Mimicking the Intestinal Host–Pathogen Interactions in a 3D In Vitro Model: The Role of the Mucus Layer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Fabrication of Villus-like Hydrogel Scaffolds
2.3. Cell Culture
2.4. Intestinal Epithelial Barrier Characterization
2.5. Cell Morphological Evaluation
2.6. PAS Staining
2.7. Bacterial Strains and Growth Conditions
2.8. Bacterial Adhesion and Invasion Assays
2.9. Scanning Electron Microscopy
2.10. Effect of Bacterial Infection on the Intestinal Barrier
2.11. Simulated Intestinal Fluid (SIF)
2.12. Statistics
3. Results
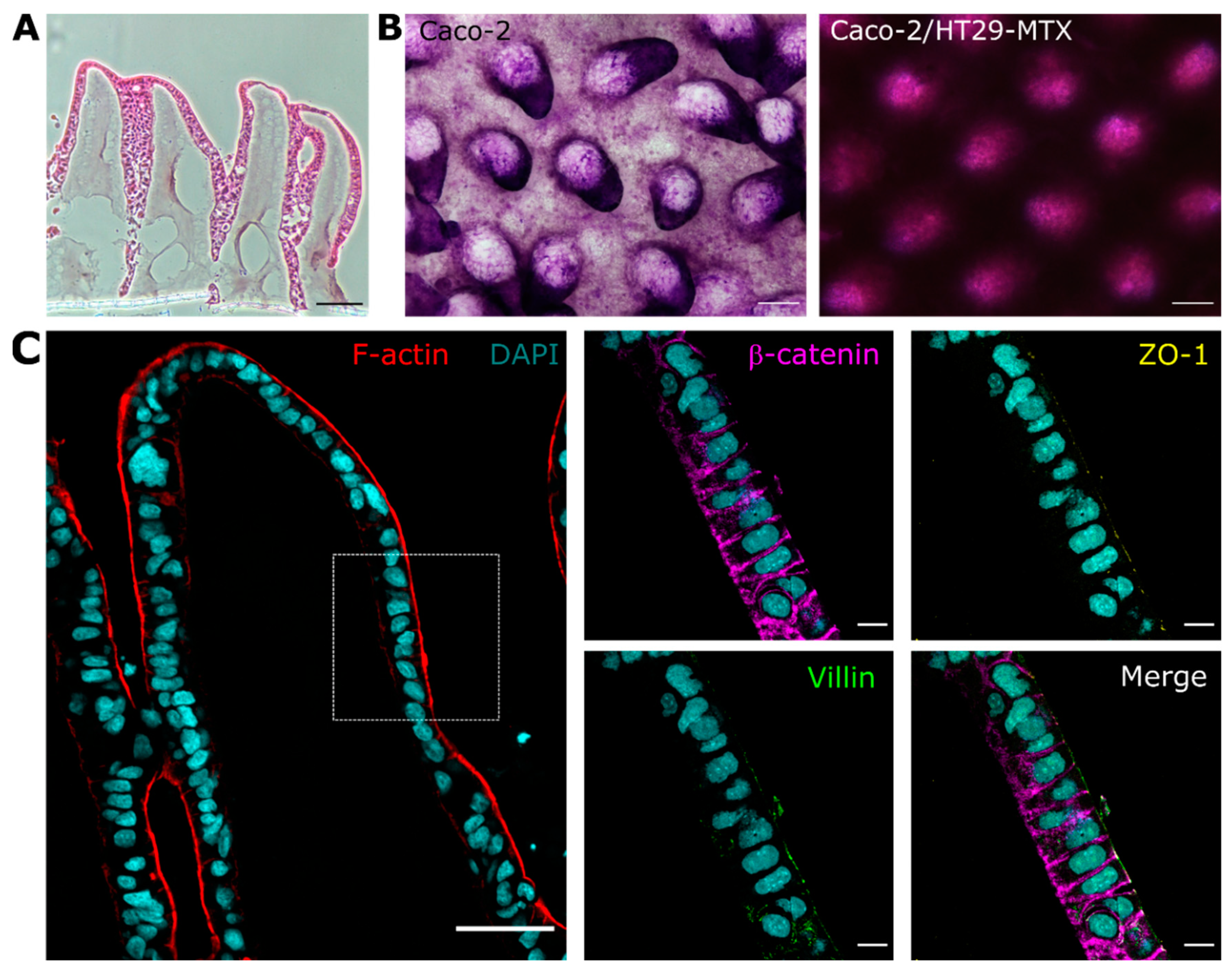
3.1. Cells Grown on 3D Villus-like Scaffolds Mimic the Native Intestinal Mucosal Barrier
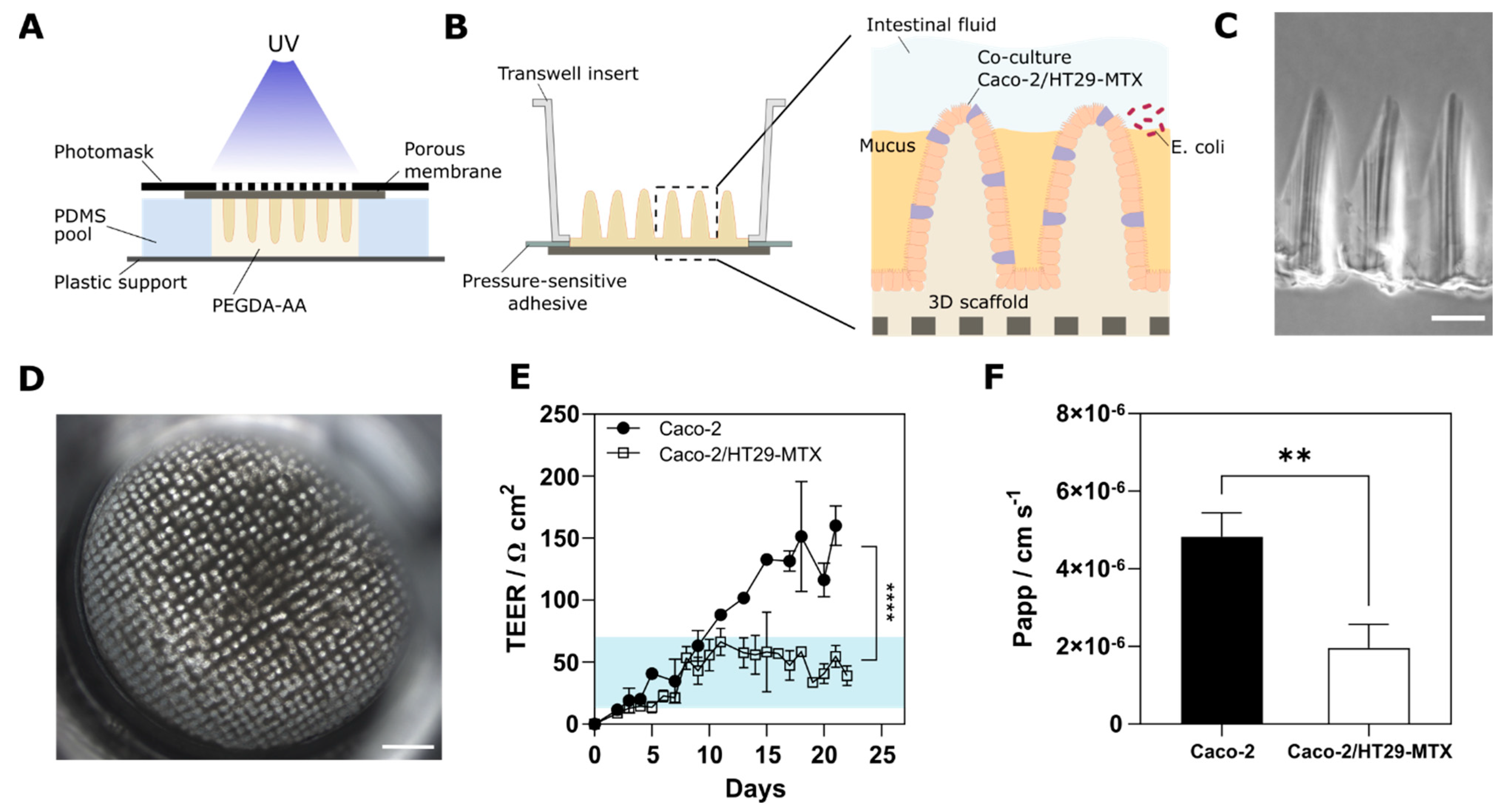
3.2. The Mucus Layer Acts as a Protective Barrier against Bacterial Invasion
3.3. The Infection with Crohn’s Disease-Associated E. coli LF82 Shows Hallmarks of the Inflamed Intestinal Mucosa
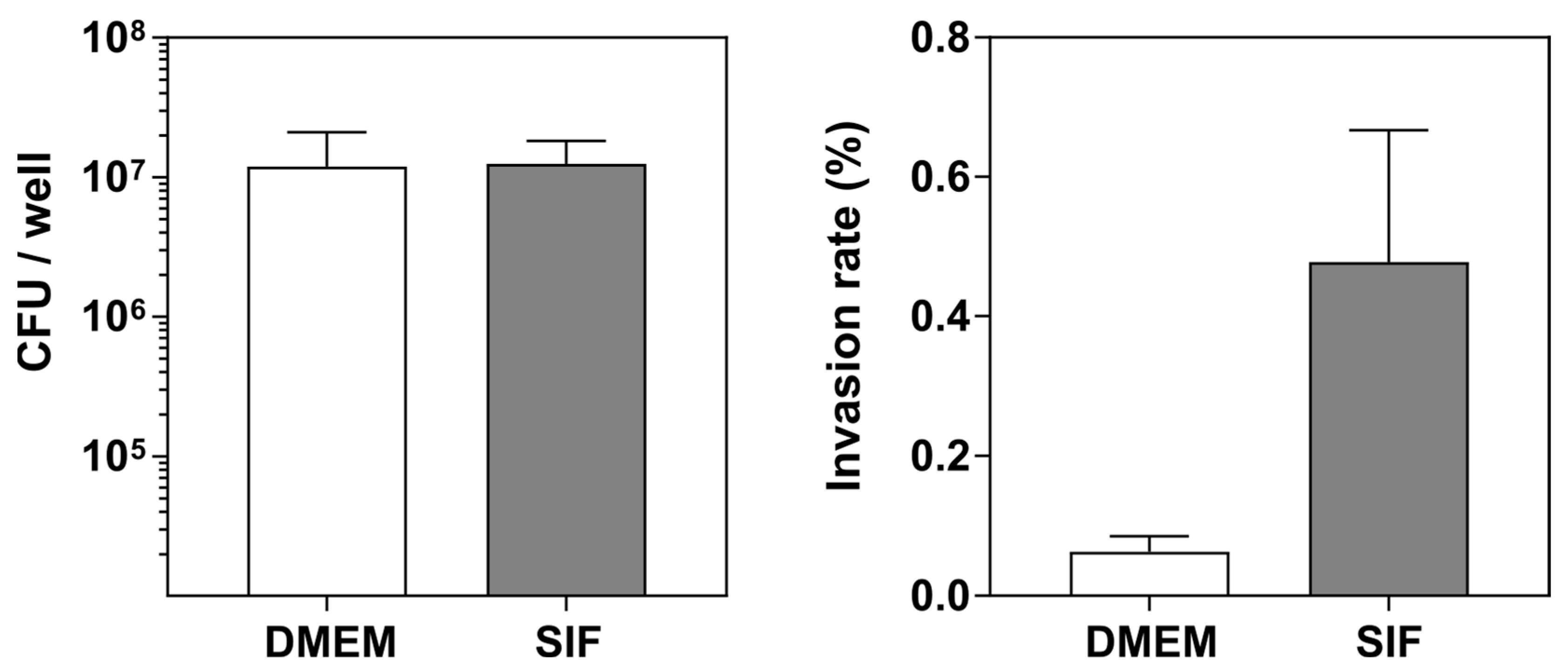
3.4. The Luminal Microenvironment Modulates E. coli LF82 Invasion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turner, J.R. Intestinal Mucosal Barrier Function in Health and Disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef] [PubMed]
- De Medina, F.S.; Romero-Calvo, I.; Mascaraque, C.; Martínez-Augustin, O. Intestinal Inflammation and Mucosal Barrier Function. Inflamm. Bowel Dis. 2014, 20, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Boegh, M.; Nielsen, H.M. Mucus as a Barrier to Drug Delivery-Understanding and Mimicking the Barrier Properties. Basic Clin. Pharmacol. Toxicol. 2014, 116, 179–186. [Google Scholar] [CrossRef]
- García-Díaz, M.; Birch, D.; Wan, F.; Nielsen, H.M. The Role of Mucus as an Invisible Cloak to Transepithelial Drug Delivery by Nanoparticles. Adv. Drug Deliv. Rev. 2018, 124, 107–124. [Google Scholar] [CrossRef]
- Sicard, J.F.; Bihan, G.L.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell. Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of Commensal and Pathogenic Microorganisms with the Intestinal Mucosal Barrier. Nat. Rev. Microbiol. 2018, 16, 457–470. [Google Scholar] [CrossRef]
- McGuckin, M.A.; Lindén, S.K.; Sutton, P.; Florin, T.H.; Linden, S.K.; Sutton, P.; Florin, T.H. Mucin Dynamics and Enteric Pathogens. Nat. Rev. Microbiol. 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V. Mucus Layers in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2014, 20, 2124–2131. [Google Scholar] [CrossRef] [PubMed]
- Kinross, J.M.; Darzi, A.W.; Nicholson, J.K. The Medical Importance of the Human Microbiome. Genome Med. 2011, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etienne-Mesmin, L.; Chassaing, B.; Desvaux, M.; De Paepe, K.; Gresse, R.; Sauvaitre, T.; Forano, E.; Van De Wiele, T.; Schüller, S.; Juge, N.; et al. Experimental Models to Study Intestinal Microbes–Mucus Interactions in Health and Disease. FEMS Microbiol. Rev. 2019, 43, 457–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiesler, P.; Fuss, I.J.; Strober, W. Experimental Models of Inflammatory Bowel Diseases. Med. Hyg. 2001, 59, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, G.S.; Park, M.H.; Shin, W.; Zhao, C.; Sheikh, S.; Oh, S.J.; Kim, H.J. Emulating Host-Microbiome Ecosystem of Human Gastrointestinal Tract in Vitro. Stem Cell Rev. Rep. 2017, 13, 321–334. [Google Scholar] [CrossRef]
- Blutt, S.E.; Crawford, S.E.; Ramani, S.; Zou, W.Y.; Estes, M.K. Engineered Human Gastrointestinal Cultures to Study the Microbiome and Infectious Diseases. Cmgh 2018, 5, 241–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torras, N.; García-Díaz, M.; Fernández-Majada, V.; Martínez, E. Mimicking Epithelial Tissues in Three-Dimensional Cell Culture Models. Front. Bioeng. Biotechnol. 2018, 6, 197. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Peng, S.; Luo, D.; March, J.C. In Vitro 3D Human Small Intestinal Villous Model for Drug Permeability Determination. Biotechnol. Bioeng. 2012, 109, 2173–2178. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Chi, M.; Yi, B.; Kim, S.H.; Oh, S.; Kim, Y.; Park, S.; Sung, J.H. Three-Dimensional Intestinal Villi Epithelium Enhances Protection of Human Intestinal Cells from Bacterial Infection by Inducing Mucin Expression. Integr. Biol. 2014, 6, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 2015, 14, 20–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urdaneta, V.; Casadesús, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef] [PubMed]
- Castaño, A.G.; García-Díaz, M.; Torras, N.; Altay, G.; Comelles, J.; Martínez, E. Dynamic Photopolymerization Produces Complex Microstructures on Hydrogels in a Moldless Approach to Generate a 3D Intestinal Tissue Model. Biofabrication 2019, 11, 025007. [Google Scholar] [CrossRef] [Green Version]
- Altay, G.; Tosi, S.; García-Díaz, M.; Martínez, E. Imaging the Cell Morphological Response to 3D Topography and Curvature in Engineered Intestinal Tissues. Front. Bioeng. Biotechnol. 2020, 8, 294. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.L.; Barnich, N.; Bringer, M.A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.F. High Prevalence of Adherent-Invasive Escherichia Coli Associated with Ileal Mucosa in Crohn’s Disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Boudeau, J.; Glasser, A.L.; Masseret, E.; Joly, B.; Darfeuille-Michaud, A. Invasive Ability of an Escherichia Coli Strain Isolated from the Ileal Mucosa of a Patient with Crohn’s Disease. Infect. Immun. 1999, 67, 4499–4509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darfeuille-Michaud, A. Adherent-Invasive Escherichia Coli: A Putative New E. Coli Pathotype Associated with Crohn’s Disease. Int. J. Med. Microbiol. 2002, 292, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Palmela, C.; Chevarin, C.; Xu, Z.; Torres, J.; Sevrin, G.; Hirten, R.; Barnich, N.; Ng, S.C.; Colombel, J.-F. Adherent-Invasive Escherichia Coli in Inflammatory Bowel Disease. Gut 2018, 67, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Gibold, L.; Garenaux, E.; Dalmasso, G.; Gallucci, C.; Cia, D.; Mottet-Auselo, B.; Faïs, T.; Darfeuille-Michaud, A.; Nguyen, H.T.T.; Barnich, N.; et al. The Vat-AIEC Protease Promotes Crossing of the Intestinal Mucus Layer by Crohn’s Disease-Associated Escherichia Coli. Cell. Microbiol. 2016, 18, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Barnich, N.; Glasser, A.; Darcha, C.; Jantscheff, P.; Allez, M.; Peeters, H.; Bommelaer, G.; Desreumaux, P.; Darfeuille-michaud, A. CEACAM6 Acts as a Receptor for Adherent-Invasive. J. Clin. Investig. 2007, 117, 1566–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Migliore, F.; Macchi, R.; Landini, P.; Paroni, M. Phagocytosis and Epithelial Cell Invasion by Crohn’s Disease-Associated Adherent-Invasive Escherichia Coli Are Inhibited by the Anti-Inflammatory Drug 6-Mercaptopurine. Front. Microbiol. 2018, 9, 964. [Google Scholar] [CrossRef] [PubMed]
- Dreux, N.; Cendra, M.d.M.; Massier, S.; Darfeuille-Michaud, A.; Barnich, N.; Torrents, E. Ribonucleotide Reductase NrdR as a Novel Regulator for Motility and Chemotaxis during Adherent-Invasive Escherichia Coli Infection. Infect. Immun. 2015, 83, 1305–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Mar Cendra, M.; Torrents, E. Differential Adaptability between Reference Strains and Clinical Isolates of Pseudomonas Aeruginosa into the Lung Epithelium Intracellular Lifestyle. Virulence 2020, 11, 862–876. [Google Scholar] [CrossRef]
- Hilgendorf, C.; Spahn-Langguth, H.; Regårdh, C.G.; Lipka, E.; Amidon, G.L.; Langguth, P. Caco-2 versus Caco-2/HT29-MTX Co-Cultured Cell Lines: Permeabilities via Diffusion, inside- and Outside-Directed Carrier-Mediated Transport. J. Pharm. Sci. 2000, 89, 63–75. [Google Scholar] [CrossRef]
- Araújo, F.; Sarmento, B. Towards the Characterization of an in Vitro Triple Co-Culture Intestine Cell Model for Permeability Studies. Int. J. Pharm. 2013, 458, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Le Ferrec, E.; Chesne, C.; Artusson, P.; Brayden, D.; Fabre, G.; Gires, P.; Guillou, F.; Rousset, M.; Rubas, W.; Scarino, M.-L. In Vitro Models of the Intestinal Barrier. Altern. Lab. Anim. 2001, 29, 649–668. [Google Scholar] [CrossRef]
- Araújo, F.; Pereira, C.; Costa, J.; Barrias, C.; Granja, P.L.; Sarmento, B. In Vitro M-like Cells Genesis through a Tissue-Engineered Triple-Culture Intestinal Model. J. Biomed. Mater. Res.-Part B Appl. Biomater. 2016, 104, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Araújo, F.; Barrias, C.C.; Granja, P.L.; Sarmento, B. Dissecting Stromal-Epithelial Interactions in a 3D Invitro Cellularized Intestinal Model for Permeability Studies. Biomaterials 2015, 56, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.; Nassra, M.; Atgié, C.; Plaisancié, P.; Géloën, A. Oleic Acid Uptake Reveals the Rescued Enterocyte Phenotype of Colon Cancer Caco-2 by HT29-MTX Cells in Co-Culture Mode. Int. J. Mol. Sci. 2017, 18, 1573. [Google Scholar] [CrossRef]
- Juge, N. Microbial Adhesins to Gastrointestinal Mucus. Trends Microbiol. 2012, 20, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Sevrin, G.; Massier, S.; Chassaing, B.; Agus, A.; Delmas, J.; Denizot, J.; Billard, E.; Barnich, N. Adaptation of Adherent-Invasive E. Coli to Gut Environment: Impact on Flagellum Expression and Bacterial Colonization Ability. Gut Microbes 2018, 11, 364–380. [Google Scholar] [CrossRef] [PubMed]
- Gokulan, K.; Williams, K.; Orr, S.; Khare, S. Human Intestinal Tissue Explant Exposure to Silver Nanoparticles Reveals Sex Dependent Alterations in Inflammatory Responses and Epithelial Cell Permeability. Int. J. Mol. Sci. 2021, 22, 9. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Mehandru, S.; Colombel, J.F.; Peyrin-Biroulet, L. Crohn’s Disease. Lancet 2017, 389, 1741–1755. [Google Scholar] [CrossRef]
- Eaves-Pyles, T.; Allen, C.A.; Taormina, J.; Swidsinski, A.; Tutt, C.B.; Eric Jezek, G.; Islas-Islas, M.; Torres, A.G. Escherichia Coli Isolated from a Crohn’s Disease Patient Adheres, Invades, and Induces Inflammatory Responses in Polarized Intestinal Epithelial Cells. Int. J. Med. Microbiol. 2008, 298, 397–409. [Google Scholar] [CrossRef]
- Wine, E.; Ossa, J.C.; Gray-Owen, S.D.; Sherman, P.M. Adherent-Invasive Escherichia Coli, Strain LF82 Disrupts Apical Junctional Complexes in Polarized Epithelia. BMC Microbiol. 2009, 9, 180. [Google Scholar] [CrossRef] [Green Version]
- Shawki, A.; McCole, D.F. Mechanisms of Intestinal Epithelial Barrier Dysfunction by Adherent-Invasive Escherichia Coli. Cmgh 2017, 3, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzarella, G.; Perna, A.; Marano, A.; Lucariello, A.; Rotondi Aufiero, V.; Sorrentino, A.; Melina, R.; Guerra, G.; Taccone, F.S.; Iaquinto, G.; et al. Pathogenic Role of Associated Adherent-Invasive Escherichia Coli in Crohn’s Disease. J. Cell. Physiol. 2017, 232, 2860–2868. [Google Scholar] [CrossRef]
- Holm, R.; Mullertz, A.; Mu, H. Bile Salts and Their Importance for Drug Absorption. Int. J. Pharm. 2013, 453, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Grüner, N.; Mattner, J. Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver–Gut Axis. Int. J. Mol. Sci. 2021, 22, 1397. [Google Scholar] [CrossRef]
- Chassaing, B.; Etienne-Mesmin, L.; Bonnet, R.; Darfeuille-Michaud, A. Bile Salts Induce Long Polar Fimbriae Expression Favouring Crohn’s Disease-Associated Adherent-Invasive Escherichia Coli Interaction with Peyer’s Patches. Environ. Microbiol. 2013, 15, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Lind, M.L.; Jacobsen, J.; Holm, R.; Müllertz, A. Development of Simulated Intestinal Fluids Containing Nutrients as Transport Media in the Caco-2 Cell Culture Model: Assessment of Cell Viability, Monolayer Integrity and Transport of a Poorly Aqueous Soluble Drug and a Substrate of Efflux Mechanisms. Eur. J. Pharm. Sci. 2007, 32, 261–270. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal Epithelial Cells: Regulators of Barrier Function and Immune Homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. Paneth Cells, Antimicrobial Peptides and Maintenance of Intestinal Homeostasis. Nat. Rev. Microbiol. 2011, 9, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Clevers, H. Organoids and Organs-on-Chips: Insights into Human Gut-Microbe Interactions. Cell Host Microbe 2021, 29, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Bartfeld, S.; Bayram, T.; Van De Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef] [Green Version]
- Mayorgas, A.; Dotti, I.; Martínez-Picola, M.; Esteller, M.; Bonet-Rossinyol, Q.; Ricart, E.; Salas, A.; Martínez-Medina, M. A Novel Strategy to Study the Invasive Capability of Adherent-Invasive Escherichia Coli by Using Human Primary Organoid-Derived Epithelial Monolayers. Front. Immunol. 2021, 12, 646906. [Google Scholar] [CrossRef]
- Nickerson, K.P.; Llanos-Chea, A.; Ingano, L.; Serena, G.; Miranda-Ribera, A.; Perlman, M.; Lima, R.; Sztein, M.B.; Fasano, A.; Senger, S.; et al. A Versatile Human Intestinal Organoid-Derived Epithelial Monolayer Model for the Study of Enteric Pathogens. Microbiol. Spectr. 2021, 9, e00003-21. [Google Scholar] [CrossRef] [PubMed]
- Roodsant, T.; Navis, M.; Aknouch, I.; Renes, I.B.; van Elburg, R.M.; Pajkrt, D.; Wolthers, K.C.; Schultsz, C.; van der Ark, K.C.H.; Sridhar, A.; et al. A Human 2D Primary Organoid-Derived Epithelial Monolayer Model to Study Host-Pathogen Interaction in the Small Intestine. Front. Cell. Infect. Microbiol. 2020, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Bozzetti, V.; Senger, S. Organoid Technologies for the Study of Intestinal Microbiota–Host Interactions. Trends Mol. Med. 2022, 28, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O. Fight Them or Feed Them: How the Intestinal Mucus Layer Manages the Gut Microbiota. Gastroenterol. Rep. 2019, 7, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lechanteur, A.; das Neves, J.; Sarmento, B. The Role of Mucus in Cell-Based Models Used to Screen Mucosal Drug Delivery. Adv. Drug Deliv. Rev. 2018, 124, 50–63. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kim, R.; Sims, C.E.; Allbritton, N.L. Building a Thick Mucus Hydrogel Layer to Improve the Physiological Relevance of In Vitro Primary Colonic Epithelial Models. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 653–655.e5. [Google Scholar] [CrossRef] [Green Version]
- Krndija, D.; El Marjou, F.; Guirao, B.; Richon, S.; Leroy, O.; Bellaiche, Y.; Hannezo, E.; Matic Vignjevic, D. Active Cell Migration Is Critical for Steady-State Epithelial Turnover in the Gut. Science 2019, 365, 705–710. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Y.; Davis, K.M.; Wang, Q.; Rnjak-Kovacina, J.; Li, C.; Isberg, R.R.; Kumamoto, C.A.; Mecsas, J.; Kaplan, D.L. Robust Bioengineered 3D Functional Human Intestinal Epithelium. Sci. Rep. 2015, 5, 13708. [Google Scholar] [CrossRef]
- Cornick, S.; Tawiah, A.; Chadee, K.; Cornick, S.; Tawiah, A.; Chadee, K. Roles and Regulation of the Mucus Barrier in the Gut Roles and Regulation of the Mucus Barrier in the Gut. Tissue Barriers 2015, 3, e982426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Raman, V.; Lee, J.; Forbes, N.S. Mucus Blocks Probiotics but Increases Penetration of Motile Pathogens and Induces TNF-α and IL-8 Secretion. Biotechnol. Bioeng. 2020, 117, 2540–2555. [Google Scholar] [CrossRef] [PubMed]
- Furter, M.; Sellin, M.E.; Hansson, G.C.; Hardt, W.D. Mucus Architecture and Near-Surface Swimming Affect Distinct Salmonella Typhimurium Infection Patterns along the Murine Intestinal Tract. Cell Rep. 2019, 27, 2665–2678.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alzheimer, M.; Svensson, S.L.; König, F.; Schweinlin, M.; Metzger, M.; Walles, H.; Sharma, C.M. A Three-Dimensional Intestinal Tissue Model Reveals Factors and Small Regulatory RNAs Important for Colonization with Campylobacter Jejuni. PLoS Pathog. 2020, 16, e1008304. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Díaz, M.; Cendra, M.d.M.; Alonso-Roman, R.; Urdániz, M.; Torrents, E.; Martínez, E. Mimicking the Intestinal Host–Pathogen Interactions in a 3D In Vitro Model: The Role of the Mucus Layer. Pharmaceutics 2022, 14, 1552. https://doi.org/10.3390/pharmaceutics14081552
García-Díaz M, Cendra MdM, Alonso-Roman R, Urdániz M, Torrents E, Martínez E. Mimicking the Intestinal Host–Pathogen Interactions in a 3D In Vitro Model: The Role of the Mucus Layer. Pharmaceutics. 2022; 14(8):1552. https://doi.org/10.3390/pharmaceutics14081552
Chicago/Turabian StyleGarcía-Díaz, María, Maria del Mar Cendra, Raquel Alonso-Roman, María Urdániz, Eduard Torrents, and Elena Martínez. 2022. "Mimicking the Intestinal Host–Pathogen Interactions in a 3D In Vitro Model: The Role of the Mucus Layer" Pharmaceutics 14, no. 8: 1552. https://doi.org/10.3390/pharmaceutics14081552
APA StyleGarcía-Díaz, M., Cendra, M. d. M., Alonso-Roman, R., Urdániz, M., Torrents, E., & Martínez, E. (2022). Mimicking the Intestinal Host–Pathogen Interactions in a 3D In Vitro Model: The Role of the Mucus Layer. Pharmaceutics, 14(8), 1552. https://doi.org/10.3390/pharmaceutics14081552







