. 2014, 5, 618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamal, M.; Hussain, T.; Das, C.R.; Andleeb, S. Characterization of Siphoviridae phage Z and studying its efficacy against multidrug-resistant Klebsiella pneumoniae planktonic cells and biofilm. J. Med. Microbiol. 2015, 64, 454–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateus, L.; Costa, L.; Silva, Y.; Pereira, C.; Cunha, A.; Almeida, A. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 2014, 424–425, 167–173. [Google Scholar] [CrossRef]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Cunha, Â.; Delgadillo, I.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Bacteriophages with potential to inactivate Salmonella Typhimurium: Use of single phage suspensions and phage cocktails. Virus Res. 2016, 220, 179–192. [Google Scholar] [CrossRef]
- Lima, R.; Del Fiol, F.S.; Balcão, V.M. Prospects for the Use of New Technologies to Combat Multidrug-Resistant Bacteria. Front. Pharmacol. 2019, 10, 692. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Liu, X.; Li, Y.; Han, W.; Lei, L.; Yang, Y.; Zhao, H.; Gao, Y.; Song, J.; Lu, R.; et al. A Method for Generation Phage Cocktail with Great Therapeutic Potential. PLoS ONE 2012, 7, e31698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chadha, P.; Katare, O.P.; Chhibber, S. In vivo efficacy of single phage versus phage cocktail in resolving burn wound infection in BALB/c mice. Microb. Pathog. 2016, 99, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Gomes, A.T.P.C.; Almeida, A. Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella Typhimurium: An In Vitro Preliminary Study. Microorganisms 2019, 7, 94. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, L.A.M.; Pereira, C.; Barreal, M.E.; Gallego, P.P.; Balcão, V.M.; Almeida, A. Use of phage ϕ6 to inactivate Pseudomonas syringae pv. actinidiae in kiwifruit plants: In vitro and ex vivo experiments. Appl. Microbiol. Biotechnol. 2019, 104, 1319–1330. [Google Scholar] [CrossRef]
- Emeaden, S.; Ekoskella, B. Exploring the risks of phage application in the environment. Front. Microbiol. 2013, 4, 358. [Google Scholar] [CrossRef] [Green Version]
- Ross, A.; Ward, S.; Hyman, P. More Is Better: Selecting for Broad Host Range Bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyman, P.; Abedon, S.T. Bacteriophage Host Range and Bacterial Resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]


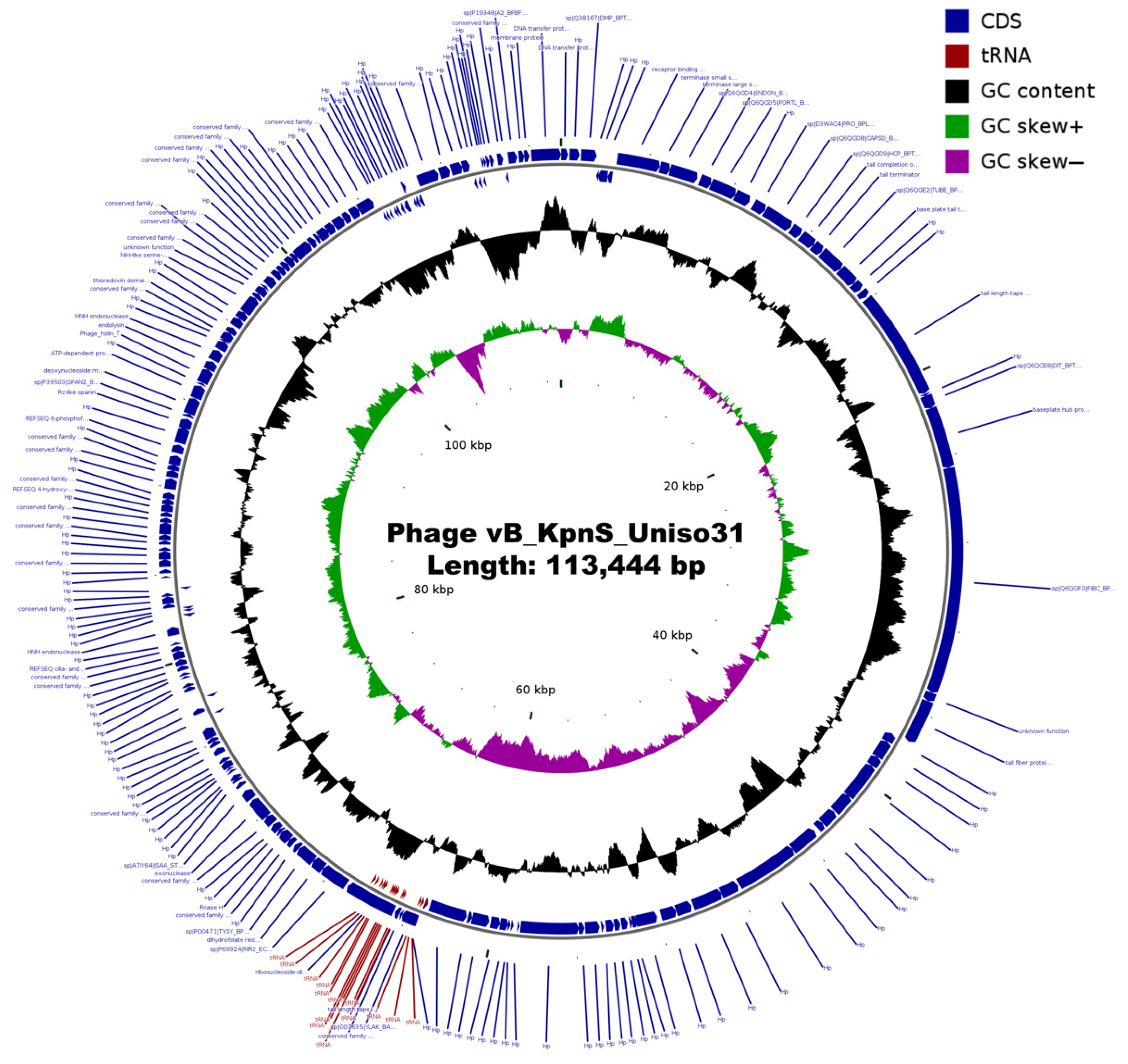
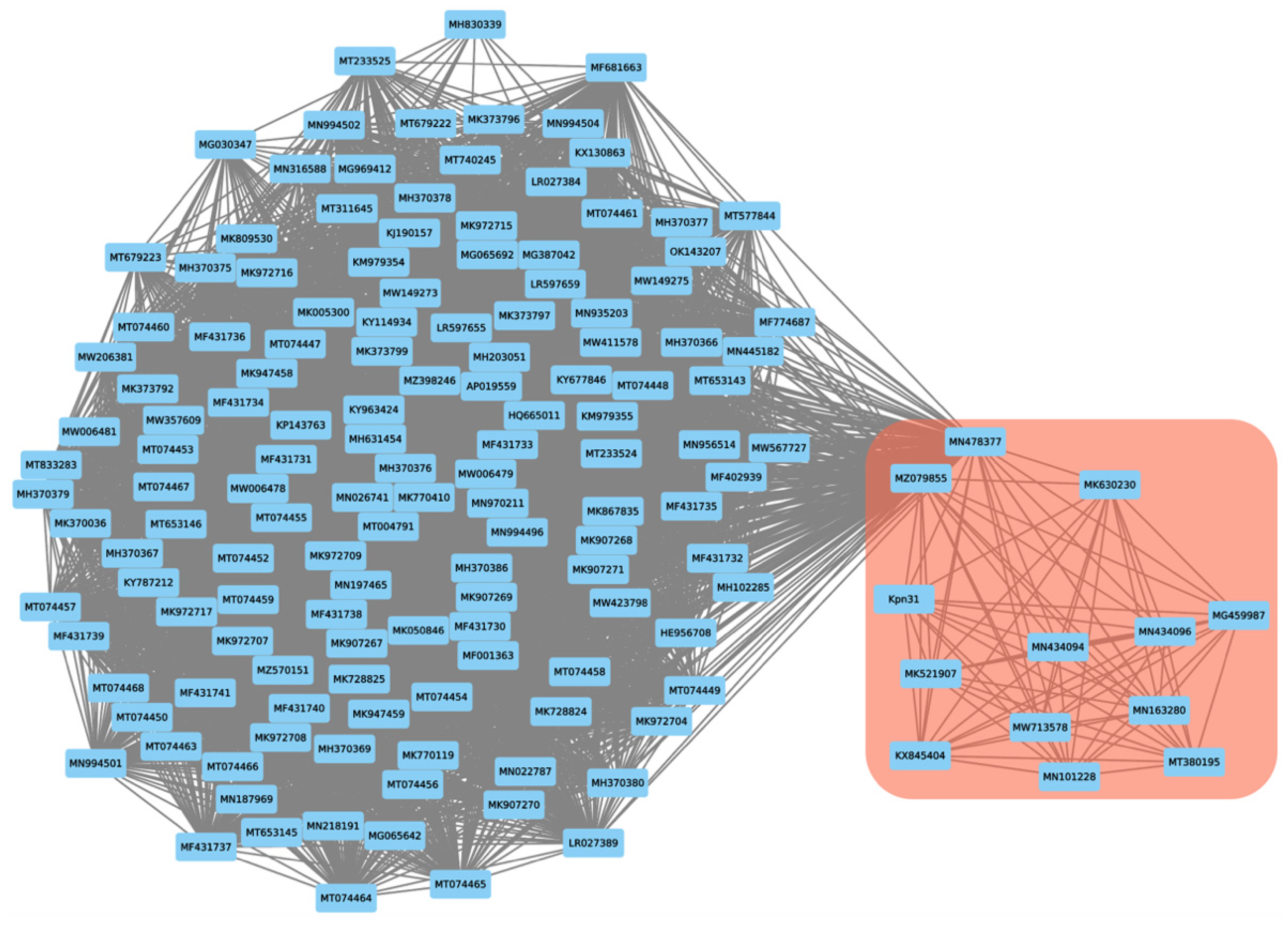


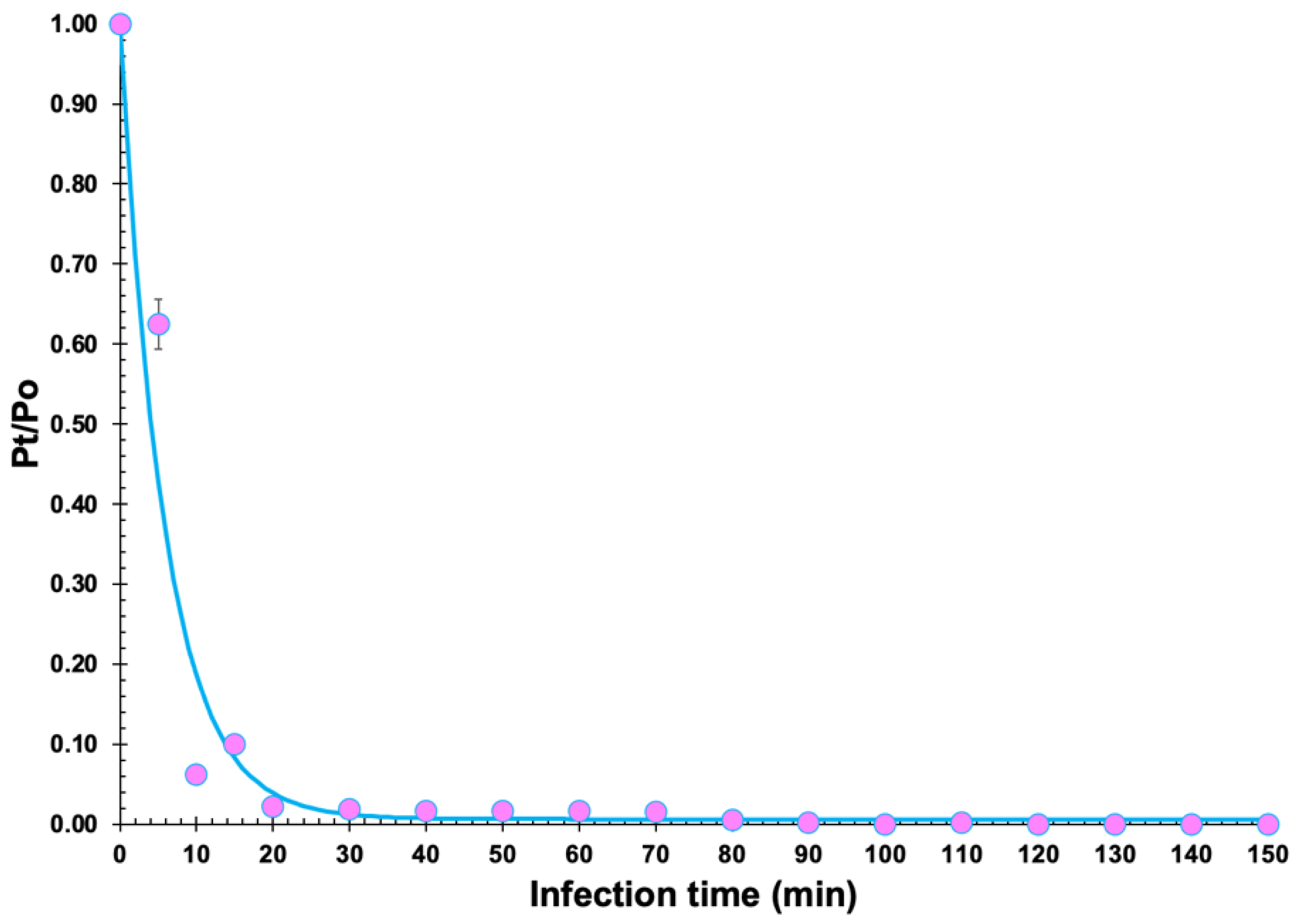

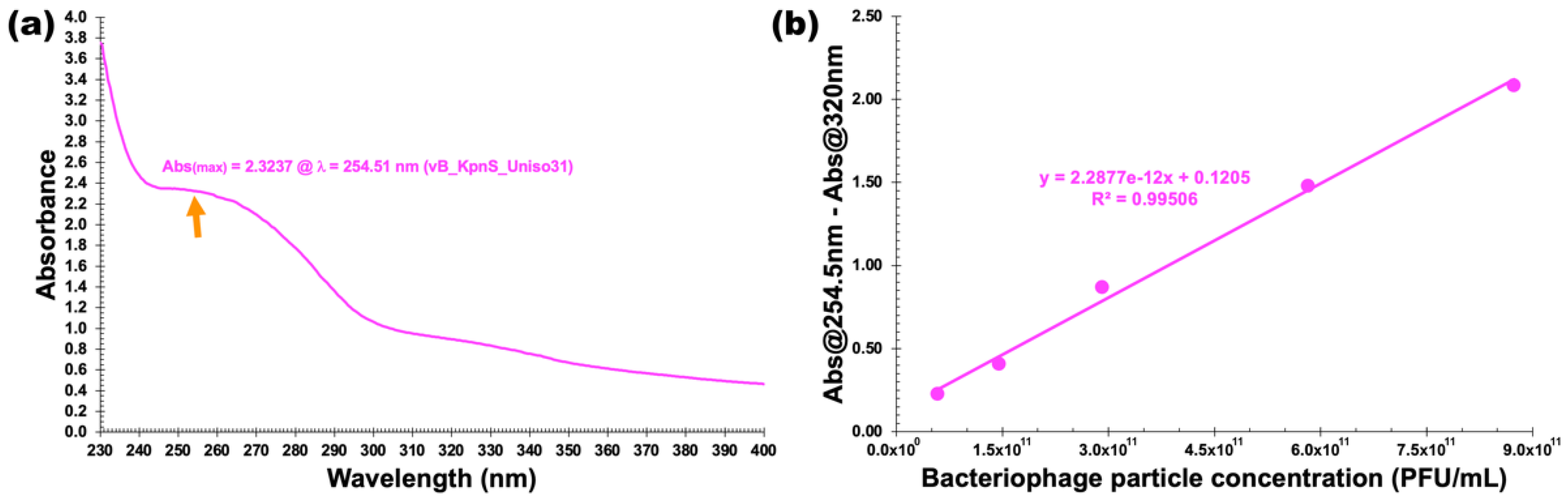

| Feature | Phage vB_KpnS_Uniso31 (Phage Kpn31) |
|---|---|
| NCBI/Genbank accession number | ON637170 |
| Genome size | 113,444 bp |
| Number of PE reads in the final assembly | 491,866 |
| Average sequencing coverage | 1000× |
| GC content | 45.3% |
| tRNA genes | 14 |
| Protein-coding genes (CDS) predicted | 188 |
| 44/188 (23.4%) |
| 144/188 (76.6%) |
| 54/144 (37.5%) |
| NCBI Accession | Description | Genome Size (bp) | % Nucleotide Identity | Query Cover | Reference |
|---|---|---|---|---|---|
| Relative to Knp31 Genome | |||||
| KX845404 | Klebsiella phage vB_Kpn_IME260 | 123,490 | 95.89 | 95 | [49] |
| MG459987 | Klebsiella phage Sugarland | 111,103 | 96.72 | 85 | [50] |
| MK521907 | Klebsiella phage vB_KpnS_FZ41 | 106,104 | 95.74 | 87 | [51] |
| MK630230 | Klebsiella phage Spivey | 110,659 | 93.84 | 84 | unpublished |
| MN101228 | Klebsiella phage KPN4 | 108,916 | 96.82 | 89 | unpublished |
| MN163280 | Klebsiella phage KpGranit | 122,710 | 96.68 | 95 | [52] |
| MN434094 | Klebsiella phage AmPh_EK80 | 112,215 | 95.82 | 82 | [53] |
| MN434096 | Klebsiella phage JIPh_Kp127 | 113,671 | 95.35 | 93 | [53] |
| MN478377 | Bacteriophage Eos | 109,015 | 76.89 | 24 | unpublished |
| MT380195 | Klebsiella phage vB_Kpn_B01 | 113,227 | 96.14 | 96 | [54] |
| MW713578 | Klebsiella phage Shaphc-TDM-1124-4 | 112,841 | 95.93 | 96 | unpublished |
| MZ079855 | Klebsiella phage vB_Kpn_3 | 112,003 | 89.56 | 77 | unpublished |
| Bacterial Strains | Source | Phage Kpn31 | |
|---|---|---|---|
| Spot Test | EOP (%) | ||
| Escherichia coli CCCD—E003 | Collection | − | n.d. |
| Salmonella enterica CCCD—S004 | Collection | − | n.d. |
| Pseudomonas aeruginosa CCCD—P004 | Collection | − | n.d. |
| Proteus mirabilis CCCD—P001 | Collection | − | n.d. |
| Enterococcus faecalis CCCD—E002 | Collection | − | n.d. |
| Bacillus subtilis CCCD—B010 | Collection | − | n.d. |
| Staphylococcus epidermidis CCCD—S010 | Collection | − | n.d. |
| Staphylococcus aureus CCCD—S009 | Collection | − | n.d. |
| Acinetobacter baumanii ATCC 16606 | Collection | − | n.d. |
| K. pneumoniae CCCD—K001 | Collection (Urine) | + | 100 |
| K. pneumoniae strain 1 | Anal swab | + | 84.64 |
| K. pneumoniae strain 2 | Urine | + | 71.23 |
| K. pneumoniae strain 3 | Urine | − | n.d. |
| K. pneumoniae strain 4 | Anal swab | + | 89.21 |
| K. pneumoniae strain 5 | Urine | + | 70.18 |
| K. pneumoniae strain 6 | Urine | − | n.d. |
| K. pneumoniae strain 7 | Urine | − | n.d. |
| K. pneumoniae strain 8 | Anal swab | + | 45.36 |
| K. pneumoniae strain 9 | Anal swab | − | n.d. |
| K. pneumoniae strain 10 | Anal swab | + | 9.54 |
| K. pneumoniae strain 12 | Femur secretion | + | 0.90 |
| K. pneumoniae strain 13 | Urine | + | 59.64 |
| K. pneumoniae strain 14 | Urine | − | n.d. |
| K. pneumoniae strain 15 | Hemoculture | − | n.d. |
| K. pneumoniae strain 16 | Catheter tip | + | 12.89 |
| K. pneumoniae strain 17 | Urine | − | n.d. |
| K. pneumoniae strain 18 | Anal swab | − | n.d. |
| K. pneumoniae strain 19 | Urine | − | n.d. |
| K. pneumoniae strain 20 | Urine | − | n.d. |
| K. pneumoniae strain 21 | Anal swab | − | n.d. |
| K. pneumoniae strain 22 | Urine | − | n.d. |
| K. pneumoniae strain 23 | Anal swab | − | n.d. |
| K. pneumoniae strain 24 | Anal swab | + | 55.12 |
| K. pneumoniae strain 26 | Urine | − | n.d. |
| K. pneumoniae strain 29 | Anal swab | + | 23.66 |
| K. pneumoniae strain 30 | Anal swab | − | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balcão, V.M.; Moreli, F.C.; Silva, E.C.; Belline, B.G.; Martins, L.F.; Rossi, F.P.N.; Pereira, C.; Vila, M.M.D.C.; da Silva, A.M. Isolation and Molecular Characterization of a Novel Lytic Bacteriophage That Inactivates MDR Klebsiella pneumoniae Strains. Pharmaceutics 2022, 14, 1421. https://doi.org/10.3390/pharmaceutics14071421
Balcão VM, Moreli FC, Silva EC, Belline BG, Martins LF, Rossi FPN, Pereira C, Vila MMDC, da Silva AM. Isolation and Molecular Characterization of a Novel Lytic Bacteriophage That Inactivates MDR Klebsiella pneumoniae Strains. Pharmaceutics. 2022; 14(7):1421. https://doi.org/10.3390/pharmaceutics14071421
Chicago/Turabian StyleBalcão, Victor M., Fernanda C. Moreli, Erica C. Silva, Bianca G. Belline, Layla F. Martins, Fernando P. N. Rossi, Carla Pereira, Marta M. D. C. Vila, and Aline M. da Silva. 2022. "Isolation and Molecular Characterization of a Novel Lytic Bacteriophage That Inactivates MDR Klebsiella pneumoniae Strains" Pharmaceutics 14, no. 7: 1421. https://doi.org/10.3390/pharmaceutics14071421







