Use of Nanoparticles to Prevent Resistance to Antibiotics—Synthesis and Characterization of Gold Nanosystems Based on Tetracycline
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Bacterial Lines and Culture Conditions
2.1.2. Synthesis of 1,3-Propanediyl-bis-(dimethylhexadecylammonium bromide), 16-3-16
2.1.3. Synthesis of Au@16-3-16 Gold Nanoparticles
2.1.4. Synthesis of Au@16-3-16/DNA-TC Nanocomplexes
2.2. Methods
2.2.1. UV/Vis Spectroscopy
2.2.2. Circular Dichroism (CD) Spectroscopy
2.2.3. Atomic Force Microscopy (AFM) Experiments
2.2.4. Dynamic Light Scattering (DLS) and Zeta Potential Measurements
2.2.5. Transmission Electron Microscopy (TEM)
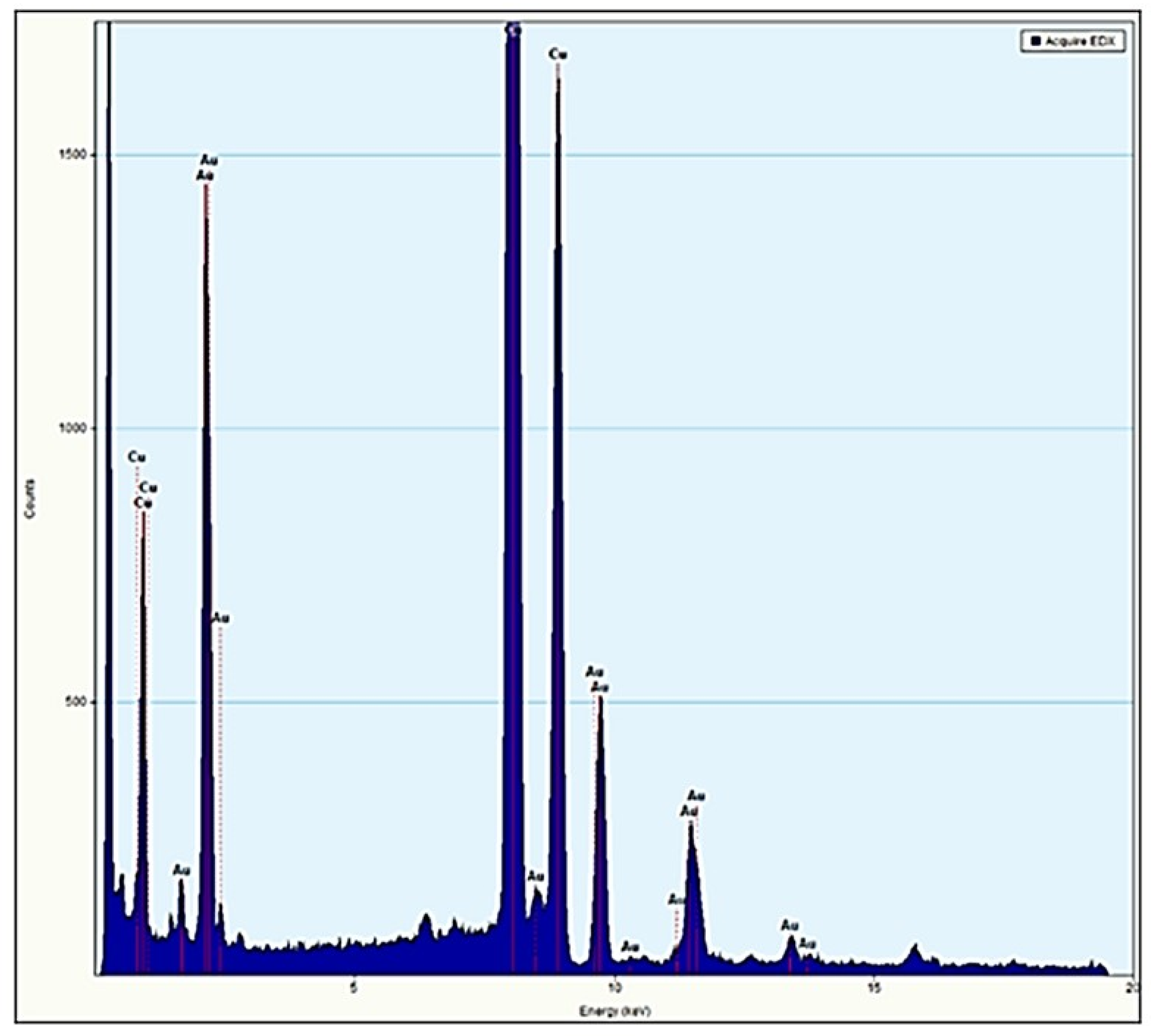
2.2.6. Energy Dispersive Spectroscopy (EDS) Measurements
2.2.7. Gold Nanosystems Susceptibility Tests against Gram + and Gram − Reference Strains
3. Results and Discussion
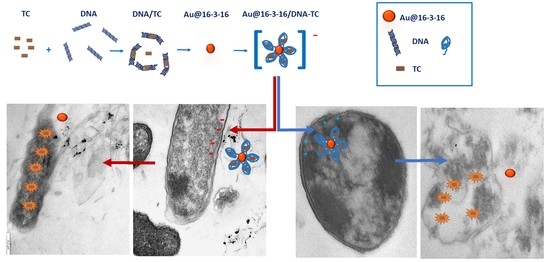
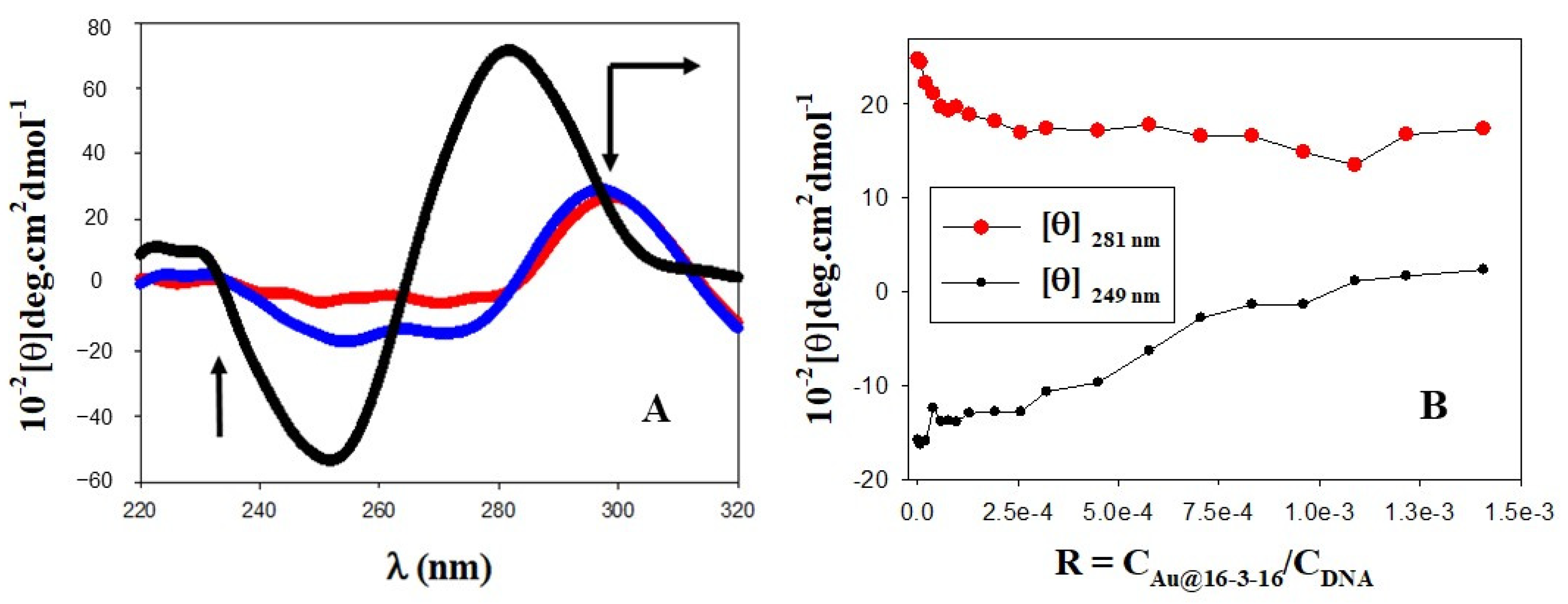
3.1. Conformational Changes in DNA/TC Complexes Induced by Au@16-3-16 Cationic Nanoparticles: Au@16-3-16/DNA-TC Complex Formation
3.2. Charge, Size and Stability of Au@16-3-16 and Au@16-3-16/DNA-TC Nanosystems
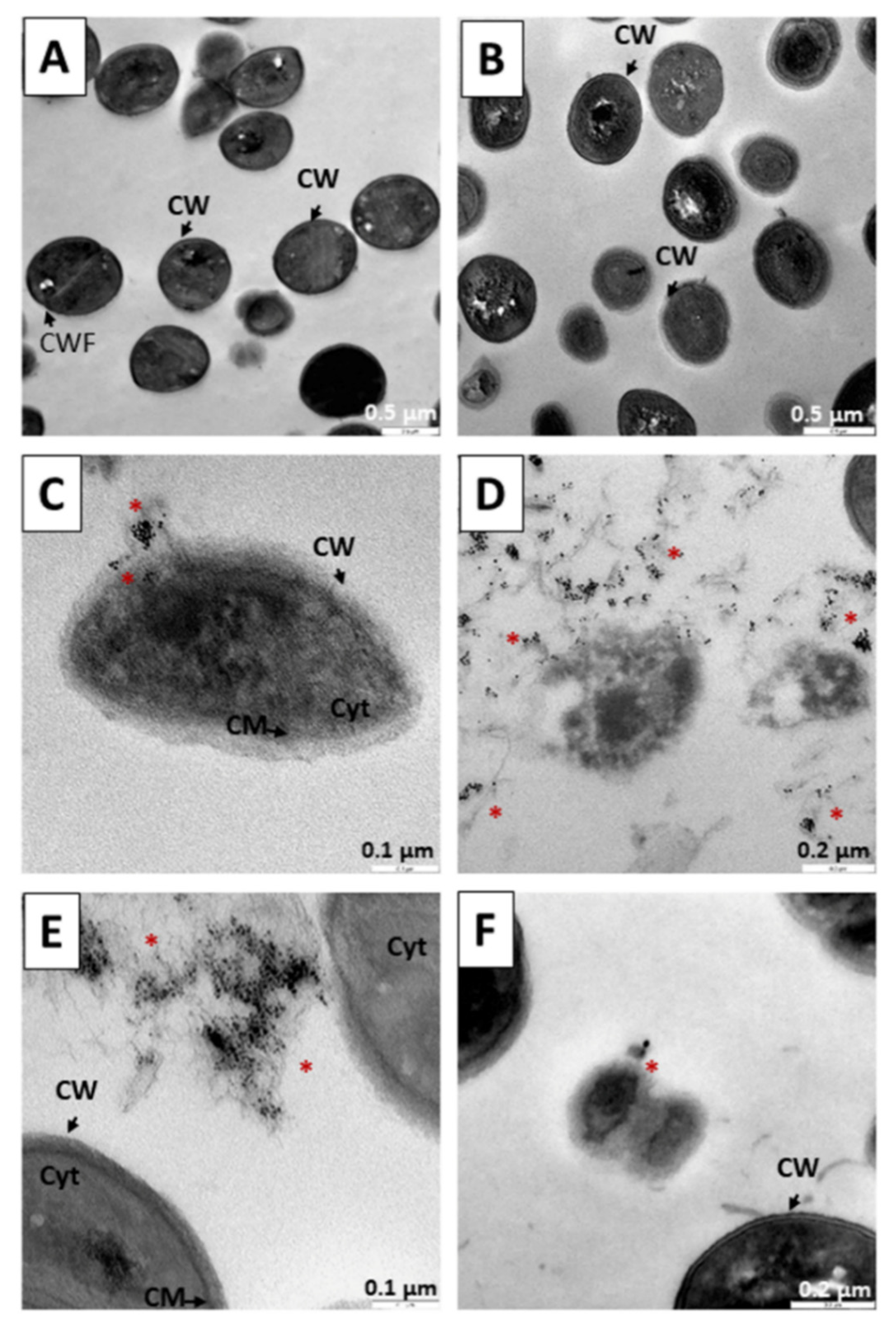
3.3. Results of Gold Nanosystems Susceptibility Tests against Gram + and Gram − Reference Strains
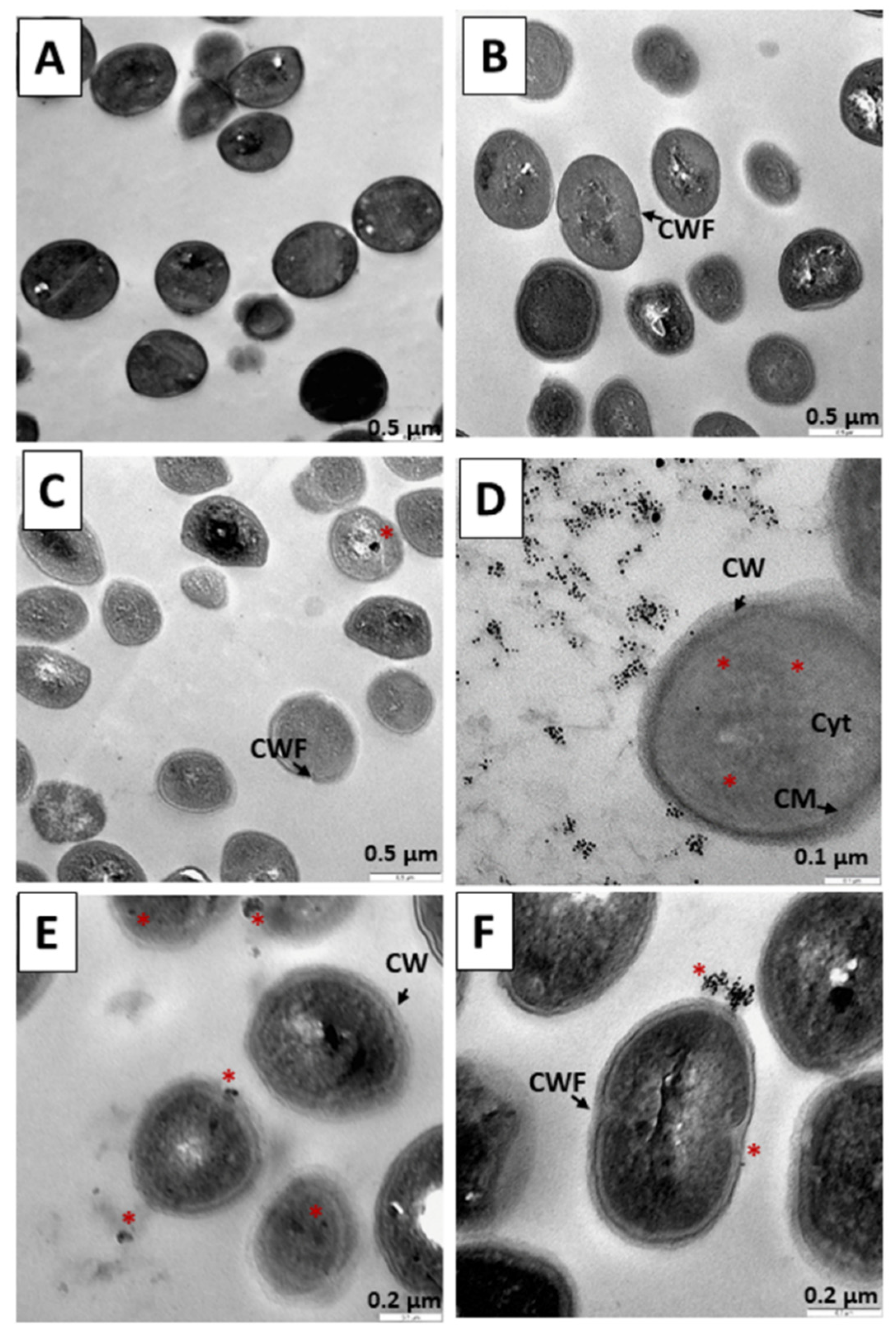
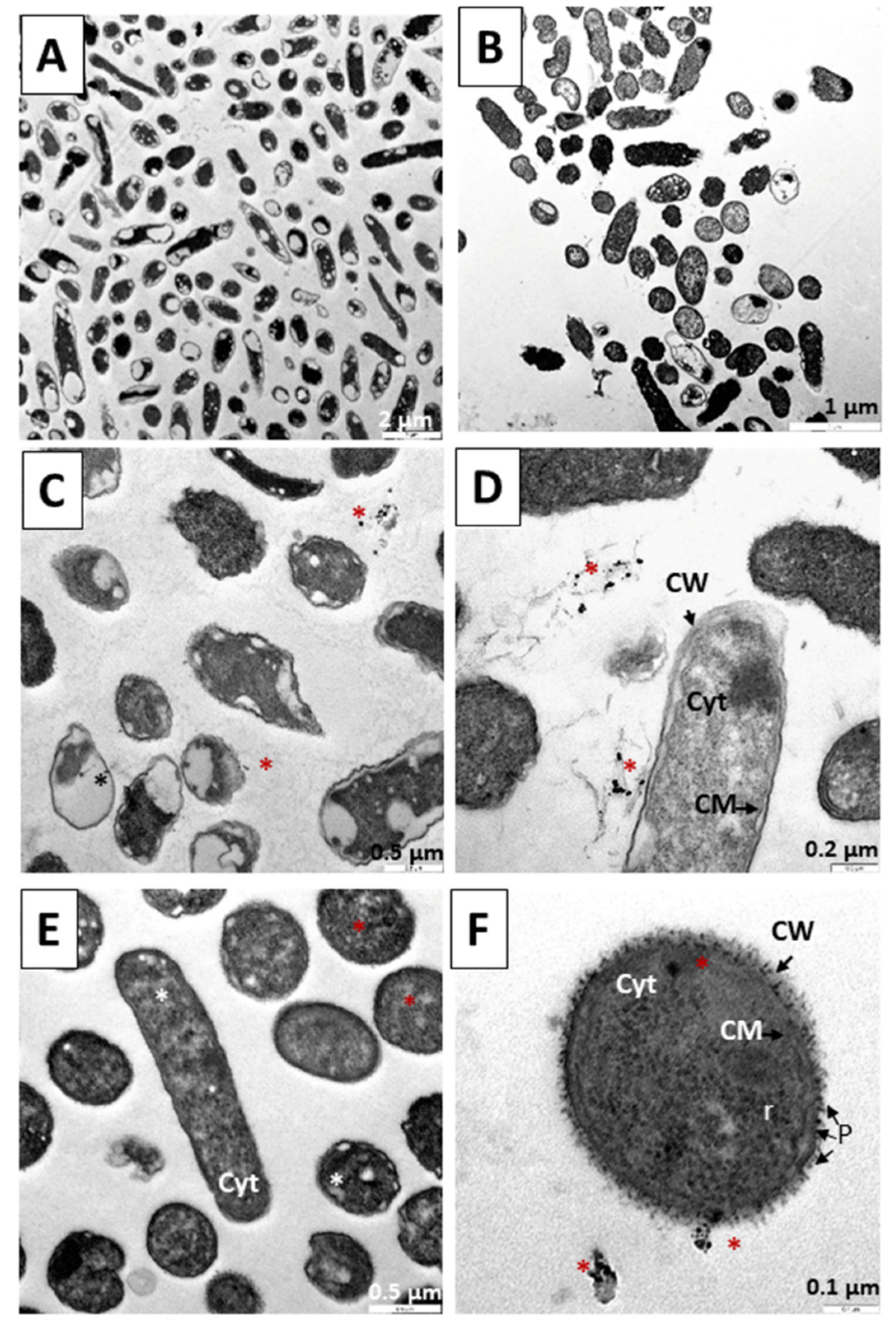
3.4. Internalization of Au@16-3-16 and Au@16-3-16/DNA-TC Nanosystems
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Organización Mundial de la Salud Resistencia a Los Antibióticos. Available online: https://www.who.int/es/news-room/fact-sheets/detail/resistencia-a-los-antibi%C3%B3ticos (accessed on 17 December 2021).
- Ejecutivo, C. 144 Resistencia a Los Antimicrobianos; Organización Mundial de la Salud: Geneva, Switzerland, 2019. [Google Scholar]
- Organización Mundial de la Salud Plan de Acción Mundial Sobre la Resistencia a Los Antimicrobianos. Available online: https://apps.who.int/iris/handle/10665/255204 (accessed on 30 May 2022).
- Organización Mundial de la Salud Lista OMS de Patógenos Prioritarios. Available online: https://www.who.int/es/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 17 December 2021).
- Potron, A.; Poirel, L.; Nordmann, P. Emerging Broad-Spectrum Resistance in Pseudomonas Aeruginosa and Acinetobacter Baumannii: Mechanisms and Epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed]
- Dahdouh, E.; Hajjar, M.; Suarez, M.; Daoud, Z. Acinetobacter Baumannii Isolated from Lebanese Patients: Phenotypes and Genotypes of Resistance, Clonality, and Determinants of Pathogenicity. Front. Cell Infect. Microbiol. 2016, 6, 163. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Pseudomonas Aeruginosa: Resistance to the Max. Front. Microbiol. 2011, 2, 65. [Google Scholar] [CrossRef]
- Rasheed, M.U.; Thajuddin, N.; Ahamed, P.; Teklemariam, Z.; Jamil, K. Antimicrobial Drug Resistance in Strains of Escherichia Coli Isolated from Food Sources. Rev. Inst. Med. Trop. 2014, 56, 341–346. [Google Scholar] [CrossRef]
- Prieto Prieto, J.; García del Potro, M.; Gómez-Lus Centelles, M.L. Guía Para El Uso de Antibióticos En Atención Primaria. In Eficacia In Vivo, Eficacia In Vitro; Ediciones Doyma: Madrid, Spain, 1997; pp. 83–96. [Google Scholar]
- Prats, G. Microbiología y Parasitología Médicas, 1st ed.; Editorial Médica Panamericana: Madrid, Spain, 2013; Volume 1, ISBN 978-84-9835-688-5. (eBook). [Google Scholar]
- Cag, Y.; Caskurlu, H.; Fan, Y.; Cao, B.; Vahaboglu, H. Resistance Mechanisms. Ann. Transl. Med. 2016, 4, 326. [Google Scholar] [CrossRef]
- Oteo Iglesias, J. La Resistencia a Los Antibióticos. La Amenaza de Las Superbacterias; Los Libros de la Catarata: Madrid, Spain, 2016; ISBN 978-84-9097-214-4. [Google Scholar]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- American Society of Health-System Pharmacists Tetraciclina. Información Sobre Medicamentos Para el Paciente. Available online: https://medlineplus.gov/spanish/druginfo/meds/a682098-es.html (accessed on 4 June 2022).
- Pérez-Trallero, E.; Iglesias, L. Tetraciclinas, Sulfamidas y Metronidazol. Enferm. Infecc. Microbiol. Clínica 2003, 21, 520–529. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrov, A.; Schuldt, L.; Hinrichs, W.; Simonson, T. Tetracycline-Tet Repressor Binding Specificity: Insights from Experiments and Simulations. Biophys. J. 2009, 97, 2829–2838. [Google Scholar] [CrossRef]
- Malgor, L.A.; Valsecia, M.E. Antibióticos de Amplio Espectro: Tetraciclinas–Cloranfenicol-Macrólidos. In Farmacología Médica; Cátedra de Farmacología Facultad de Medicina, Universidad Nacional del Noreste: Corrientes, Argentina, 1999; Volume 3, pp. 34–45. [Google Scholar]
- El Korchi, G. Farmacocinética y Eficacia de Oxitetraciclina Tras su Administración Intramuscular en Bovino. Depleción Tisular. Ph.D. Thesis, Universitat Autònoma de Barcelona, Barcelona, Spain, 2006; p. 171. [Google Scholar]
- Gao, X.; Cui, Y.; Levenson, R.M.; Chung, L.W.K.; Nie, S. In Vivo Cancer Targeting and Imaging with Semiconductor Quantum Dots. Nat. Biotechnol. 2004, 22, 969–976. [Google Scholar] [CrossRef]
- De, M.; Ghosh, P.S.; Rotello, V.M. Applications of Nanoparticles in Biology. Adv. Mater. 2008, 20, 4225–4241. [Google Scholar] [CrossRef]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, Applications and Toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Vergara O’ Neale, L. Empleo de Nanopartículas Metálicas Para La Detección de Tensioactivos y Biomoléculas; idUS—Depósito de Investigación Universidad de Sevilla: Sevilla, Spain, 2016. [Google Scholar]
- Chen, Y.; Xianyu, Y.; Jiang, X. Surface Modification of Gold Nanoparticles with Small Molecules for Biochemical Analysis. Acc. Chem. Res. 2017, 50, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Chen, C.-P.; Wu, T.-H.; Yang, C.-H.; Lin, C.-W.; Chen, C.-Y. Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications. Nanomaterials 2019, 9, 861. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B.; Goh, C.S.; Citartan, M.; Lakshmipriya, T.; Arshad, M.K.M.; Faudzi, F.N.M.; Rahim, R.A.; Hashim, U.; Chinni, S.V.; Tang, T.-H. Micro-Encapsulation of Antibiotic in Cellulose Nanoparticle Inhibits Bacteria. Micro Nanosyst. 2016, 8, 41–46. [Google Scholar] [CrossRef]
- Valetti, S.; Xia, X.; Costa-Gouveia, J.; Brodin, P.; Bernet-Camard, M.-F.; Andersson, M.; Feiler, A. Clofazimine Encapsulation in Nanoporous Silica Particles for the Oral Treatment of Antibiotic-Resistant Mycobacterium Tuberculosis Infections. Nanomedicine 2017, 12, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Moyá, M.L.; López-López, M.; Lebrón, J.A.; Ostos, F.J.; Pérez, D.; Camacho, V.; Beck, I.; Merino-Bohórquez, V.; Camean, M.; Madinabeitia, N.; et al. Preparation and Characterization of New Liposomes. Bactericidal Activity of Cefepime Encapsulated into Cationic Liposomes. Pharmaceutics 2019, 11, 69. [Google Scholar] [CrossRef]
- Gonzalez Gomez, A.; Syed, S.; Marshall, K.; Hosseinidoust, Z. Liposomal Nanovesicles for Efficient Encapsulation of Staphylococcal Antibiotics. ACS Omega 2019, 4, 10866–10876. [Google Scholar] [CrossRef]
- Hibbitts, A.; O’Leary, C. Emerging Nanomedicine Therapies to Counter the Rise of Methicillin-Resistant Staphylococcus Aureus. Materials 2018, 11, 321. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Shi, L.; Yang, Y.; Huang, H.; Jie, Y.; Lin, M.; Liang, J.; Liu, C.; Song, H. Ultrafine Control of Cu Nanoparticles and Their Antibiotic Effect on E. Coli. Surf. Rev. Lett. 2019, 26, 1850214. [Google Scholar] [CrossRef]
- Huq, M.A.; Akter, S. Biosynthesis, Characterization and Antibacterial Application of Novel Silver Nanoparticles against Drug Resistant Pathogenic Klebsiella Pneumoniae and Salmonella Enteritidis. Molecules 2021, 26, 5996. [Google Scholar] [CrossRef] [PubMed]
- Huq, M.A. Biogenic Silver Nanoparticles Synthesized by Lysinibacillus Xylanilyticus MAHUQ-40 to Control Antibiotic-Resistant Human Pathogens Vibrio Parahaemolyticus and Salmonella Typhimurium. Front. Bioeng. Biotechnol. 2020, 8, 597502. [Google Scholar] [CrossRef] [PubMed]
- Irshad, A.; Sarwar, N.; Sadia, H.; Malik, K.; Javed, I.; Irshad, A.; Afzal, M.; Abbas, M.; Rizvi, H. Comprehensive Facts on Dynamic Antimicrobial Properties of Polysaccharides and Biomolecules-Silver Nanoparticle Conjugate. Int. J. Biol. Macromol. 2020, 145, 189–196. [Google Scholar] [CrossRef]
- Ayala-Núñez, N.V.; Lara Villegas, H.H.; del Carmen Ixtepan Turrent, L.; Rodríguez Padilla, C. Silver Nanoparticles Toxicity and Bactericidal Effect against Methicillin-Resistant Staphylococcus Aureus: Nanoscale Does Matter. Nanobiotechnology 2009, 5, 2–9. [Google Scholar] [CrossRef]
- Huh, A.J.; Kwon, Y.J. “Nanoantibiotics”: A New Paradigm for Treating Infectious Diseases Using Nanomaterials in the Antibiotics Resistant Era. J. Control. Release 2011, 156, 128–145. [Google Scholar] [CrossRef]
- Gerber, A.; Bundschuh, M.; Klingelhofer, D.; Groneberg, D.A. Gold Nanoparticles: Recent Aspects for Human Toxicology. J. Occup. Med. Toxicol. 2013, 8, 32. [Google Scholar] [CrossRef]
- Costa-Gouveia, J.; Aínsa, J.A.; Brodin, P.; Lucía, A. How Can Nanoparticles Contribute to Antituberculosis Therapy? Drug Discov. Today 2017, 22, 600–607. [Google Scholar] [CrossRef]
- Golmohamadi, M.; Ghorbani, H.R.; Otadi, M. Synthesis of Chitosan Nanoparticles Loaded with Antibiotics as Drug Carriers and the Study of Antibacterial Activity. J. Nanoanal. 2019, 6, 72–79. [Google Scholar] [CrossRef]
- Nardone, L.L.; Andrews, S.B. Cell Line A549 as a Model of the Type II Pneumocyte. Phospholipid Biosynthesis from Native and Organometallic Precursors. Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab. 1979, 573, 276–295. [Google Scholar] [CrossRef]
- Laschewsky, A.; Lunkenheimer, K.; Rakotoaly, R.H.; Wattebled, L. Spacer Effects in Dimeric Cationic Surfactants. Colloid Polym. Sci. 2005, 283, 469–479. [Google Scholar] [CrossRef]
- Chen, J.-P.; Battini, N.; Ansari, M.F.; Zhou, C.-H. Membrane Active 7-Thiazoxime Quinolones as Novel DNA Binding Agents to Decrease the Genes Expression and Exert Potent Anti-Methicillin-Resistant Staphylococcus Aureus Activity. Eur. J. Med. Chem. 2021, 217, 113340. [Google Scholar] [CrossRef] [PubMed]
- Wettig, S.D.; Verrall, R.E. Thermodynamic Studies of Aqueous m–s–m Gemini Surfactant Systems. J. Colloid Interface Sci. 2001, 235, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Zana, R.; Benrraou, M.; Rueff, R. Alkanediyl-α,ω-Bis(Dimethylalkylammonium Bromide) Surfactants. 1. Effect of the Spacer Chain Length on the Critical Micelle Concentration and Micelle Ionization Degree. Langmuir 1991, 7, 1072–1075. [Google Scholar] [CrossRef]
- Khan, M.A.; Musarrat, J. Interactions of Tetracycline and Its Derivatives with DNA in Vitro in Presence of Metal Ions. Int. J. Biol Macromol. 2003, 33, 49–56. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef] [PubMed]
- Grueso, E.; Roldan, E.; Perez-Tejeda, P.; Kuliszewska, E.; Molero, B.; Brecker, L.; Giráldez-Pérez, R.M. Reversible DNA Compaction Induced by Partial Intercalation of 16-Ph-16 Gemini Surfactants: Evidence of Triple Helix Formation. Phys. Chem. Chem. Phys. 2018, 20, 24902–24914. [Google Scholar] [CrossRef] [PubMed]
- Giráldez-Pérez, R.M.; Grueso, E.; Domínguez, I.; Pastor, N.; Kuliszewska, E.; Prado-Gotor, R.; Requena-Domenech, F. Biocompatible DNA/5-Fluorouracil-Gemini Surfactant-Functionalized Gold Nanoparticles as Promising Vectors in Lung Cancer Therapy. Pharmaceutics 2021, 13, 423. [Google Scholar] [CrossRef]
- Giráldez-Pérez, R.M.; Grueso, E.; Lhamyani, S.; Perez-Tejeda, P.; Gentile, A.-M.; Kuliszewska, E.; Roman-Perez, J.; el Bekay, R. MiR-21/Gemini Surfactant-Capped Gold Nanoparticles as Potential Therapeutic Complexes: Synthesis, Characterization and in Vivo Nanotoxicity Probes. J. Mol. Liq. 2020, 313, 113577. [Google Scholar] [CrossRef]
- Leclercq, R.; Cantón, R.; Brown, D.F.J.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST Expert Rules in Antimicrobial Susceptibility Testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef]
- EUCAST. Subcommittee on MIC distributions and ECOFFs Antimicrobial Wild Type Distributions of Microorganisms. Available online: https://mic.eucast.org/search/ (accessed on 17 December 2021).
- Pham, T.-N.; Loupias, P.; Dassonville-Klimpt, A.; Sonnet, P. Drug Delivery Systems Designed to Overcome Antimicrobial Resistance. Med. Res. Rev. 2019, 39, 2343–2396. [Google Scholar] [CrossRef]
- Kumar, V.; Palazzolo, S.; Bayda, S.; Corona, G.; Toffoli, G.; Rizzolio, F. DNA Nanotechnology for Cancer Therapeutics. Theranostics 2016, 6, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, C.; Pei, H.; Shi, J.; Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. [Google Scholar] [CrossRef]
- Chao, J.; Liu, H.; Su, S.; Wang, L.; Huang, W.; Fan, C. Structural DNA Nanotechnology for Intelligent Drug Delivery. Small 2014, 10, 4626–4635. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Nucleic Acid Structure and Recognition, 1st ed.; Oxford University Press: New York, NY, USA, 2002; ISBN 0-19-850635 X. [Google Scholar]
- Maestre, M.F.; Wang, J.C. Circular Dichroism of Superhelical DNA. Biopolymers 1971, 10, 1021–1030. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Wilson, J., Hunt, T., Eds.; W.W. Norton & Company: New York, NY, USA, 2017; ISBN 9781315735368. [Google Scholar]
- Prado-Gotor, R.; Grueso, E. A Kinetic Study of the Interaction of DNA with Gold Nanoparticles: Mechanistic Aspects of the Interaction. Phys. Chem. Chem. Phys. 2011, 13, 1479–1489. [Google Scholar] [CrossRef]
- Hansma, H.G.; Sinsheimer, R.L.; Li, M.-Q.; Hansma, P.K. Atomic Force Microscopy of Single-and Double-Stranded DNA. Nucleic Acids Res. 1992, 20, 3585–3590. [Google Scholar] [CrossRef]
- Pelta, J.; Livolant, F.; Sikorav, J.-L. DNA Aggregation Induced by Polyamines and Cobalthexamine. J. Biol. Chem. 1996, 271, 5656–5662. [Google Scholar] [CrossRef]
- Agyapong, D.A.Y.; Zhao, R.-L.; Li, C.-X.; Du, W.-J.; Zeng, H.-J. Novel Mercaptan Derivatives Nanogold Particles with Hyperchromic and Fluorescence Sensitization Effects. In Proceedings of the 2016 International Conference on Biological Sciences and Technology, Guangzhou, China, 8–10 January 2016; Atlantis Press: Amsterdam, The Netherlands, 2016; pp. 143–148. [Google Scholar]
- Gelperina, S.; Kisich, K.; Iseman, M.D.; Heifets, L. The Potential Advantages of Nanoparticle Drug Delivery Systems in Chemotherapy of Tuberculosis. Am. J. Respir. Crit. Care Med. 2005, 172, 1487–1490. [Google Scholar] [CrossRef]
- Zuber, A.; Purdey, M.; Schartner, E.; Forbes, C.; van der Hoek, B.; Giles, D.; Abell, A.; Monro, T.; Ebendorff-Heidepriem, H. Detection of Gold Nanoparticles with Different Sizes Using Absorption and Fluorescence Based Method. Sens. Actuators B Chem. 2016, 227, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Freitas, C.; Müller, R.H. Effect of Light and Temperature on Zeta Potential and Physical Stability in Solid Lipid Nanoparticle (SLN®) Dispersions. Int. J. Pharm. 1998, 168, 221–229. [Google Scholar] [CrossRef]
- Ghosh, S.; Banik, D.; Roy, A.; Kundu, N.; Kuchlyan, J.; Sarkar, N. Spectroscopic Investigation of the Binding Interactions of a Membrane Potential Molecule in Various Supramolecular Confined Environments: Contrasting Behavior of Surfactant Molecules in Relocation or Release of the Probe between Nanocarriers and DNA Surfa. Phys. Chem. Chem. Phys. 2014, 16, 25024–25038. [Google Scholar] [CrossRef] [PubMed]
- Grueso, E.; Giráldez-Pérez, R.M.; Kuliszewska, E.; Guerrero, J.A.; Prado-Gotor, R. Reversible Cationic Gemini Surfactant-Induced Aggregation of Anionic Gold Nanoparticles for Sensing Biomolecules. Colloids Surf. A Physicochem. Eng. Asp. 2021, 610, 125893. [Google Scholar] [CrossRef]
- Gagnon, Z.; Senapati, S.; Gordon, J.; Chang, H.-C. Dielectrophoretic Detection and Quantification of Hybridized DNA Molecules on Nano-Genetic Particles. Electrophoresis 2008, 29, 4808–4812. [Google Scholar] [CrossRef]
- Phan, H.T.; Haes, A.J. What Does Nanoparticle Stability Mean? J. Phys. Chem. C 2019, 123, 16495–16507. [Google Scholar] [CrossRef]
- Li, X.; Robinson, S.M.; Gupta, A.; Saha, K.; Jiang, Z.; Moyano, D.F.; Sahar, A.; Riley, M.A.; Rotello, V.M. Functional Gold Nanoparticles as Potent Antimicrobial Agents against Multi-Drug-Resistant Bacteria. ACS Nano 2014, 8, 10682–10686. [Google Scholar] [CrossRef]
- Niemirowicz, K.; Swiecicka, I.; Wilczewska, A.Z.; Misztalewska, I.; Kalska-Szostko, B.; Bienias, K.; Bucki, R.; Car, H. Gold-Functionalized Magnetic Nanoparticles Restrict Growth of Pseudomonas Aeruginosa. Int. J. Nanomed. 2014, 9, 2217–2224. [Google Scholar] [CrossRef]
- Uma Suganya, K.S.; Govindaraju, K.; Ganesh Kumar, V.; Stalin Dhas, T.; Karthick, V.; Singaravelu, G.; Elanchezhiyan, M. Blue Green Alga Mediated Synthesis of Gold Nanoparticles and Its Antibacterial Efficacy against Gram Positive Organisms. Mater. Sci. Eng. C 2015, 47, 351–356. [Google Scholar] [CrossRef]
- Ahmad, A.; Wei, Y.; Ullah, S.; Shah, S.I.; Nasir, F.; Shah, A.; Iqbal, Z.; Tahir, K.; Khan, U.A.; Yuan, Q. Synthesis of Phytochemicals-Stabilized Gold Nanoparticles and Their Biological Activities against Bacteria and Leishmania. Microb. Pathog. 2017, 110, 304–312. [Google Scholar] [CrossRef]
- Chamakura, K.; Perez-Ballestero, R.; Luo, Z.; Bashir, S.; Liu, J. Comparison of Bactericidal Activities of Silver Nanoparticles with Common Chemical Disinfectants. Colloids Surf. B BioInterfaces 2011, 84, 88–96. [Google Scholar] [CrossRef]
- Brown, S.D.; Traczewski, M.M. Comparative in Vitro Antimicrobial Activity of Tigecycline, a New Glycylcycline Compound, in Freshly Prepared Medium and Quality Control. J. Clin. Microbiol. 2007, 45, 2173–2179. [Google Scholar] [CrossRef] [PubMed]
- Ewnetu, Y.; Lemma, W.; Birhane, N. Antibacterial Effects of Apis Mellifera and Stingless Bees Honeys on Susceptible and Resistant Strains of Escherichia Coli, Staphylococcus Aureus and Klebsiella Pneumoniae in Gondar, Northwest Ethiopia. BMC Complement Altern. Med. 2013, 13, 269. [Google Scholar] [CrossRef]
- Chaalal, W.; Aggad, H.; Zidane, K.; Saidi, N.; Kihal, M. Antimicrobial Susceptibility Profiling of Staphylococcus Aureus Isolates from Milk. Br. Microbiol. Res. J. 2016, 13, 1–7. [Google Scholar] [CrossRef]
- Kłodzińska, E.; Szumski, M.; Dziubakiewicz, E.; Hrynkiewicz, K.; Skwarek, E.; Janusz, W.; Buszewski, B. Effect of Zeta Potential Value on Bacterial Behavior during Electrophoretic Separation. Electrophoresis 2010, 31, 1590–1596. [Google Scholar] [CrossRef] [PubMed]
- Nirmala Grace, A.; Pandian, K. Antibacterial Efficacy of Aminoglycosidic Antibiotics Protected Gold Nanoparticles—A Brief Study. Colloids Surf. A Physicochem. Eng. Asp. 2007, 297, 63–70. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Wang, P.; Robertson, I.; Abraham, S.; Sahibzada, S.; Habib, I. Antimicrobial Resistance and Genomic Characterisation of Escherichia Coli Isolated from Caged and Non-Caged Retail Table Eggs in Western Australia. Int. J. Food Microbiol. 2021, 340, 109054. [Google Scholar] [CrossRef]
- Karahutová, L.; Mandelík, R.; Bujňáková, D. Antibiotic Resistant and Biofilm-Associated Escherichia Coli Isolates from Diarrheic and Healthy Dogs. Microorganisms 2021, 9, 1334. [Google Scholar] [CrossRef]
- McDougall, S.; Williamson, J.; Gohary, K.; Lacy-Hulbert, J. Detecting Intramammary Infection at the End of Lactation in Dairy Cows. J. Dairy Sci. 2021, 104, 10232–10249. [Google Scholar] [CrossRef]
- Martini, C.L.; Lange, C.C.; Brito, M.A.V.P.; Ribeiro, J.B.; Mendonça, L.C.; Vaz, E.K. Characterisation of Penicillin and Tetracycline Resistance in Staphylococcus Aureus Isolated from Bovine Milk Samples in Minas Gerais, Brazil. J. Dairy Res. 2017, 84, 202–205. [Google Scholar] [CrossRef]
- Akter, S.; Huq, M.A. Biologically Rapid Synthesis of Silver Nanoparticles by Sphingobium Sp. MAH-11T and Their Antibacterial Activity and Mechanisms Investigation against Drug-Resistant Pathogenic Microbes. Artif. Cells Nanomed. Biotechnol. 2020, 48, 672–682. [Google Scholar] [CrossRef] [Green Version]
- Rehman, S.; Jermy, B.R.; Akhtar, S.; Borgio, J.F.; Abdul Azeez, S.; Ravinayagam, V.; al Jindan, R.; Alsalem, Z.H.; Buhameid, A.; Gani, A. Isolation and Characterization of a Novel Thermophile; Bacillus Haynesii, Applied for the Green Synthesis of ZnO Nanoparticles. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, J.; Yang, L.; Shi, F.; Yang, L.; Ye, M. Antibacterial Activity and a Membrane Damage Mechanism of Lachnum YM30 Melanin against Vibrio Parahaemolyticus and Staphylococcus Aureus. Food Control. 2017, 73, 1445–1451. [Google Scholar] [CrossRef]
- Akter, S.; Lee, S.-Y.; Siddiqi, M.Z.; Balusamy, S.R.; Ashrafudoulla, M.; Rupa, E.J.; Huq, M.A. Ecofriendly Synthesis of Silver Nanoparticles by Terrabacter Humi Sp. Nov. and Their Antibacterial Application against Antibiotic-Resistant Pathogens. Int. J. Mol. Sci. 2020, 21, 9746. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, A.; Perumal, E. Metal Oxide Nanoparticles as Antimicrobial Agents: A Promise for the Future. Int. J. Antimicrob. Agents 2017, 49, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.-C.; Su, Y.-P.; Chen, C.-C.; Jia, G.; Wang, H.-L.; Wu, J.C.G.; Lin, J.-G. Relationship between Antibacterial Activity of Chitosan and Surface Characteristics of Cell Wall. Acta Pharmacol. Sin. 2004, 25, 932–936. [Google Scholar]
- Beveridge, T.J. Structures of Gram-Negative Cell Walls and Their Derived Membrane Vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [CrossRef]
- Nel, A.; Xia, T.; Mädler, L.; Li, N. Toxic Potential of Materials at the Nanolevel. Science 2006, 311, 622–627. [Google Scholar] [CrossRef]
- Padil, V.V.T.; Černík, M. Green Synthesis of Copper Oxide Nanoparticles Using Gum Karaya as a Biotemplate and Their Antibacterial Application. Int. J. Nanomed. 2013, 8, 889–898. [Google Scholar] [CrossRef]
- Hajipour, M.J.; Fromm, K.M.; Akbar Ashkarran, A.; Jimenez de Aberasturi, D.; Larramendi, I.R.D.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial Properties of Nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Kumar, S.R.; Imlay, J.A. How Escherichia Coli Tolerates Profuse Hydrogen Peroxide Formation by a Catabolic Pathway. J. Bacteriol. 2013, 195, 4569–4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gram, C. The Differential Staining of Schizomycetes in Tissue Sections and in Dried Preparations. Fortschitte Med. 1884, 2, 185–189. [Google Scholar]
- Ghuysen, J.M.; Hakenbeck, R. (Eds.) Bacterial Cell Wall, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1994; ISBN 9780080860879. [Google Scholar]









| Sample | Sample Composition | Zeta Potential (mV) | Size (nm); Population % | |
|---|---|---|---|---|
| 1 | Au@16-3-16 | (51.3 ± 1.9) | (2.6 ± 0.3) | |
| Region 1 | 2 | DNA/TC | (−80 ± 2) | d1 = (81 ± 13); 81.2% d2 = (831 ± 63); 18.8% |
| 3 | Au@16-3-16/DNA-TC (CAu@16-3-16 = 22 nM) | (−62.1 ± 1.0) | d1 = (74 ± 11); 93.1% d2 = (606 ± 75); 6.9% | |
| 4 | Au@16-3-16/DNA-TC (CAu@16-3-16 = 40 nM) | (−42.9 ± 0.8) | d1 = (70 ± 10); 93.1% d2 = (463 ± 67); 6.9% | |
| Region 2 | 5 | C1 (CAu@16-3-16 = 51 nM) | (−36.2 ± 1.6) | d1 = (50 ± 10); 99.2% d2 = (143 ± 11); 0.8% |
| 6 | Au@16-3-16/DNA-TC (CAu@16-3-16 = 62 nM) | (−36.2 ± 1.4) | d1 = (52 ± 14); 99.2% d2 = (363 ± 15); 0.8% | |
| 7 | C2 (CAu@16-3-16 = 74 nM) | (−35 ± 2) | d1 = (61 ± 15); 99.3% d2 = (332 ± 21); 0.7% | |
| 8 | Au@16-3-16/DNA-TC (CAu@16-3-16 = 96 nM) | (−33.3 ± 0.5) | d1 = (57 ± 10); 99.2% d2 = (455 ± 60); 0.8% | |
| 9 | C3 (CAu@16-3-16 = 130 nM) | (−31.1 ± 0.7) | d1 = (68 ± 7); 99.3% d2 = (461 ± 54); 0.7% |
| System | MIC in E. coli | MIC in S. aureus |
|---|---|---|
| TC | 5.05 mg/L (25µM) | 5.05 mg/L (25 µM) |
| N1 | 25.5 nM * | 25.5 nM * |
| N2 | 37.0 nM * | 37.0 nM * |
| N3 | 65.0 nM * | 16.2 nM * |
| C1 | 5.05 mg/L (25µM) | 5.05 mg/L (25 µM) |
| C2 | 5.05 mg/L (25µM) | 5.05 mg/L (25 µM) |
| C3 | 5.05 mg/L (25µM) | 5.05 mg/L (25 µM) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giráldez-Pérez, R.M.; Grueso, E.M.; Jiménez-Aguayo, R.; Carbonero, A.; González-Bravo, M.; Kuliszewska, E.; Prado-Gotor, R. Use of Nanoparticles to Prevent Resistance to Antibiotics—Synthesis and Characterization of Gold Nanosystems Based on Tetracycline. Pharmaceutics 2022, 14, 1941. https://doi.org/10.3390/pharmaceutics14091941
Giráldez-Pérez RM, Grueso EM, Jiménez-Aguayo R, Carbonero A, González-Bravo M, Kuliszewska E, Prado-Gotor R. Use of Nanoparticles to Prevent Resistance to Antibiotics—Synthesis and Characterization of Gold Nanosystems Based on Tetracycline. Pharmaceutics. 2022; 14(9):1941. https://doi.org/10.3390/pharmaceutics14091941
Chicago/Turabian StyleGiráldez-Pérez, Rosa M., Elia M. Grueso, Raquel Jiménez-Aguayo, Alfonso Carbonero, Marina González-Bravo, Edyta Kuliszewska, and Rafael Prado-Gotor. 2022. "Use of Nanoparticles to Prevent Resistance to Antibiotics—Synthesis and Characterization of Gold Nanosystems Based on Tetracycline" Pharmaceutics 14, no. 9: 1941. https://doi.org/10.3390/pharmaceutics14091941