Molecular Mechanisms of Cassia fistula against Epithelial Ovarian Cancer Using Network Pharmacology and Molecular Docking Approaches
Abstract
1. Introduction
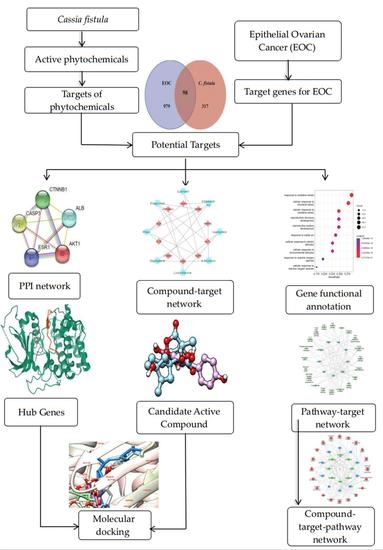
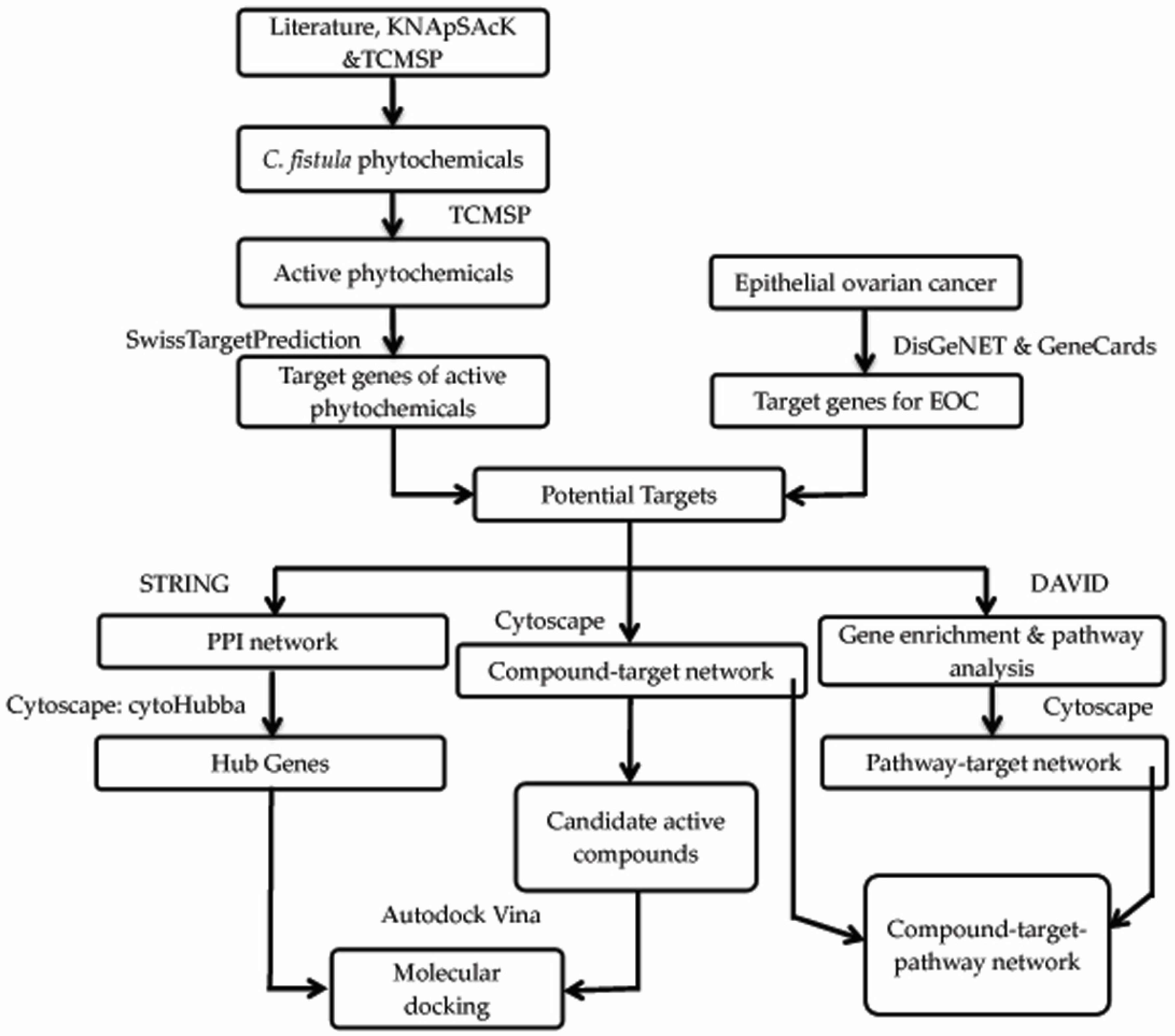
2. Materials and Methods
2.1. Phytochemical Library Construction
2.2. Drug Target Profiles for C. fistula
2.3. Candidate Targets of C. fistula for EOC
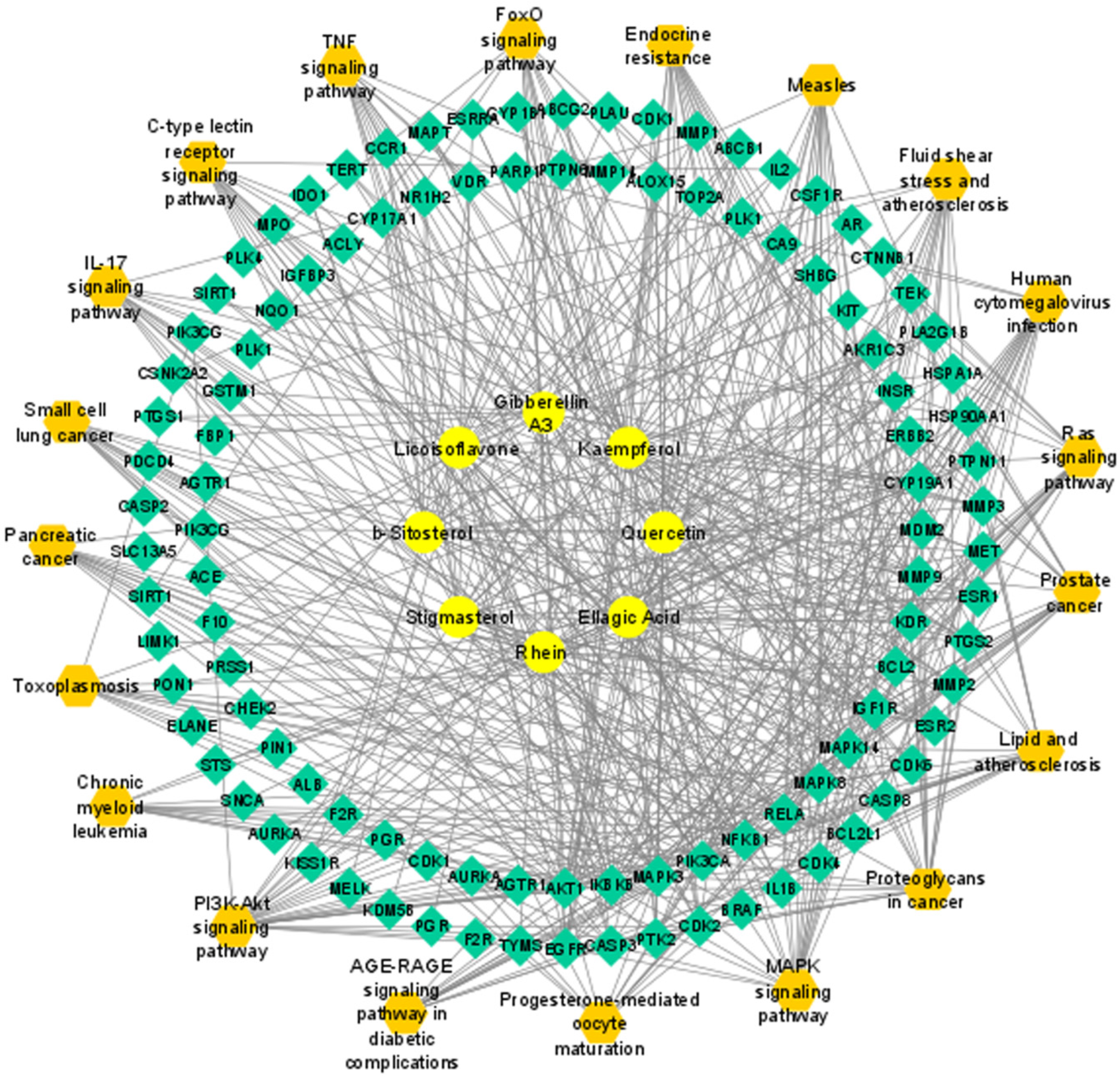
2.4. Compound-Target Network
2.5. Protein–Protein Interaction (PPI) Network
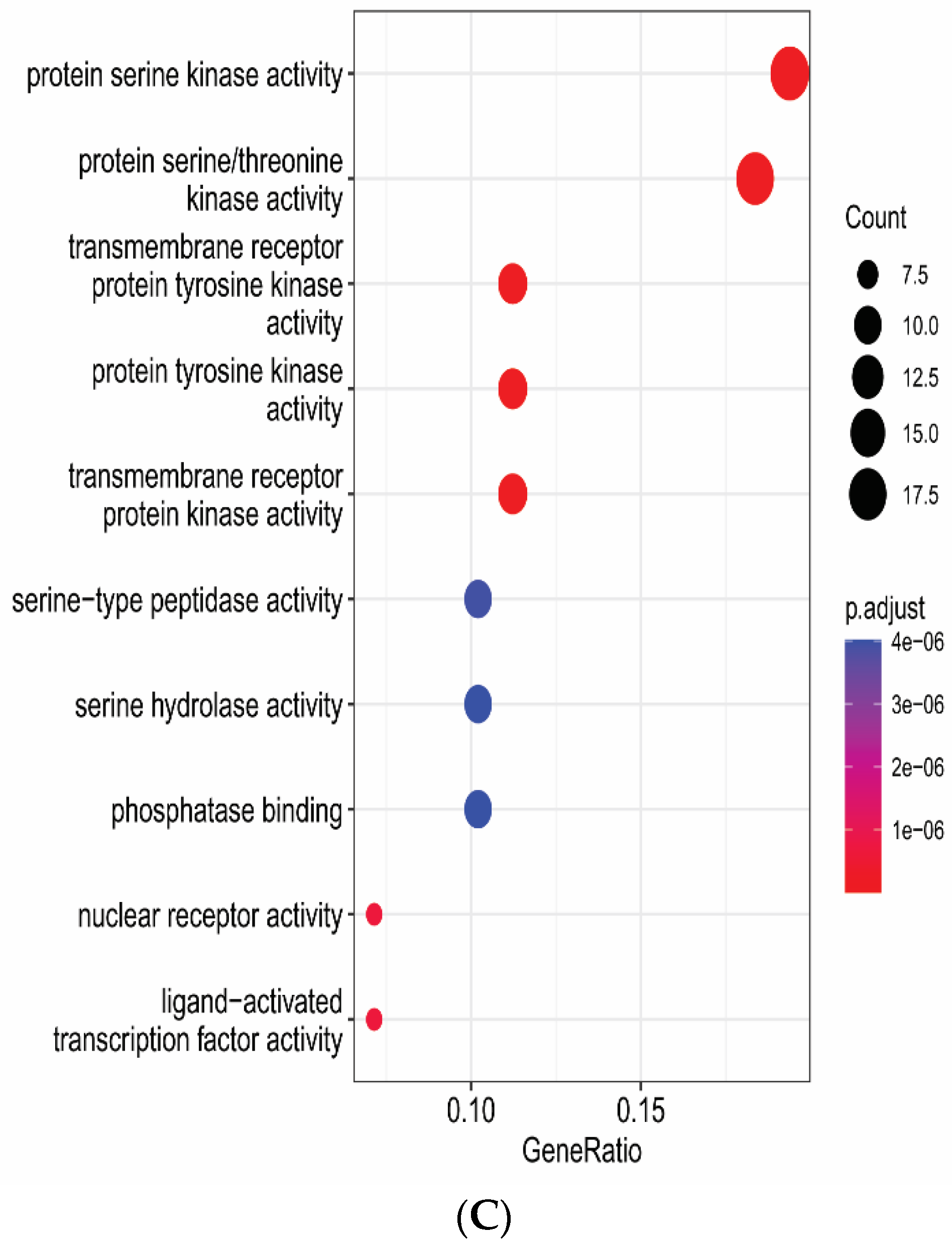
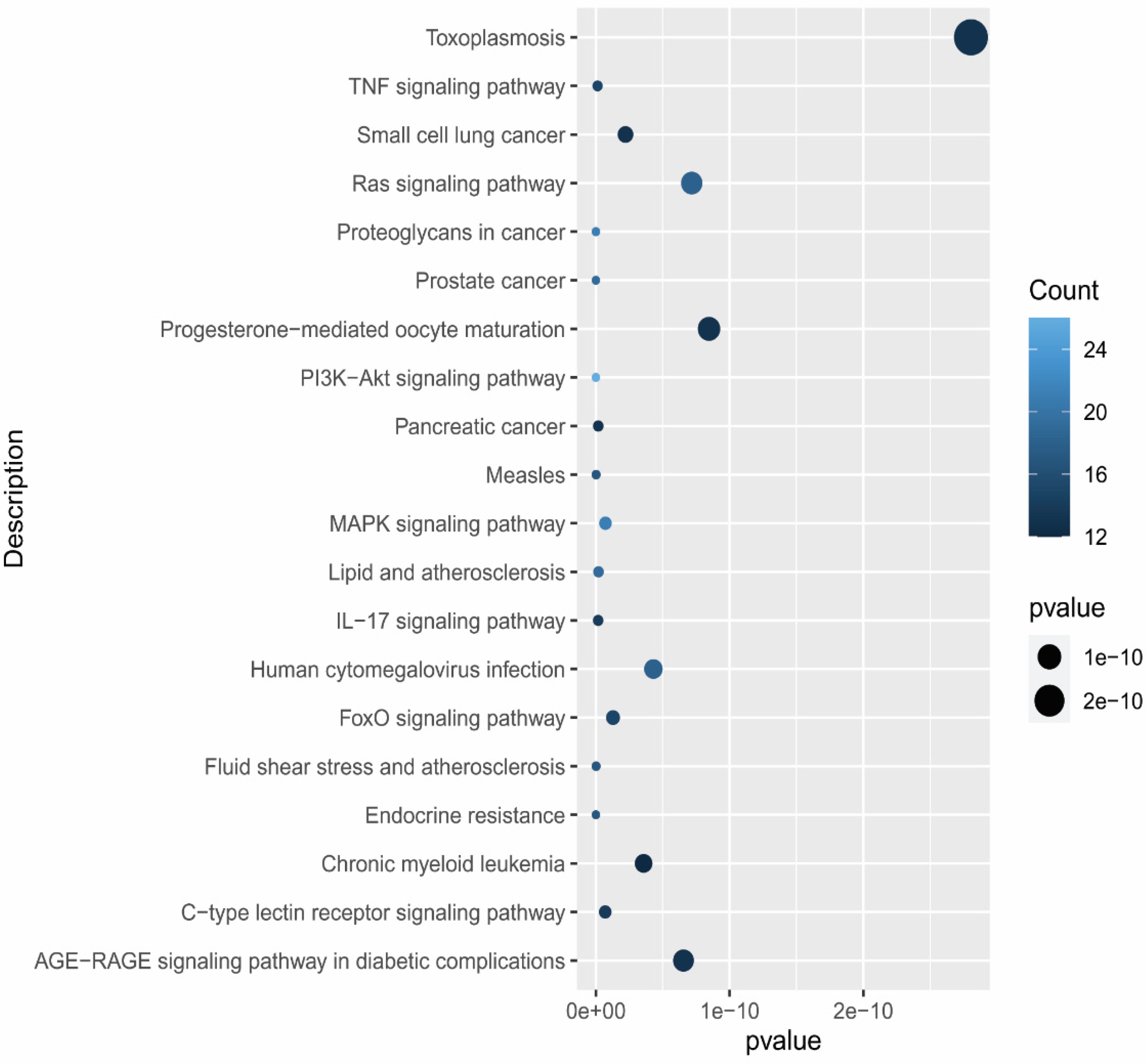
2.6. Gene Functional Annotation
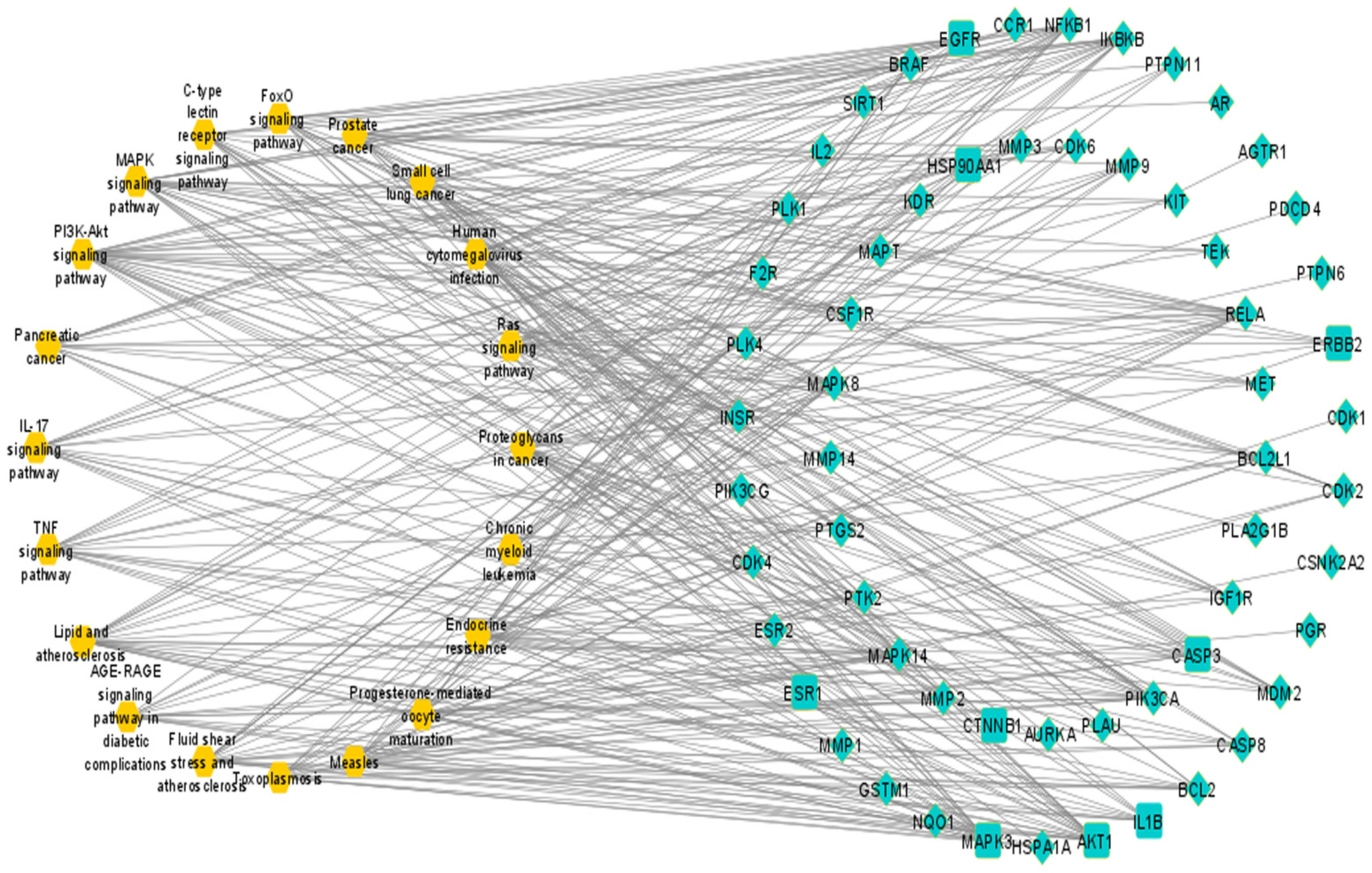
2.7. Compound–Target–Signaling Pathway Network
2.8. Molecular Docking
3. Results
3.1. Active Constituents of C. fistula
3.2. Drug Target Profiling for C. fistula
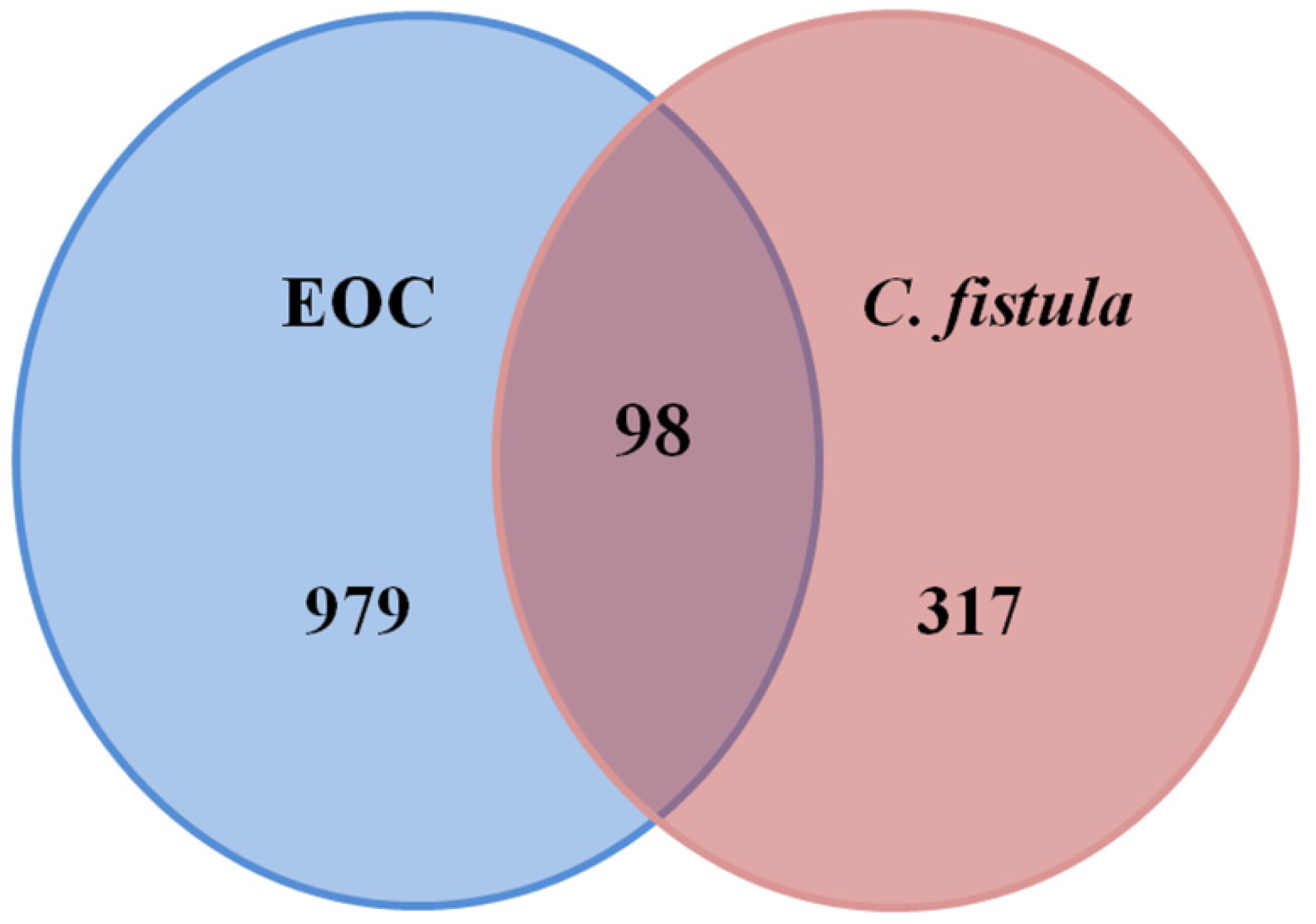
3.3. Potential Targets of C. fistula for EOC
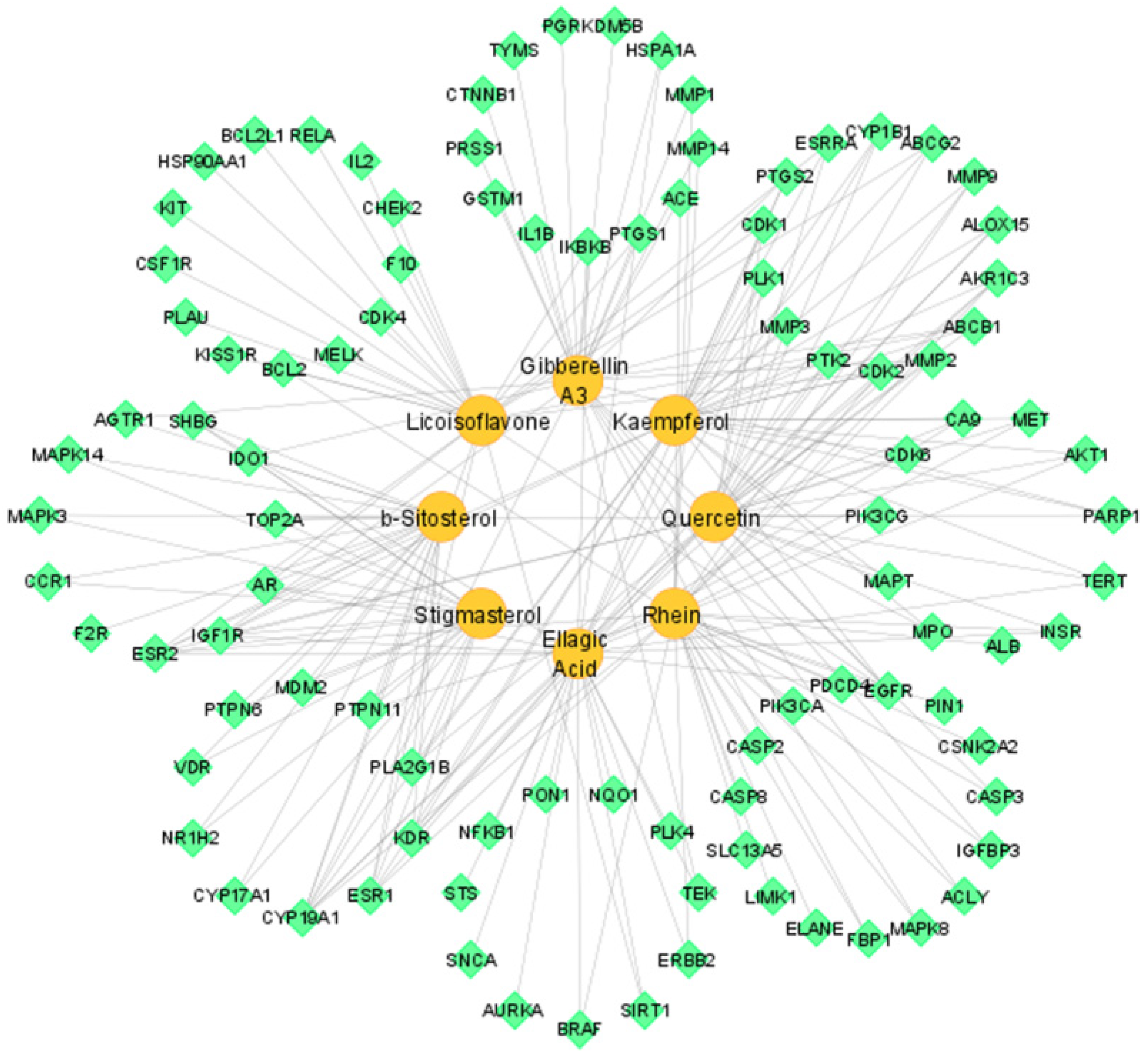
3.4. Compound–Target Network
3.5. Protein–Protein Interaction (PPI) Network
3.6. Gene Functional Annotation
3.7. Compound–Target–Signaling Pathway Network
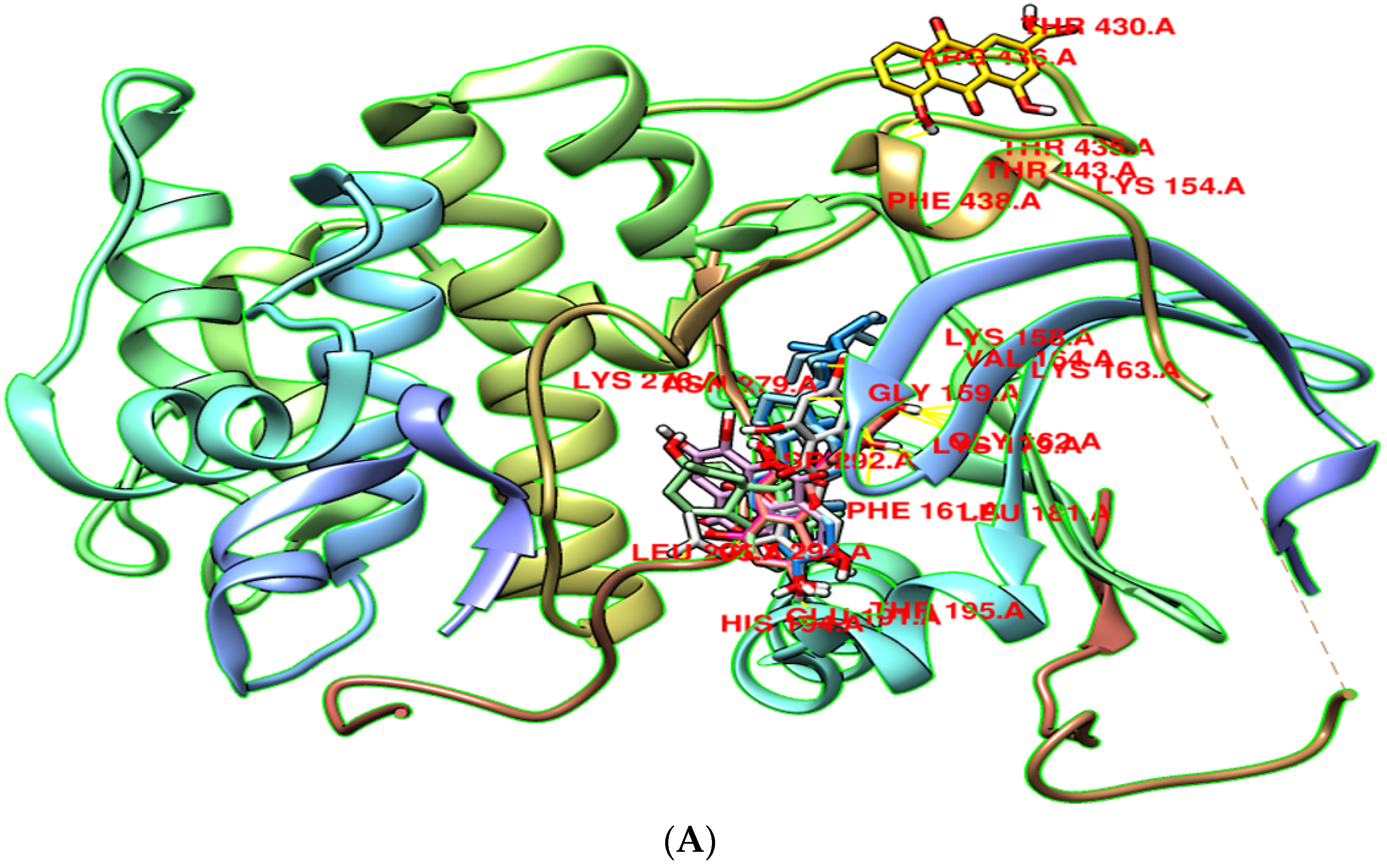
3.8. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeSantis, C.E.; Miller, K.D.; Dale, W.; Mohile, S.G.; Cohen, H.J.; Leach, C.R.; Sauer, A.G.; Jemal, A.; Siegel, R.L. Cancer statistics for adults aged 85 years and older, 2019. CA Cancer J. Clin. 2019, 69, 452–467. [Google Scholar] [CrossRef] [PubMed]
- Oronsky, B.; Ray, C.M.; Spira, A.I.; Trepel, J.B.; Carter, C.A.; Cottrill, H.M. A brief review of the management of platinum-resistant–platinum-refractory ovarian cancer. Med. Oncol. 2017, 34, 103. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. The origin and pathogenesis of epithelial ovarian cancer-a proposed unifying theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef]
- Desai, A.; Xu, J.; Aysola, K.; Qin, Y.; Okoli, C.; Hariprasad, R.; Chinemerem, U.; Gates, C.; Reddy, A.; Danner, O.; et al. Epithelial ovarian cancer: An overview. World J. Transl. Med. 2014, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, J.; Louie, E.; Pike, M.; Roy, S.; Ross, R.J.L.K.; Henderson, B.E. Incessant ovulation and ovarian cancer. Lancet 1979, 28, 170–173. [Google Scholar] [CrossRef]
- Fathalla, M.J.L. Incessant ovulation—A factor in ovarian neoplasia. Lancet 1971, 2, 163. [Google Scholar] [CrossRef]
- Cramer, D.W.; Wrjjotnci, W. Determinants of ovarian cancer risk. II. Inferences regarding pathogenesis. J. Natl. Cancer Inst. 1983, 71, 717–721. [Google Scholar]
- Cramer, D.; Barbieri, R.; Fraer, A.; Harlow, B. Determinants of early follicular phase gonadotrophin and estradiol concentrations in women of late reproductive age. Hum. Reprod. 2002, 17, 221–227. [Google Scholar] [CrossRef][Green Version]
- Rahmani, A.H.; Smjajpcr, A. Nigella sativa and its active constituents thymoquinone shows pivotal role in the diseases prevention and treatment. Asian J. Pharm. Clin. Res. 2015, 8, 48–53. [Google Scholar]
- Rahmani, A.H.; Aly, S.M.; Ali, H.; Babiker, A.Y.; Srikar, S.; Khan, A.A. Therapeutic effects of date fruits (Phoenix dactylifera) in the prevention of diseases via modulation of anti-inflammatory, anti-oxidant and anti-tumour activity. Int. J. Clin. Exp. Med. 2014, 7, 483–491. [Google Scholar]
- Rahmani, A.; Pathophysiology, P. Active ingredients of ginger as potential candidates in the prevention and treatment of diseases via modulation of biological activities. Int. J. Phys. Pathophysiol. Pharm. 2014, 6, 125–136. [Google Scholar]
- Hopkins, A. Network pharmacology. Nat. Biotechnol. 2007, 25, 1110–1111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, X.; Li, S. An integrative platform of TCM network pharmacology and its application on a herbal formula, Qing-Luo-Yin. Altern. Med. 2013, 2013, 456747. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.; Iyengar, R. Network analyses in systems pharmacology. Bioinformatics 2009, 25, 2466–2472. [Google Scholar] [CrossRef] [PubMed]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, Y.; Jain, C.G. Effect of Petroleum ether extract of Cassia fistula Seeds on Uterine Histoarchitecture of Ovariectomized Female Rats. Indian J. Fundam. Appl. Life Sci. 2009, 1, 438–444. [Google Scholar]
- Yadava, R.; Verma, V. A new biologically active flavone glycoside from the seeds of Cassia fistula (Linn.). J. Asian Nat. Prod. Res. 2003, 5, 57–61. [Google Scholar] [CrossRef]
- Bhakta, T.; Mukherjee, P.; Saha, K.; Pal, M.; Saha, B. Hypoglycemic activity of Cassia fistula Linn.(Leguminosae) leaf (Methanol extract) in alloxan-induced diabetic rats. Indian J. Pharm. Sci. 1997, 9, 35–38. [Google Scholar]
- Sangita, D.; Sarkar, P.; Sengupta, A.; Chattopadhyay, A. Clinical study of Aragvadha (Cassia fistula Linn) on Vicharchika (Eczema). Ayu 2008, 39, 9–15. [Google Scholar]
- Karthikeyan, S.; Gobianand, K. Antiulcer activity of ethanol leaf extract of Cassia fistula. Pharm. Biol. 2010, 48, 869–877. [Google Scholar] [CrossRef]
- Silawat, N.; Jarald, E.E.; Jain, N.; Yadav, A.; Deshmukh, P. The mechanism of hypoglycemic and antidiabetic action of hydroalcholic extract of Cassia fistula Linn. in rats. Biol. Chem. 2009, 1, 82–92. [Google Scholar]
- Singh, K.; Bharadwaj, U. Hypoglycaemic activity of Albizzia stipulata, Albizzia moluccana and Cassia fistula leguminous seed diets on normal young rats. Indian J. Pharm. Sci. 1975, 7, 47–49. [Google Scholar]
- Manonmani, G.; Bhavapriya, V.; Kalpana, S.; Govindasamy, S.; Apparanantham, T. Antioxidant activity of Cassia fistula (Linn.) flowers in alloxan induced diabetic rats. J. Ethnopharmacol. 2005, 97, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Duraipandiyan, V.; Ignacimuthu, S. Antibacterial and antifungal activity of Cassia fistula L.: An ethnomedicinal plant. J. Ethnopharmacol. 2007, 112, 590–594. [Google Scholar] [CrossRef]
- Bhatnagar, M.; Vimal, S.; Vyas, Y.; Sharma, D.; Sharma, K. Antioxidant activity of fruit pulp powder of Cassia fistula. Pharmacogn. J. 2010, 2, 219–228. [Google Scholar] [CrossRef]
- Abo, K.; Lasaki, S.; Adeyemi, A. Laxative and antimicrobial properties of Cassia species growing in Ibadan. Niger. J. Nat. Prod. Med. 1999, 3, 47–50. [Google Scholar] [CrossRef]
- Dutta, A.; Bratati, D. Seasonal variation in the content of sennosides and rhein in leaves and pods of Cassia fistula. Pharm. Sci. 1998, 60, 388. [Google Scholar]
- Aminah, N.; Tun, K.; Kristanti, A.; Aung, H.; Takaya, Y.; Choudhary, M. Chemical constituents and their biological activities from Taunggyi (Shan state) medicinal plants. Heliyon 2021, 7, e06173. [Google Scholar] [CrossRef]
- Ali, M. Cassia fistula Linn: A review of phytochemical and pharmacological studies. Int. J. Pharm. Sci. Res. 2014, 5, 2125–2130. [Google Scholar]
- Mwangi, R.W.; Macharia, J.M.; Wagara, I.N.; Bence, R.L. The medicinal properties of Cassia fistula L.: A review. Biomed. Pharmacother. 2021, 144, 112240. [Google Scholar] [CrossRef]
- Wijaya, S.H.; Tanaka, Y.; Amin, A.U.; Morita, A.H.; Afendi, F.M.; Batubara, I.; Ono, N.; Darusman, L.K.; Kanaya, S. Utilization of KNApSAcK family databases for developing herbal medicine systems. J. Comput. Aided Chem. 2016, 17, 1–7. [Google Scholar] [CrossRef]
- Li, J.; Huang, Y.; Zhao, S.; Guo, Q.; Zhou, J.; Han, W.; Xu, Y. Based on network pharmacology to explore the molecular mechanisms of Astragalus membranaceus for treating T2 diabetes mellitus. Ann. Transl. Med. 2019, 7, 633. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Stein, I.T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, 2017, bax028. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 1–7. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- De Jong, H.; Geiselmann, J.; Hernandez, C.; Page, M. Genetic Network Analyzer: Qualitative simulation of genetic regulatory networks. Bioinformatics 2003, 19, 336–344. [Google Scholar] [CrossRef]
- Li, H.; Hung, A.; Yang, A.W.H. Herb-target virtual screening and network pharmacology for prediction of molecular mechanism of Danggui Beimu Kushen Wan for prostate cancer. Sci. Rep. 2021, 11, 6656. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Lu, X.; Zheng, Y.; Wen, F.; Huang, W.; Chen, X.; Ruan, S.; Gu, S.; Hu, Y.; Teng, Y.; Shu, P. Study of the active ingredients and mechanism of Sparganii rhizoma in gastric cancer based on HPLC-Q-TOF–MS/MS and network pharmacology. Sci. Rep. 2021, 11, 1905. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Bittrich, S.; Rose, Y.; Segura, J.; Lowe, R.; Westbrook, J.D.; Duarte, J.M.; Burley, S.K. RCSB Protein Data Bank: Improved annotation, search and visualization of membrane protein structures archived in the PDB. Bioinformatics 2022, 38, 1452–1454. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Mirzaei, H.; Zarbafian, S.; Villar, E.; Mottarella, S.; Beglov, D.; Vajda, S.; Paschalidis, I.C.; Vakili, P.; Kozakov, D. Energy minimization on manifolds for docking flexible molecules. J. Chem. Theory Comput. 2015, 11, 1063–1076. [Google Scholar] [CrossRef]
- Nobili, S.; Lippi, D.; Witort, E.; Donnini, M.; Bausi, L.; Mini, E.; Capaccioli, S. Natural compounds for cancer treatment and prevention. Pharmacol. Res. 2009, 59, 365–378. [Google Scholar] [CrossRef]
- Chen, L.; Cheng, X.; Tu, W.; Qi, Z.; Li, H.; Liu, F.; Yang, F.; Zhang, Z.; Wang, Z. Apatinib inhibits glycolysis by suppressing the VEGFR2/AKT1/SOX5/GLUT4 signaling pathway in ovarian cancer cells. Cell Oncol. 2019, 42, 679–690. [Google Scholar] [CrossRef]
- Etemadmoghadam, D.; Bowtell, D. AKT1 gene amplification as a biomarker of treatment response in ovarian cancer: Mounting evidence of a therapeutic target. Gynecol. Oncol. 2014, 135, 409–410. [Google Scholar] [CrossRef]
- Hua, G.; He, C.; Lv, X.; Fan, L.; Wang, C.; Remmenga, S.W.; Rodabaugh, K.J.; Yang, L.; Lele, S.M.; Yang, P.; et al. The four and a half LIM domains 2 (FHL2) regulates ovarian granulosa cell tumor progression via controlling AKT1 transcription. Cell Death Dis. 2016, 7, e2297. [Google Scholar] [CrossRef]
- Chen, Q.; He, Q.; Zhuang, L.; Wang, K.; Yin, C.; He, L. IP10-CDR3 Reduces the Viability and Induces the Apoptosis of Ovarian Cancer Cells by Down-Regulating the Expression of Bcl-2 and Caspase 3. Onco Targets Ther. 2019, 12, 9697. [Google Scholar] [CrossRef] [PubMed]
- Gregoraszczuk, E.; Rak-Mardyla, A.; Rys, J.; Jakubowicz, J.; Urbański, K. Effect of chemotherapeutic drugs on caspase-3 activity, as a key biomarker for apoptosis in ovarian tumor cell cultured as monolayer. A pilot study. Iran. J. Pharm. Res. 2015, 14, 1153–1161. [Google Scholar] [PubMed]
- Kleinberg, L.; Dong, H.P.; Holth, A.; Risberg, B.; Trope, C.G.; Nesland, J.M.; Flørenes, V.A.; Davidson, B. Cleaved caspase-3 and nuclear factor-κB p65 are prognostic factors in metastatic serous ovarian carcinoma. Hum. Pathol. 2009, 40, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zheng, F.; Xing, H.; Gao, Q.; Wei, W.; Lu, Y.; Wang, S.; Zhou, J.; Hu, W.; Ma, D. Resistance to chemotherapy-induced apoptosis via decreased caspase-3 activity and overexpression of antiapoptotic proteins in ovarian cancer. J. Cancer Res. Clin. Oncol. 2004, 130, 423–428. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, D.; Zhao, Z.; Xiao, Y.; Sengupta, S.; Xiao, Y.; Zhang, R.; Lauber, K.; Wesselborg, S.; Feng, L.; et al. Caspase-3-dependent activation of calcium-independent phospholipase A2 enhances cell migration in non-apoptotic ovarian cancer cells. J. Biol. Chem. 2006, 281, 29357–29368. [Google Scholar] [CrossRef]
- Zhou, C.; Qiu, L.; Sun, Y.; Healey, S.; Wanebo, H.; Kouttab, N.; Di, W.; Yan, B.; Wan, Y. Inhibition of EGFR/PI3K/AKT cell survival pathway promotes TSA’s effect on cell death and migration in human ovarian cancer cells. Int. J. Oncol. 2006, 29, 269–278. [Google Scholar] [CrossRef]
- Sheppard, K.E.; Cullinane, C.; Hannan, K.M.; Wall, M.; Chan, J.; Barber, F.; Foo, J.; Cameron, D.; Neilsen, A.; Ng, P.; et al. Synergistic inhibition of ovarian cancer cell growth by combining selective PI3K/mTOR and RAS/ERK pathway inhibitors. Eur. J. Cancer 2013, 49, 3936–3944. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, X.; Chen, M.; Zheng, S.; Li, J.; Lin, S.; Wang, X. ST3Gal3 confers paclitaxel—mediated chemoresistance in ovarian cancer cells by attenuating caspase-8/3 signaling. Mol. Med. Rep. 2019, 20, 4499–4506. [Google Scholar] [CrossRef]
- Ediriweera, M.K.; Tennekoon, K.H.; Samarakoon, S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Sanderson, R.D. Heparan sulfate proteoglycans in invasion and metastasis. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2001. [Google Scholar]
- Gupta, M.; Babic, A.; Beck, A.H.; Terry, K. TNF-α expression, risk factors, and inflammatory exposures in ovarian cancer: Evidence for an inflammatory pathway of ovarian carcinogenesis? Hum. Pathol. 2016, 54, 82–91. [Google Scholar] [CrossRef]
- Thaklaewphan, P.; Ruttanapattanakul, J.; Monkaew, S.; Buatoom, M.; Sookkhee, S.; Nimlamool, W.; Potikanond, S. Kaempferia parviflora extract inhibits TNF-α-induced release of MCP-1 in ovarian cancer cells through the suppression of NF-κB signaling. Biomed. Pharmacother. 2021, 141, 111911. [Google Scholar] [CrossRef]
- Girondel, C.; Lévesque, K.; Langlois, M.-J.; Pasquin, S.; Saba-El-Leil, M.K.; Rivard, N.; Friesel, R.; Servant, M.J.; Gauchat, J.-F.; Lesage, S.; et al. Loss of interleukin-17 receptor D promotes chronic inflammation-associated tumorigenesis. Oncogene 2021, 40, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]




| Sr.No. | Phytochemicals | Molecular Formula | Molecular Weight (Dalton) | Drug Likeness (>0.18) | Oral Bioavailability (>30%) | 2D Structures | PubChem ID |
|---|---|---|---|---|---|---|---|
| 01 | Rhein | C15H8O6 | 284.22 | 0.28 | 47.07 | 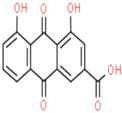 | 10168 |
| 02 | Ellagic Acid | C14H6O8 | 302.19 | 0.43 | 43.06 |  | 5281855 |
| 03 | Quercetin | C15H10O7 | 302.23 | 0.28 | 46.43 | 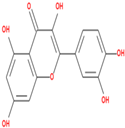 | 5280343 |
| 04 | Kaempferol | C15H10O6 | 286.24 | 0.24 | 41.88 |  | 5280863 |
| 05 | Gibberellin A3 | C19H22O6 | 346.4 | 0.53 | 81.59 |  | 6466 |
| 06 | Licoisoflavone | C20H18O6 | 354.4 | 0.42 | 41.61 |  | 5281789 |
| 07 | β-Sitosterol | C29H50O | 414.7 | 0.75 | 36.91 |  | 222284 |
| 08 | Stigmasterol | C29H48O | 412.7 | 0.76 | 43.83 |  | 5280794 |
| Sr. No. | Phytochemical Name | Categories | Degree |
|---|---|---|---|
| 01 | Rhein | Anthraquinone | 32 |
| 02 | Ellagic Acid | Polyphenol | 32 |
| 03 | Quercetin | Flavonoid | 30 |
| 04 | Kaempferol | Flavonoid | 29 |
| 05 | Gibberellin A3 | Hormone | 28 |
| 06 | Licoisoflavone | Flavonoid | 26 |
| 07 | ß-Sitosterol | Phytosterols | 21 |
| 08 | Stigmasterol | Phytosterols | 19 |
| Sr. No | Compound | Binding Affinities (kcal/mol) | |||
|---|---|---|---|---|---|
| AKT1 | CTNNB1 | ESR1 | CASP3 | ||
| 01 | Rhein | −7.5 | −6.9 | −6.8 | −6.2 |
| 02 | Ellagic acid | −7.9 | −7.2 | −6.6 | −6.4 |
| 03 | Quercetin | −7.7 | −6.5 | −7.7 | −6.1 |
| 04 | Kaempferol | −7.6 | −6.7 | −7.3 | −5.8 |
| 05 | Gibberelin A3 | −8.2 | −6.7 | −6.6 | −6.7 |
| 06 | Licoisoflavone | −7.9 | −6.1 | −6.6 | −6.5 |
| 07 | β- Sitosterol | −8.8 | −7.0 | −6.7 | −6.8 |
| 08 | Stigmasterol | −9.2 | −6.7 | −7.0 | −6.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanwal, A.; Azeem, F.; Nadeem, H.; Ashfaq, U.A.; Aadil, R.M.; Kober, A.K.M.H.; Rajoka, M.S.R.; Rasul, I. Molecular Mechanisms of Cassia fistula against Epithelial Ovarian Cancer Using Network Pharmacology and Molecular Docking Approaches. Pharmaceutics 2022, 14, 1970. https://doi.org/10.3390/pharmaceutics14091970
Kanwal A, Azeem F, Nadeem H, Ashfaq UA, Aadil RM, Kober AKMH, Rajoka MSR, Rasul I. Molecular Mechanisms of Cassia fistula against Epithelial Ovarian Cancer Using Network Pharmacology and Molecular Docking Approaches. Pharmaceutics. 2022; 14(9):1970. https://doi.org/10.3390/pharmaceutics14091970
Chicago/Turabian StyleKanwal, Aqsa, Farrukh Azeem, Habibullah Nadeem, Usman Ali Ashfaq, Rana Muhammad Aadil, A. K. M. Humayun Kober, Muhammad Shahid Riaz Rajoka, and Ijaz Rasul. 2022. "Molecular Mechanisms of Cassia fistula against Epithelial Ovarian Cancer Using Network Pharmacology and Molecular Docking Approaches" Pharmaceutics 14, no. 9: 1970. https://doi.org/10.3390/pharmaceutics14091970
APA StyleKanwal, A., Azeem, F., Nadeem, H., Ashfaq, U. A., Aadil, R. M., Kober, A. K. M. H., Rajoka, M. S. R., & Rasul, I. (2022). Molecular Mechanisms of Cassia fistula against Epithelial Ovarian Cancer Using Network Pharmacology and Molecular Docking Approaches. Pharmaceutics, 14(9), 1970. https://doi.org/10.3390/pharmaceutics14091970