Determination of Mutational Timing of Colistin-Resistance Genes through Klebsiella pneumoniae Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Strain and Culture Media
2.2. Bacterial Transfer and Resistance Selection
2.3. MIC Measurement
2.4. Whole-Genome DNA Sequencing
2.5. Comprehensive Mutation Analysis
2.6. Mutation Selection Criteria
2.7. String Test for Hypervirulence
3. Results
3.1. Rapid MIC Increase in Colistin-Treated KP Populations
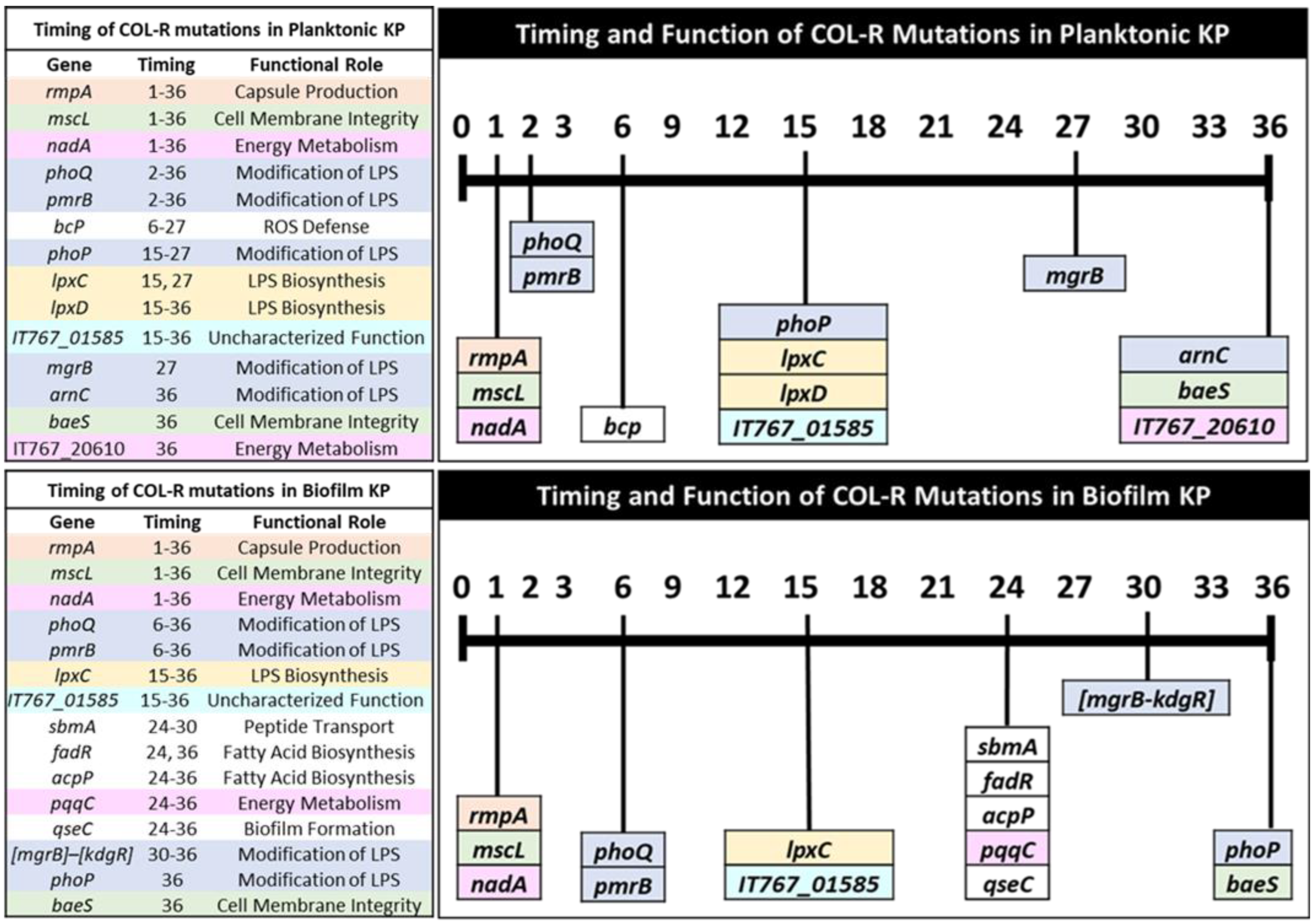
3.2. Temporal Regulation of COL-R Mutations in Planktonic KP
3.3. Temporal Regulation of COL-R Mutations in Biofilm KP
3.4. Timing of Colistin Resistance Mutations by Functional Roles
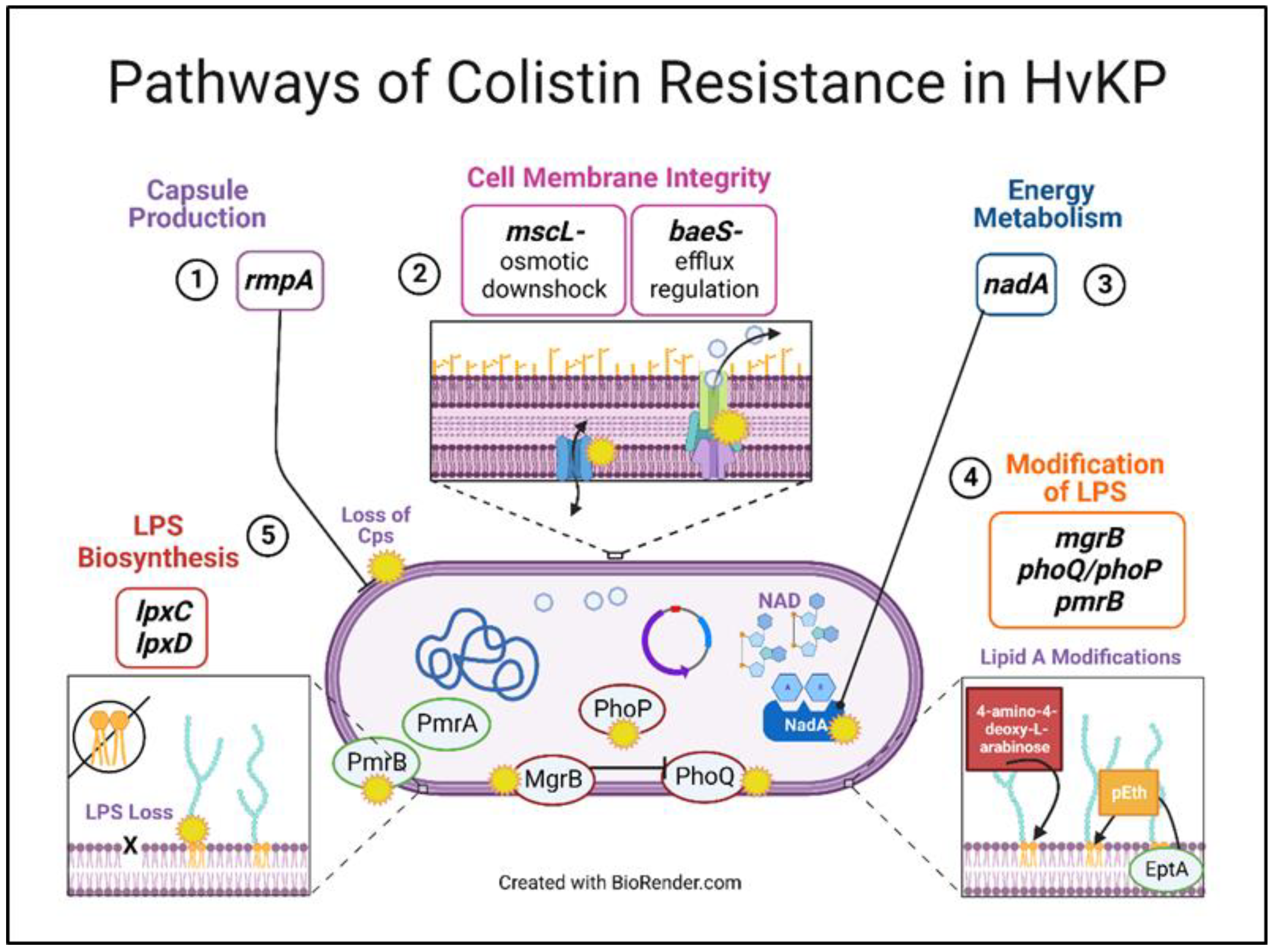
3.4.1. Capsule Production
3.4.2. Cell Membrane Integrity
3.4.3. Energy Metabolism
3.4.4. Modification of LPS
3.4.5. LPS Biosynthesis
3.4.6. ROS Defense
3.4.7. Peptide Transport
3.4.8. Fatty-Acid Biosynthesis
3.4.9. Biofilm Formation
3.4.10. Mutations with Uncharacterized Function
3.5. Theoretical Pathways of Colistin Resistance in K. pneumoniae
3.6. COL-R Isolates Remain Susceptible to Dual-Inhibitor Antibiotics
3.7. Loss of Hypermucoviscous Phenotype with Colistin Selection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jean, S.-S.; Harnod, D.; Hsueh, P.-R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Bradford, P.A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, G.; Del Giacomo, P.; Posteraro, B.; Sanguinetti, M.; Tumbarello, M. Molecular Mechanisms, Epidemiology, and Clinical Importance of β-Lactam Resistance in Enterobacteriaceae. Int. J. Mol. Sci. 2020, 21, 5090. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Podschun, R.; Ullmann, U. Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [Green Version]
- Lan, P.; Jiang, Y.; Zhou, J.; Yu, Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 25, 26–34. [Google Scholar] [CrossRef]
- Jorge, P.; Pérez-Pérez, M.; Pérez-Rodríguez, G.; Pereira, M.O.; Lourenço, A. A network perspective on antimicrobial peptide combination therapies: The potential of colistin, polymyxin B and nisin. Int. J. Antimicrob. Agents 2017, 49, 668–676. [Google Scholar] [CrossRef] [Green Version]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5311054, Polymyxin E. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5311054 (accessed on 12 December 2022).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 4868, Polymyxin b. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/4868 (accessed on 12 December 2022).
- Jorge, P.; Lourenço, A.; Pereira, M.O. New trends in peptide-based anti-biofilm strategies: A review of recent achievements and bioinformatic approaches. Biofouling 2012, 28, 1033–1061. [Google Scholar] [CrossRef] [Green Version]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Nation, R.L.; Milne, R.W.; Turnidge, J.D.; Coulthard, K. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 2005, 25, 11–25. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Pozo, J.L.; Patel, R. The Challenge of Treating Biofilm-associated Bacterial Infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, G.; Chao, X.; Xie, L.; Wang, H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 2020, 17, 6278. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Kobayashi, K. Role of Glutamate Synthase in Biofilm Formation by Bacillus subtilis. J. Bacteriol. 2020, 202, e00120-20. [Google Scholar] [CrossRef]
- Palwe, S.; Bakthavatchalam, Y.D.; Khobragadea, K.; Kharat, A.S.; Walia, K.; Veeraraghavan, B. In-Vitro Selection of Ceftazidime/Avibactam Resistance in OXA-48-Like-Expressing Klebsiella pneumoniae: In-Vitro and In-Vivo Fitness, Genetic Basis and Activities of β-Lactam Plus Novel β-Lactamase Inhibitor or β-Lactam Enhancer Combinations. Antibiotics 2021, 10, 1318. [Google Scholar] [CrossRef]
- O’Rourke, D.; FitzGerald, C.E.; Traverse, C.C.; Cooper, V.S. There and back again: Consequences of biofilm specialization under selection for dispersal. Front. Genet. 2015, 6, 18. [Google Scholar] [CrossRef]
- Cooper, V.S. Experimental Evolution as a High-Throughput Screen for Genetic Adaptations. mSphere 2018, 3, e00121-18. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Pilewski, J.M.; Di, Y.P. Acidic Microenvironment Determines Antibiotic Susceptibility and Biofilm Formation of Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 747834. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. CLSI Document M07-A10: Methods for Dilution of Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—10th Edition; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015.
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Budnick, J.A.; Bina, X.R.; Bina, J.E. Complete Genome Sequence of Klebsiella pneumoniae Strain ATCC 43816. Microbiol. Resour. Announc. 2021, 10, e01441-20. [Google Scholar] [CrossRef] [PubMed]
- Deatherage, D.E.; Barrick, J.E. Identification of Mutations in Laboratory-Evolved Microbes from Next-Generation Sequencing Data Using breseq. In Engineering and Analyzing Multicellular Systems; Sun, L., Shou, W., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2014; Volume 1151, pp. 165–188. ISBN 9781493905539. [Google Scholar]
- Fang, C.-T.; Chuang, Y.-P.; Shun, C.-T.; Chang, S.-C.; Wang, J.-T. A Novel Virulence Gene in Klebsiella pneumoniae Strains Causing Primary Liver Abscess and Septic Metastatic Complications. J. Exp. Med. 2004, 199, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.-W.; Zheng, J.-X.; Bai, B.; Xu, G.-J.; Lin, F.-J.; Chen, Z.; Sun, X.; Qu, D.; Yu, Z.-J.; Deng, Q.-W. Characteristics of Hypervirulent Klebsiella pneumoniae: Does Low Expression of rmpA Contribute to the Absence of Hypervirulence? Front. Microbiol. 2020, 11, 436. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.-C.; Peng, H.-L.; Chang, H.-Y. RmpA2, an Activator of Capsule Biosynthesis in Klebsiella pneumoniae CG43, Regulates K2 cps Gene Expression at the Transcriptional Level. J. Bacteriol. 2003, 185, 788–800. [Google Scholar] [CrossRef] [Green Version]
- Ni, L.; Tonthat, N.K.; Chinnam, N.; Schumacher, M.A. Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: An AraC DNA-binding family member with a LacI/GalR ligand-binding domain. Nucleic Acids Res. 2013, 41, 1998–2008. [Google Scholar] [CrossRef] [Green Version]
- Magnusson, O.T.; Toyama, H.; Saeki, M.; Rojas, A.; Reed, J.C.; Liddington, R.C.; Klinman, J.P.; Schwarzenbacher, R. Quinone biogenesis: Structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proc. Natl. Acad. Sci. USA 2004, 101, 7913–7918. [Google Scholar] [CrossRef] [Green Version]
- Herrera, C.M.; Hankins, J.V.; Trent, M.S. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 2010, 76, 1444–1460. [Google Scholar] [CrossRef]
- Leung, L.M.; Cooper, V.S.; Rasko, D.A.; Guo, Q.; Pacey, M.P.; McElheny, C.L.; Mettus, R.T.; Yoon, S.H.; Goodlett, D.R.; Ernst, R.K.; et al. Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2017, 72, 3035–3042. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Bontron, S.; Villegas, M.-V.; Ozdamar, M.; Türkoglu, S.; Nordmann, P. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 75–80. [Google Scholar] [CrossRef]
- Gunn, J.S.; Lim, K.B.; Krueger, J.; Kim, K.; Guo, L.; Hackett, M.; Miller, S.I. PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998, 27, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.; Cha, M.-K.; Kim, I.-H. Thioredoxin-dependent Hydroperoxide Peroxidase Activity of Bacterioferritin Comigratory Protein (BCP) as a New Member of the Thiol-specific Antioxidant Protein (TSA)/Alkyl Hydroperoxide Peroxidase C (AhpC) Family. J. Biol. Chem. 2000, 275, 2924–2930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghilarov, D.; Inaba-Inoue, S.; Stepien, P.; Qu, F.; Michalczyk, E.; Pakosz, Z.; Nomura, N.; Ogasawara, S.; Walker, G.C.; Rebuffat, S.; et al. Molecular mechanism of SbmA, a promiscuous transporter exploited by antimicrobial peptides. Sci. Adv. 2021, 7, eabj5363. [Google Scholar] [CrossRef]
- Ghosal, A.; Vitali, A.; Stach, J.E.M.; Nielsen, P.E. Role of SbmA in the Uptake of Peptide Nucleic Acid (PNA)-Peptide Conjugates in E. coli. ACS Chem. Biol. 2013, 8, 360–367. [Google Scholar] [CrossRef]
- Cronan, J.E. The Escherichia coli FadR transcription factor: Too much of a good thing? Mol. Microbiol. 2021, 115, 1080–1085. [Google Scholar] [CrossRef]
- Byers, D.M.; Gong, H. Acyl carrier protein: Structure-function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 2007, 85, 649–662. [Google Scholar] [CrossRef]
- Lv, J.; Zhu, J.; Wang, T.; Xie, X.; Wang, T.; Zhu, Z.; Chen, L.; Zhong, F.; Du, H. The Role of the Two-Component QseBC Signaling System in Biofilm Formation and Virulence of Hypervirulent Klebsiella pneumoniae ATCC43816. Front. Microbiol. 2022, 13, 817974. [Google Scholar] [CrossRef]
- Boinett, C.J.; Cain, A.K.; Hawkey, J.; Do Hoang, N.T.; Khanh, N.N.T.; Thanh, D.P.; Dordel, J.; Campbell, J.I.; Lan, N.P.H.; Mayho, M.; et al. Clinical and laboratory-induced colistin-resistance mechanisms in Acinetobacter baumannii. Microb. Genom. 2019, 5, e000246. [Google Scholar] [CrossRef]
- Masood, K.I.; Umar, S.; Hasan, Z.; Farooqi, J.; Razzak, S.A.; Jabeen, N.; Rao, J.; Shakoor, S.; Hasan, R. Lipid A-Ara4N as an alternate pathway for (colistin) resistance in Klebsiella pneumonia isolates in Pakistan. BMC Res. Notes 2021, 14, 449. [Google Scholar] [CrossRef]
- Nummila, K.; Kilpeläinen, I.; Zähringer, U.; Vaara, M.; Helander, I.M. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coii are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 1995, 16, 271–278. [Google Scholar] [CrossRef]
- Moskowitz, S.M.; Brannon, M.K.; Dasgupta, N.; Pier, M.; Sgambati, N.; Miller, A.K.; Selgrade, S.E.; Miller, S.I.; Denton, M.; Conway, S.P.; et al. PmrB Mutations Promote Polymyxin Resistance of Pseudomonas aeruginosa Isolated from Colistin-Treated Cystic Fibrosis Patients. Antimicrob. Agents Chemother. 2012, 56, 1019–1030. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Chen, Q.; Shen, F.; Jiang, Y.; Wu, X.; Hua, X.; Fu, Y.; Yu, Y. Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg. Microbes Infect. 2021, 10, 1129–1136. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.F.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St. Michael, F.; Cox, A.D.; et al. Colistin Resistance in Acinetobacter baumannii Is Mediated by Complete Loss of Lipopolysaccharide Production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Li, X.; Chen, Y.; Jiang, Y.; Zhou, Z.; Zhang, H.; Sun, L.; Ruan, Z.; Feng, Y.; Akova, M.; et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: A multicentre longitudinal study. Lancet Infect. Dis. 2017, 17, 400–410. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 119–141. [Google Scholar] [CrossRef]
- Cantó, C.; Menzies, K.J.; Auwerx, J. NAD+ Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef] [Green Version]
- Booth, I.R.; Blount, P. The MscS and MscL Families of Mechanosensitive Channels Act as Microbial Emergency Release Valves. J. Bacteriol. 2012, 194, 4802–4809. [Google Scholar] [CrossRef] [Green Version]
- Wray, R.; Wang, J.; Iscla, I.; Blount, P. Novel MscL agonists that allow multiple antibiotics cytoplasmic access activate the channel through a common binding site. PLoS ONE 2020, 15, e0228153. [Google Scholar] [CrossRef]
- Moosavian, M.; Emam, N.; Pletzer, D.; Savari, M. Rough-type and loss of the LPS due to lpx genes deletions are associated with colistin resistance in multidrug-resistant clinical Escherichia coli isolates not harbouring mcr genes. PLoS ONE 2020, 15, e0233518. [Google Scholar] [CrossRef]
- He, L.; Dai, K.; Wen, X.; Ding, L.; Cao, S.; Huang, X.; Wu, R.; Zhao, Q.; Huang, Y.; Yan, Q.; et al. QseC Mediates Osmotic Stress Resistance and Biofilm Formation in Haemophilus parasuis. Front. Microbiol. 2018, 9, 212. [Google Scholar] [CrossRef]
- Leblanc, S.K.D.; Oates, C.W.; Raivio, T.L. Characterization of the Induction and Cellular Role of the BaeSR Two-Component Envelope Stress Response of Escherichia coli. J. Bacteriol. 2011, 193, 3367–3375. [Google Scholar] [CrossRef] [Green Version]
- Ehmann, D.E.; Jahić, H.; Ross, P.L.; Gu, R.-F.; Hu, J.; Durand-Réville, T.F.; Lahiri, S.; Thresher, J.; Livchak, S.; Gao, N.; et al. Kinetics of Avibactam Inhibition against Class A, C, and D β-Lactamases. J. Biol. Chem. 2013, 288, 27960–27971. [Google Scholar] [CrossRef] [Green Version]
- Ku, Y.-H.; Lee, M.-F.; Chuang, Y.-C.; Chen, C.-C.; Yu, W.-L. In vitro activity of colistin sulfate against Enterobacteriaceae producing extended-spectrum β-lactamases. J. Microbiol. Immunol. Infect. 2015, 48, 699–702. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, X.; Wang, S.; Sun, S.; Li, H.; Chen, H.; Wang, Q.; Wang, H. Emergence of Colistin Resistance in Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Under the Pressure of Tigecycline. Front. Microbiol. 2021, 12, 756580. [Google Scholar] [CrossRef]
- Muhammad, J.S.; Khan, N.A.; Maciver, S.K.; Alharbi, A.M.; Alfahemi, H.; Siddiqui, R. Epigenetic-Mediated Antimicrobial Resistance: Host versus Pathogen Epigenetic Alterations. Antibiotics 2022, 11, 809. [Google Scholar] [CrossRef]
- McConville, T.H.; Giddins, M.J.; Uhlemann, A.-C. An efficient and versatile CRISPR-Cas9 system for genetic manipulation of multi-drug resistant Klebsiella pneumoniae. STAR Protoc. 2021, 2, 100373. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Chen, W.; Song, L.; Zhang, Y.; Shen, Z.; Yu, F.; Li, M.; Ji, Q. CRISPR-Cas9 and CRISPR-Assisted Cytidine Deaminase Enable Precise and Efficient Genome Editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 2018, 84, e01834-18. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Wang, Y.; Dong, N.; Shen, L.; Zhou, H.; Hu, Y.; Gu, D.; Chen, S.; Zhang, R.; Ji, Q. Application of CRISPR/Cas9-Based Genome Editing in Studying the Mechanism of Pandrug Resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2019, 63, e00113-19. [Google Scholar] [CrossRef] [PubMed]



| Population 1 | Position | Mutation | Gene | Day 1: 64 | Day 2: 64 | Day 3: 64 | Day 6: 128 | Day 15: 512 | Day 27: 512 | Day 36: 2048 |
| 796,797 | SNP | mscL | 94.20% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,898,145 | SNP | nadA | 94.50% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 4,782,985:1 | INS | rmpA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 3,366,007 | SNP | phoQ | 26.30% | 9.80% | 92.50% | 100% | 100% | |||
| 5,228,037 | SNP | bcp | 24.80% | 100% | 100% | |||||
| 2,287,052 | SNP | lpxD | 15.00% | 100% | ||||||
| 4,585,314 | SNP | mgrB | 70.80% | |||||||
| 3,366,111 | SNP | phoQ | 100% | |||||||
| 2,949,548 | SNP | pmrB | 68.60% | |||||||
| 363,465 | SNP | mdtO | 12.20% | |||||||
| Population 2 | Position | Mutation | Gene | Day 1: 32 | Day 2: 256 | Day 3: 512 | Day 6: 512 | Day 15: 512 | Day 27: 512 | Day 36: 2048 |
| 796,797 | SNP | mscL | 94.20% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,898,145 | SNP | nadA | 94.50% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 4,782,985:1 | INS | rmpA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 3,367,207 | SNP | phoQ | 12.60% | 79.30% | 100% | 100% | ||||
| 3,366,550 | SNP | phoQ | 25.10% | 14.70% | ||||||
| 5,228,086 | SNP | bcp | 85.00% | 85.50% | ||||||
| 2,287,052 | SNP | lpxD | 10.40% | |||||||
| 2,287,023 | SNP | lpxD | 100% | |||||||
| 3,366,222 | SNP | phoQ | 100% | |||||||
| 5,228,407 | DEL | bcp | 100% | |||||||
| Population 3 | Position | Mutation | Gene | Day 1: 32 | Day 2: 64 | Day 3: 64 | Day 6: 64 | Day 15: 512 | Day 27: 512 | Day 36: 2048 |
| 796,797 | SNP | mscL | 94.20% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,898,145 | SNP | nadA | 94.50% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 4,782,985:1 | INS | rmpA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,949,368 | SNP | pmrB | 12.60% | 86.30% | 63.60% | |||||
| 3,367,778 | SNP | phoP | 33.00% | |||||||
| 3,366,214 | SNP | phoQ | 50.30% | |||||||
| 2,287,022 | SNP | lpxD | 37.60% | |||||||
| 3,367,218 | SNP | phoQ | 22.40% | |||||||
| 3,366,222 | SNP | phoQ | 72.90% | |||||||
| 796,974 | SNP | mscL | 21.00% | |||||||
| 3,366,013 | SNP | phoQ | 15.90% |
| Population 1 | Position | Mutation | Gene | Day 1: 2 | Day 2: 8 | Day 3: 128 | Day 6: 128 | Day 15: 256 | Day 24: 256 | Day 30: 512 | Day 36: 1024 |
| 796,797 | SNP | mscL | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,898,145 | SNP | nadA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 4,782,985:1 | INS | rmpA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,949,706 | SNP | pmrB | 47.60% | ||||||||
| 2,949,547 | SNP | pmrB | 40.70% | ||||||||
| 2,949,548 | SNP | pmrB | 38.60% | ||||||||
| 2,950,483 | SNP | pmrA | 36.40% | ||||||||
| 3,367,219 | SNP | phoQ | 13.10% | 52.20% | |||||||
| 2,178,392 | SNP | lpxC | 100% | ||||||||
| 2,178,666 | SNP | lpxC | 36.70% | ||||||||
| 4,585,299 | DEL | [mgrB]–[kdgR] | 100% | 100% | |||||||
| 2,950,399 | SNP | pmrA | 100% | ||||||||
| Population 2 | Position | Mutation | Gene | Day 1: 2 | Day 2: 2 | Day 3: 16 | Day 6: 64 | Day 15: 96 | Day 24: 512 | Day 30: 512 | Day 36: 2048 |
| 796,797 | SNP | mscL | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,898,145 | SNP | nadA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 4,782,985:1 | INS | rmpA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | ||
| 3,366,550 | SNP | phoQ | 74.60% | ||||||||
| 2,949,916 | SNP | pmrB | 75.90% | ||||||||
| 3,366,423 | SNP | phoQ | 33.90% | ||||||||
| 554,800 | DEL | qseC | 100% | 100% | 100% | ||||||
| 3,365,950 | SNP | phoQ | 100% | 100% | 100% | ||||||
| 4,903,958 | SNP | baeS | 100% | ||||||||
| Population 3 | Position | Mutation | Gene | Day 1: 2 | Day 2: 16 | Day 3: 64 | Day 6: 64 | Day 15: 128 | Day 24: 256 | Day 30: 1024 | Day 36: 2048 |
| 796,797 | SNP | mscL | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 2,898,145 | SNP | nadA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 4,782,985:1 | INS | rmpA | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| 864,664 | SNP | envZ | 100% | ||||||||
| 2,178,740 | SNP | lpxC | 100% | 100% | 100% | ||||||
| 2,949,703 | SNP | pmrB | 100% | 100% | 100% | ||||||
| 864,325 | SNP | envZ | 61.80% | ||||||||
| 3,367,440 | SNP | phoP | 45.40% |
| Planktonic KP Colistin-Resistance Mutations by Functional Role and MIC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population 1 | Population 2 | Population 3 | |||||||||
| Gene | Description | Function | Day | Freq % | MIC µg/mL | Day | Freq % | MIC µg/mL | Day | Freq % | MIC µg/mL |
| rmpA | mucoid phenotype A regulator | Capsule production | 1–36 | 100 | 64–2048 | 1–36 | 100 | 32–2048 | 1–36 | 100 | 32–2048 |
| mscL | large-conductance mechanosensitive channel protein | Cell Membrane Integrity | 1–36 | 94–100 | 64–2048 | 1–36 | 92–100 | 32–2048 | 1–36 | 100 | 32–2048 |
| baeS | two-component system sensor histidine kinase | - | - | - | - | - | - | 36 | 75 | 2048 | |
| nadA | quinolinate synthase | Energy Metabolism | 1–36 | 95–100 | 64–2048 | 1–36 | 91–100 | 32–2048 | 1–36 | 95–100 | 32–2048 |
| IT767_20610 | XylR family transcriptional regulator | - | - | - | 36 | 100 | 2048 | 36 | 83 | 2048 | |
| mgrB | PhoP/PhoQ regulator | Modification of LPS | 27 | 71 | 512 | - | - | - | - | - | - |
| phoQ | two-component system sensor histidine kinase | 2–27 36 | 26–100 | 64-2048 | 3–6, 3–27 36 | 13–100 | 512–2048 | 27 36 | 22 73 | 512 2048 | |
| phoP | two-component system response regulator | 27 | 10–15 | 512 | - | - | - | 15–27 | 8–33 | 512 | |
| pmrB | two-component system sensor histidine kinase | 36 | 69 | 2048 | - | - | - | 2–27 | 7–86 | 64–512 | |
| arnC | undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase | - | - | - | - | - | - | 36 | 81 | 2048 | |
| lpxD | UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | LPS biosynthesis | 15–27 | 15–100 | 512–2048 | 27 36 | 10 100 | 512 2048 | 27 | 38 | 512 |
| lpxC | UDP-3-O-acyl-N-acetylglucosamine deacetylase | - | - | - | 27 | 22 | 512 | 15 | 14 | 512 | |
| IT767_01585 | DUF3413 domain-containing protein | Unknown | 15 | 16 | 512 | 27 | 30 | 512 | 27–36 | 100 | 512–2048 |
| bcp | thioredoxin-dependent thiol peroxidase | ROS Defense | 6–27 | 25–100 | 128–512 | 15–27 36 | 8–100 | 512–2048 | - | - | - |
| Biofilm KP Colistin-Resistance Mutations by Functional Role and MIC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population 1 | Population 2 | Population 3 | |||||||||
| Gene | Description | Function | Day | Freq % | MIC µg/mL | Day | Freq % | MIC µg/mL | Day | Freq % | MIC µg/mL |
| rmpA | mucoid phenotype A regulator | Capsule production | 1–36 | 100 | 2–1024 | 1–36 | 100 | 2–2048 | 1–36 | 100 | 2–2048 |
| mscL | large-conductance mechanosensitive channel protein | Cell Membrane Integrity | 1–36 | 100 | 2–1024 | 1–36 | 100 | 2–2048 | 1–36 | 100 | 2–2048 |
| baeS | two-component system sensor histidine kinase | - | - | - | 36 | 100 | 2048 | - | - | - | |
| nadA | quinolinate synthase | Energy Metabolism | 1–36 | 100 | 2–1024 | 1–36 | 100 | 2–2048 | 1–36 | 100 | 2–2048 |
| pqqC | pyrroloquinoline-quinone synthase | - | - | - | - | - | - | 24–36 | 100 | 256–2048 | |
| [mgrB]–[kdgR] | PhoP/PhoQ regulator - DNA-binding transcriptional repressor | Modification of LPS | 30–36 | 100 | 512–1024 | - | - | - | - | - | - |
| phoQ | two-component system sensor histidine kinase | 6, 24 | 13, 52 | 128, 256 | 6, 15, 24–36 | 34–100 | 64–2048 | - | - | - | |
| phoP | two-component system response regulator | - | - | - | - | - | - | 36 | 45 | 2048 | |
| pmrB | two-component system sensor histidine kinase | 6, 6, 6 | 39–48 | 128 | 15 | 76 | 96 | 24–36 | 100 | 256–2048 | |
| lpxC | UDP-3-O-acyl-N-acetylglucosamine deacetylase | LPS Biosynthesis | 15, 24 | 100, 37 | 256 | - | - | - | 24–36 | 100 | 256–2048 |
| IT767_01585 | DUF3413 domain-containing protein | Unknown | - | - | - | 15, 24–36 | 63, 100 | 96, 512–2048 | 15 | 94 | 128 |
| fadR | fatty acid metabolism transcriptional regulator | Fatty Acid Biosynthesis | 24, 36 | 34, 100 | 256, 1024 | - | - | - | - | - | - |
| acpP | acyl carrier protein | 24–36 | 46–100 | 256–1024 | - | - | - | - | - | - | |
| qseC | two-component system sensor histidine kinase | Biofilm Formation | - | - | - | 24–36 | 100 | 512–2048 | - | - | - |
| sbmA | peptide antibiotic transporter | Peptide Transport | - | - | - | - | - | - | 24–30 | 52–70 | 256–1024 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuhn, J.M.; Di, Y.P. Determination of Mutational Timing of Colistin-Resistance Genes through Klebsiella pneumoniae Evolution. Pharmaceutics 2023, 15, 270. https://doi.org/10.3390/pharmaceutics15010270
Kuhn JM, Di YP. Determination of Mutational Timing of Colistin-Resistance Genes through Klebsiella pneumoniae Evolution. Pharmaceutics. 2023; 15(1):270. https://doi.org/10.3390/pharmaceutics15010270
Chicago/Turabian StyleKuhn, Jenna M., and Yuanpu Peter Di. 2023. "Determination of Mutational Timing of Colistin-Resistance Genes through Klebsiella pneumoniae Evolution" Pharmaceutics 15, no. 1: 270. https://doi.org/10.3390/pharmaceutics15010270
APA StyleKuhn, J. M., & Di, Y. P. (2023). Determination of Mutational Timing of Colistin-Resistance Genes through Klebsiella pneumoniae Evolution. Pharmaceutics, 15(1), 270. https://doi.org/10.3390/pharmaceutics15010270







