Quantitative Analysis of a Pilot Transwell Barrier Model with Automated Sampling and Mathematical Modeling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents, Buffers, and Media
2.2. Transwell Culture
2.3. Transepithelial Electrical Resistance
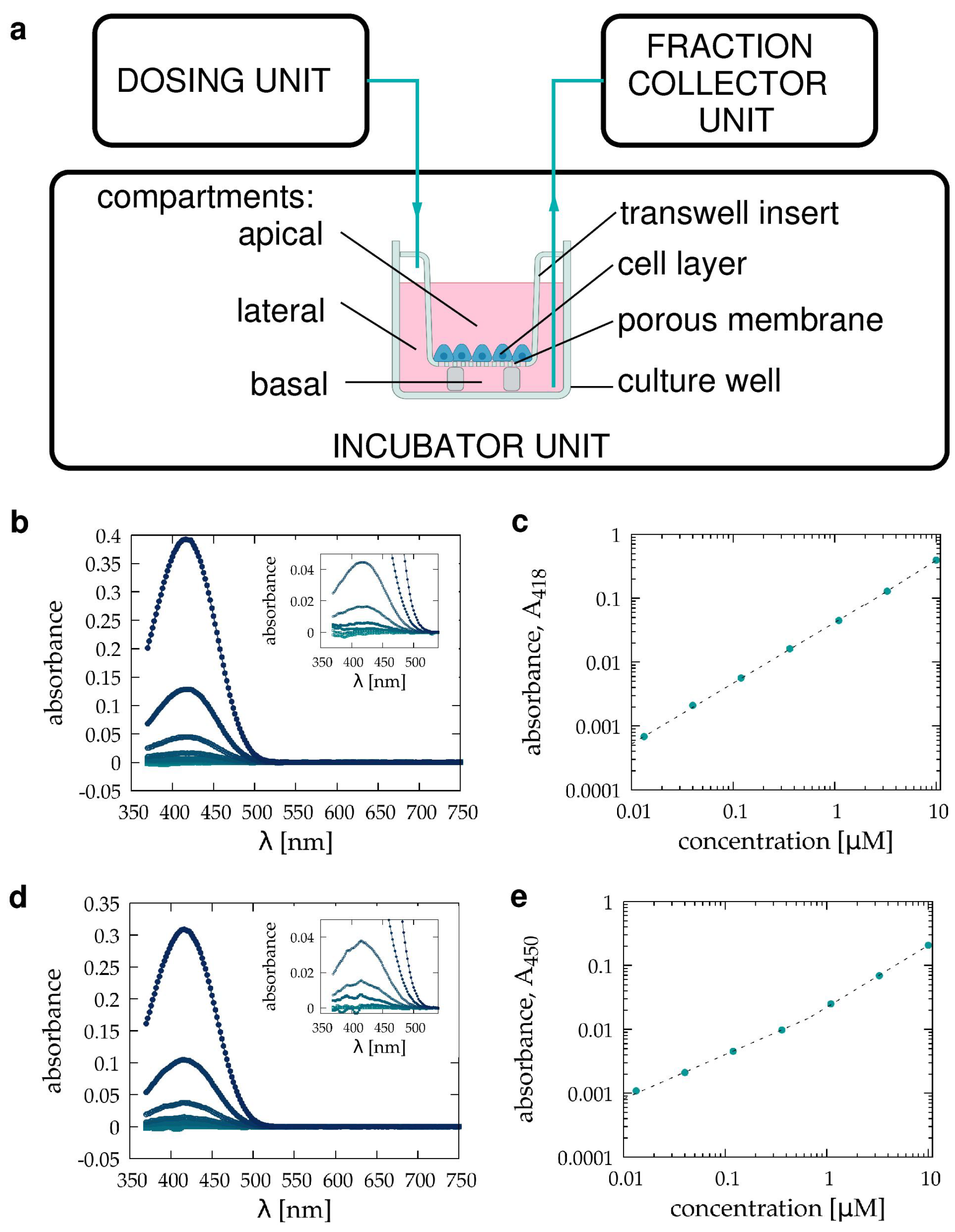
2.4. Spectroscopy
2.5. Automated Sample Handling
2.6. Computational Methods
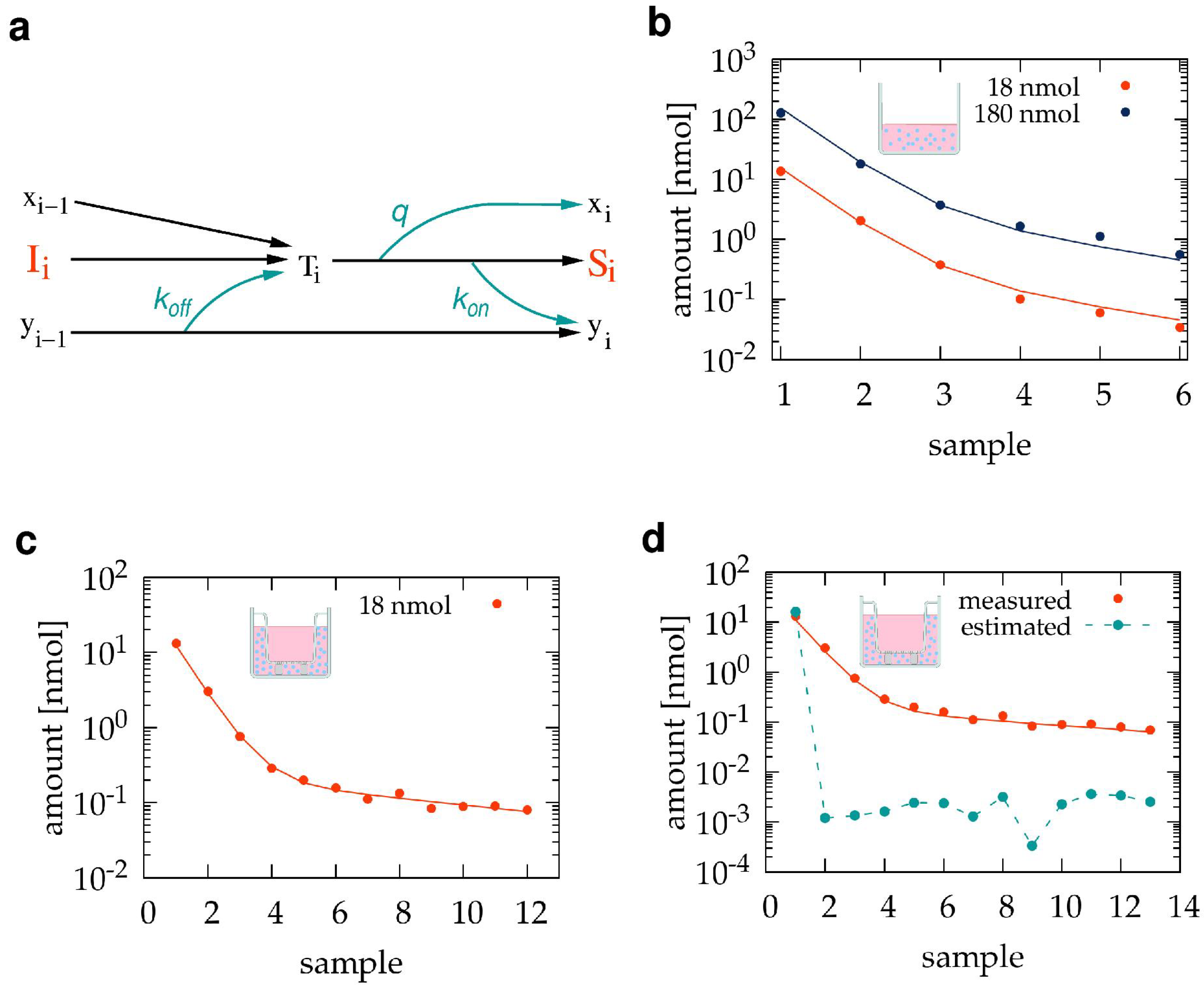
2.6.1. Adsorption Effects during Sample Collection
| Parameter | Value | |
|---|---|---|
| Without Transwell | With Transwell | |
| 0.034 | 0.088 | |
| 0.400 | 0.110 | |
| q | 0.110 | 0.240 |
2.6.2. Adsorption Model Distinguishing Sub-Compartments

| Basal Sub-Volume | Lateral Sub-Volume | ||
|---|---|---|---|
| Parameter | Value | Parameter | Value |
3. Results
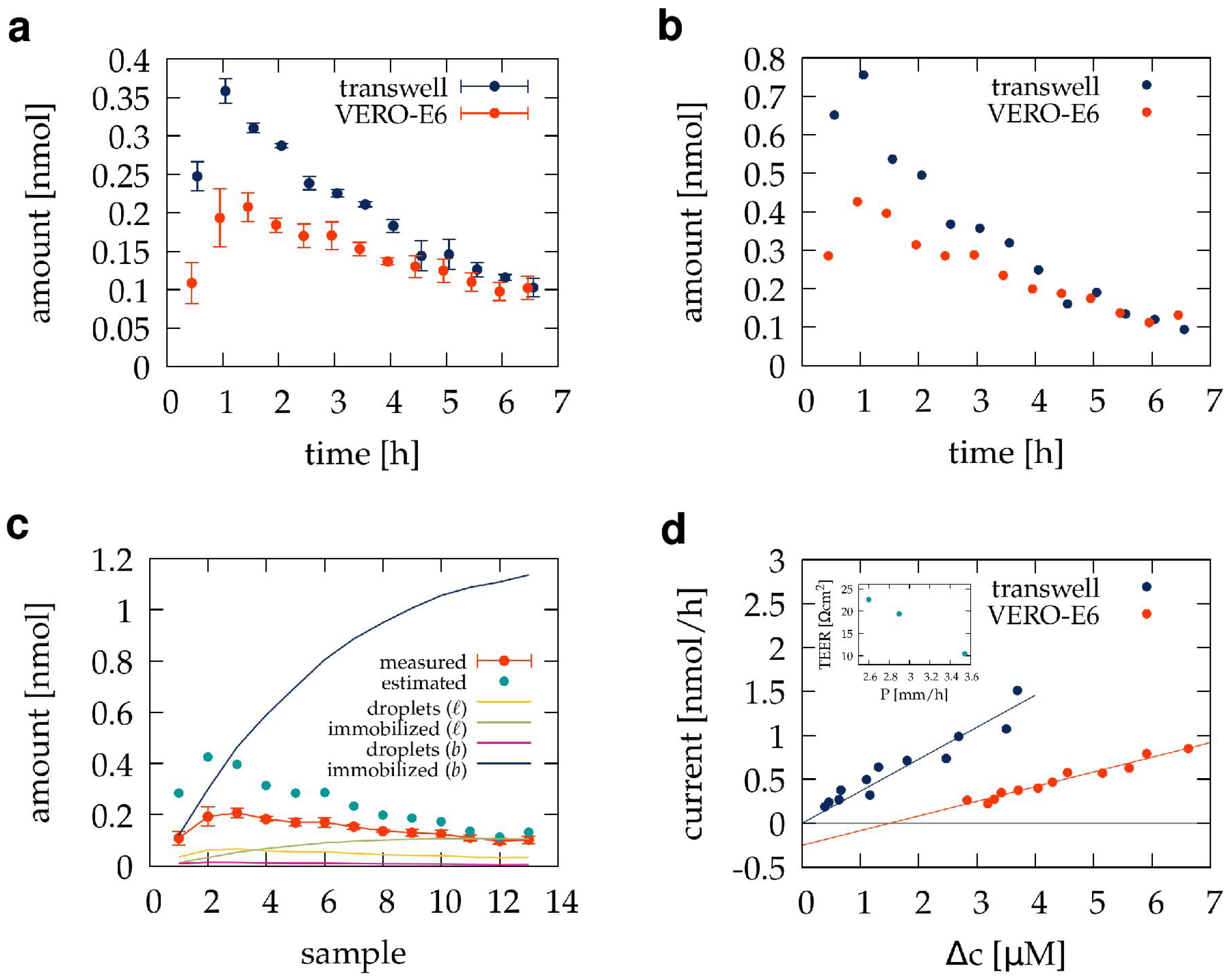
3.1. Pilot Transwell Transport Measurement
3.2. Estimating and Correcting Sampling Artifacts
3.3. The Sub-Compartment Mathematical Model of Transwell Transport Measurements
3.4. Transport through the VERO E6 Barrier
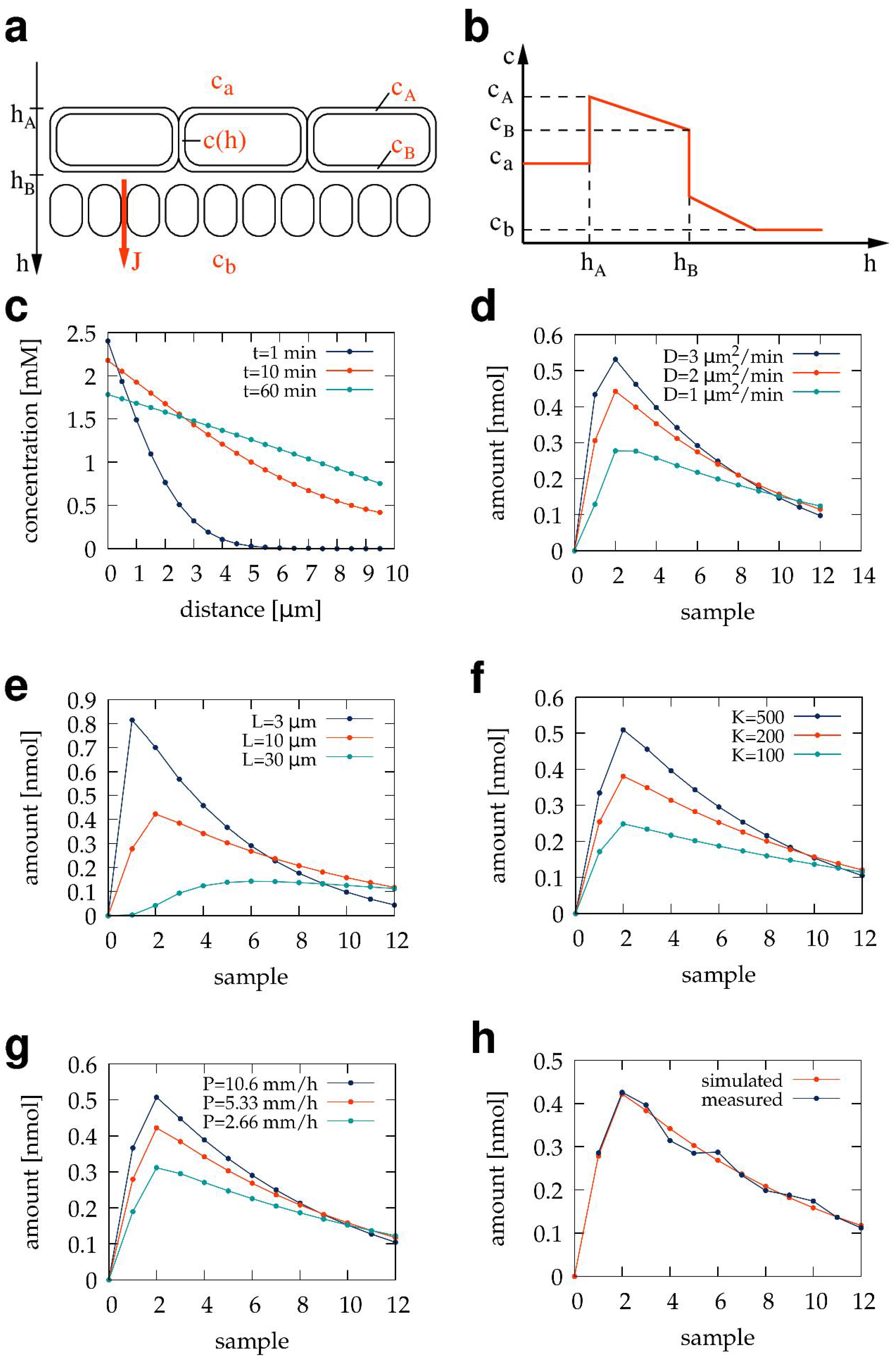
3.5. Mathematical Model of Diffusion through a Transwell Barrier Culture
| Parameter | Value |
|---|---|
| 250 | |
| D | 1.8 μm/min |
| 5.33 mm/h | |
| H | 10 μm |
| M | 0.81 nmol/h cm |
4. Discussion
4.1. Permeability Coefficient and Kinetic Parameters
4.2. Comparison of Transwell Inserts and Franz Chambers
4.3. Adsorption of the Test Compound
4.4. Diffusion through a Barrier Culture
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuehn, A.; Kletting, S.; de Souza Carvalho-Wodarz, C.; Repnik, U.; Griffiths, G.; Fischer, U.; Meese, E.; Huwer, H.; Wirth, D.; May, T.; et al. Human alveolar epithelial cells expressing tight junctions to model the air-blood barrier. ALTEX 2016, 33, 251–260. [Google Scholar] [CrossRef]
- Tavelin, S.; Gråsjö, J.; Taipalensuu, J.; Ocklind, G.; Artursson, P. Applications of epithelial cell culture in studies of drug transport. Methods Mol. Biol. 2002, 188, 233–272. [Google Scholar] [CrossRef]
- Forbes, B.; Ehrhardt, C. Human respiratory epithelial cell culture for drug delivery applications. Eur. J. Pharm. Biopharm. 2005, 60, 193–205. [Google Scholar] [CrossRef]
- Santa-Maria, A.R.; Heymans, M.; Walter, F.R.; Culot, M.; Gosselet, F.; Deli, M.A.; Neuhaus, W. Transport Studies Using Blood-Brain Barrier In Vitro Models: A Critical Review and Guidelines. Handb. Exp. Pharmacol. 2022, 273, 187–204. [Google Scholar]
- Horváti, K.; Fodor, K.; Pályi, B.; Henczkó, J.; Balka, G.; Gyulai, G.; Kiss, É.; Biri-Kovács, B.; Senoner, Z.; Bősze, S. Novel Assay Platform to Evaluate Intracellular Killing of Mycobacterium tuberculosis: In Vitro and In Vivo Validation. Front. Immunol. 2021, 12, 750496. [Google Scholar] [CrossRef]
- Salmon, D.; Gilbert, E.; Gioia, B.; Haftek, M.; Pivot, C.; Verrier, B.; Pirot, F. New easy handling and sampling device for bioavailability screening of topical formulations. Eur. J. Dermatol. 2015, 25 (Suppl. S1), 23–29. [Google Scholar] [CrossRef]
- Ng, S.F.; Rouse, J.J.; Sanderson, F.D.; Meidan, V.; Eccleston, G.M. Validation of a static Franz diffusion cell system for in vitro permeation studies. AAPS PharmSciTech 2010, 11, 1432–1441. [Google Scholar] [CrossRef]
- Addicks, W.J.; Flynn, G.L.; Weiner, N. Validation of a flow-through diffusion cell for use in transdermal research. Pharm. Res. 1987, 4, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Neri, I.; Laneri, S.; Lorenzo, R.D.; Dini, I.; Russo, G.; Grumetto, L. Parabens Permeation through Biological Membranes: A Comparative Study Using Franz Cell Diffusion System and Biomimetic Liquid Chromatography. Molecules 2022, 27, 4263. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, D.S.; Volpe, V.; Insausti, M.; Grünhut, M. On line kinetic analysis of permeation profiles for UV filter loaded microemulsions using an automatic system with spectroscopic detection and a chemometric approach. RSC Adv. 2021, 11, 15528–15538. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Patel, A.; Sinko, B.; Bell, M.; Wibawa, J.; Hadgraft, J.; Lane, M.E. A comparative study of the in vitro permeation of ibuprofen in mammalian skin, the PAMPA model and silicone membrane. Int. J. Pharm. 2016, 505, 14–19. [Google Scholar] [CrossRef]
- Michel, M.; Germain, L.; Bélanger, P.M.; Auger, F.A. Functional evaluation of anchored skin equivalent cultured in vitro: Percutaneous absorption studies and lipid analysis. Pharm. Res. 1995, 12, 455–458. [Google Scholar] [CrossRef]
- Labouta, H.I.; Thude, S.; Schneider, M. Setup for investigating gold nanoparticle penetration through reconstructed skin and comparison to published human skin data. J. Biomed. Opt. 2013, 18, 061218. [Google Scholar] [CrossRef] [PubMed]
- Pulsoni, I.; Lubda, M.; Aiello, M.; Fedi, A.; Marzagalli, M.; von Hagen, J.; Scaglione, S. Comparison Between Franz Diffusion Cell and a novel Micro-physiological System for In Vitro Penetration Assay Using Different Skin Models. SLAS Technol. 2022, 27, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.R.; Xiao, X.; Brener, I.; Burckel, D.B.; Narayan, R.; Polsky, R. Microneedle-based transdermal sensor for on-chip potentiometric determination of K(+). Adv. Healthc. Mater. 2014, 3, 876–881. [Google Scholar] [CrossRef] [PubMed]
- Ponmozhi, J.; Dhinakaran, S.; Varga-Medveczky, Z.; Fónagy, K.; Bors, L.A.; Iván, K.; Erdő, F. Development of Skin-On-A-Chip Platforms for Different Utilizations: Factors to Be Considered. Micromachines 2021, 12, 294. [Google Scholar] [CrossRef]
- Lukács, B.; Bajza, Á.; Kocsis, D.; Csorba, A.; Antal, I.; Iván, K.; Laki, A.J.; Erdő, F. Skin-on-a-Chip Device for Ex Vivo Monitoring of Transdermal Delivery of Drugs-Design, Fabrication, and Testing. Pharmaceutics 2019, 11, 445. [Google Scholar] [CrossRef]
- Bajza, Á.; Kocsis, D.; Berezvai, O.; Laki, A.J.; Lukács, B.; Imre, T.; Iván, K.; Szabó, P.; Erdő, F. Verification of P-Glycoprotein Function at the Dermal Barrier in Diffusion Cells and Dynamic “Skin-On-A-Chip” Microfluidic Device. Pharmaceutics 2020, 12, 804. [Google Scholar] [CrossRef]
- Alberti, M.; Dancik, Y.; Sriram, G.; Wu, B.; Teo, Y.L.; Feng, Z.; Bigliardi-Qi, M.; Wu, R.G.; Wang, Z.P.; Bigliardi, P.L. Multi-chamber microfluidic platform for high-precision skin permeation testing. Lab Chip 2017, 17, 1625–1634. [Google Scholar] [CrossRef]
- Tárnoki-Zách, J.; Mehes, E.; Varga-Medveczky, Z.; Isai, D.G.; Barany, N.; Bugyik, E.; Revesz, Z.; Paku, S.; Erdo, F.; Czirok, A. Development and Evaluation of a Human Skin Equivalent in a Semiautomatic Microfluidic Diffusion Chamber. Pharmaceutics 2021, 13, 910. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Horváth, L.B.; Krátký, M.; Pflégr, V.; Méhes, E.; Gyulai, G.; Kohut, G.; Babiczky, Á.; Biri-Kovács, B.; Baranyai, Z.; Vinšová, J.; et al. Host cell targeting of novel antimycobacterial 4-aminosalicylic acid derivatives with tuftsin carrier peptides. Eur. J. Pharm. Biopharm. 2022, 174, 111–130. [Google Scholar] [CrossRef] [PubMed]
- Baranyai, Z.; Biri-Kovács, B.; Krátký, M.; Szeder, B.; Debreczeni, M.L.; Budai, J.; Kovács, B.; Horváth, L.; Pári, E.; Németh, Z.; et al. Cellular Internalization and Inhibition Capacity of New Anti-Glioma Peptide Conjugates: Physicochemical Characterization and Evaluation on Various Monolayer- and 3D-Spheroid-Based in Vitro Platforms. J. Med. Chem. 2021, 64, 2982–3005. [Google Scholar] [CrossRef] [PubMed]
- Scheich, C.; Szabadka, Z.; Vértessy, B.; Pütter, V.; Grolmusz, V.; Schade, M. Discovery of novel MDR-Mycobacterium tuberculosis inhibitor by new FRIGATE computational screen. PLoS ONE 2011, 6, e28428. [Google Scholar] [CrossRef]
- Horváti, K.; Bacsa, B.; Szabó, N.; Dávid, S.; Mező, G.; Grolmusz, V.; Vértessy, B.; Hudecz, F.; Bősze, S. Enhanced cellular uptake of a new, in silico identified antitubercular candidate by peptide conjugation. Bioconjug. Chem. 2012, 23, 900–907. [Google Scholar] [CrossRef]
- Kósa, N.; Zolcsák, Á.; Voszka, I.; Csík, G.; Horváti, K.; Horváth, L.; Bősze, S.; Herenyi, L. Comparison of the Efficacy of Two Novel Antitubercular Agents in Free and Liposome-Encapsulated Formulations. Int. J. Mol. Sci. 2021, 22, 2457. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef]
- Mészáros, B.; Tóth, J.; Vértessy, B.G.; Dosztányi, Z.; Simon, I. Proteins with complex architecture as potential targets for drug design: A case study of Mycobacterium tuberculosis. PLoS Comput. Biol. 2011, 7, e1002118. [Google Scholar] [CrossRef]
- Nagy, G.N.; Leveles, I.; Vértessy, B.G. Preventive DNA repair by sanitizing the cellular (deoxy)nucleoside triphosphate pool. FEBS J. 2014, 281, 4207–4223. [Google Scholar] [CrossRef] [PubMed]
- Varga, B.; Barabás, O.; Takács, E.; Nagy, N.; Nagy, P.; Vértessy, B.G. Active site of mycobacterial dUTPase: Structural characteristics and a built-in sensor. Biochem. Biophys. Res. Commun. 2008, 373, 8–13. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model 2005, 45, 177–182. [Google Scholar] [CrossRef]
- Almada, P.; Pereira, P.M.; Culley, S.; Caillol, G.; Boroni-Rueda, F.; Dix, C.L.; Charras, G.; Baum, B.; Laine, R.F.; Leterrier, C.; et al. Automating multimodal microscopy with NanoJ-Fluidics. Nat. Commun. 2019, 10, 1223. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pang, X.; Jiang, H.; Duan, M.; Liu, H.; Yang, Z.; Xi, Y.; Russell, T.P. Open millifluidics based on powder-encased channels. Proc. Natl. Acad. Sci. USA 2023, 120, e2302907120. [Google Scholar] [CrossRef]
- Konagaya, S.; Ando, T.; Yamauchi, T.; Suemori, H.; Iwata, H. Long-term maintenance of human induced pluripotent stem cells by automated cell culture system. Sci. Rep. 2015, 5, 16647. [Google Scholar] [CrossRef] [PubMed]
- Saalfrank, D.; Konduri, A.K.; Latifi, S.; Habibey, R.; Golabchi, A.; Martiniuc, A.V.; Knoll, A.; Ingebrandt, S.; Blau, A. Incubator-independent cell-culture perfusion platform for continuous long-term microelectrode array electrophysiology and time-lapse imaging. R. Soc. Open Sci. 2015, 2, 150031. [Google Scholar] [CrossRef]
- Wong, B.G.; Mancuso, C.P.; Kiriakov, S.; Bashor, C.J.; Khalil, A.S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 2018, 36, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Kapus, A.; Grinstein, S.; Wasan, S.; Kandasamy, R.; Orlowski, J. Functional characterization of three isoforms of the Na+/H+ exchanger stably expressed in Chinese hamster ovary cells. ATP dependence, osmotic sensitivity, and role in cell proliferation. J. Biol. Chem. 1994, 269, 23544–23552. [Google Scholar] [CrossRef] [PubMed]
- Mizusawa, H.; Simizu, B.; Terasima, T. Cell line vero deposited to Japanese Cancer Research Resources Bank. In VERO Cell Origin, Properties Biomedical Application; Simizu, B., Terasima, T., Eds.; Chiba University: Chiba, Japan, 1988; pp. 24–25. [Google Scholar]
- Terasima, T.; Yasukawa, M.; Simizu, B. History of Vero Cells in Japan. In VERO Cell Origin, Properties Biomedical Application; Simizu, B., Terasima, T., Eds.; Chiba University: Chiba, Japan, 1988; pp. 22–23. [Google Scholar]
- Earley, E.; Johnson, K. The Lineage of the Vero, Vero 76 and its Clone C1008 in the United States. In VERO Cell Origin, Properties Biomedical Application; Simizu, B., Terasima, T., Eds.; Chiba University: Chiba, Japan, 1988; pp. 26–29. [Google Scholar]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and maintenance of Vero cell lines. Curr. Protoc. Microbiol. 2008, 11, A.4E.1–A.4E.7. [Google Scholar] [CrossRef]
- Nocedal, J.; Wright, S.J. Numerical Optimization; Springer: New York, NY, USA, 2006. [Google Scholar]
- Zsila, F.; Bősze, S.; Beke-Somfai, T. Interaction of antitubercular drug candidates with α1-acid glycoprotein produced in pulmonary granulomas. Int. J. Biol. Macromol. 2020, 147, 1318–1327. [Google Scholar] [CrossRef]
- Masel, R. Principles of Adsorption and Reaction on Solid Surfaces.; Wiley Interscience: Hoboken, NJ, USA, 1996. [Google Scholar]
- Lechner, C.; Mönning, U.; Reichel, A.; Fricker, G. Potential and Limits of Kidney Cells for Evaluation of Renal Excretion. Pharmaceuticals 2014, 9, 908. [Google Scholar] [CrossRef]
- Li, X.; Ding, X.; Bian, C.; Wu, S.; Chen, M.; Wang, W.; Wang, J.; Cheng, L. Hydrophobic drug adsorption loss to syringe filters from a perspective of drug delivery. J. Pharmacol. Toxicol. Methods 2019, 95, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.A. Reduced dynamic models in epithelial transport. J. Biophys. 2013, 2013, 654543. [Google Scholar] [CrossRef] [PubMed]
- Kadakia, E.; Shah, L.; Amiji, M.M. Mathematical Modeling and Experimental Validation of Nanoemulsion-Based Drug Transport across Cellular Barriers. Pharm. Res. 2017, 34, 1416–1427. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tárnoki-Zách, J.; Bősze, S.; Czirók, A. Quantitative Analysis of a Pilot Transwell Barrier Model with Automated Sampling and Mathematical Modeling. Pharmaceutics 2023, 15, 2646. https://doi.org/10.3390/pharmaceutics15112646
Tárnoki-Zách J, Bősze S, Czirók A. Quantitative Analysis of a Pilot Transwell Barrier Model with Automated Sampling and Mathematical Modeling. Pharmaceutics. 2023; 15(11):2646. https://doi.org/10.3390/pharmaceutics15112646
Chicago/Turabian StyleTárnoki-Zách, Júlia, Szilvia Bősze, and András Czirók. 2023. "Quantitative Analysis of a Pilot Transwell Barrier Model with Automated Sampling and Mathematical Modeling" Pharmaceutics 15, no. 11: 2646. https://doi.org/10.3390/pharmaceutics15112646
APA StyleTárnoki-Zách, J., Bősze, S., & Czirók, A. (2023). Quantitative Analysis of a Pilot Transwell Barrier Model with Automated Sampling and Mathematical Modeling. Pharmaceutics, 15(11), 2646. https://doi.org/10.3390/pharmaceutics15112646





