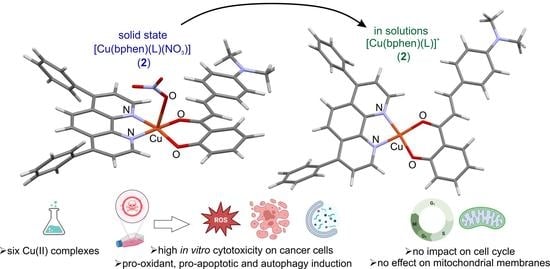
Heteroleptic Copper(II) Complexes Containing 2′-Hydroxy-4-(Dimethylamino)Chalcone Show Strong Antiproliferative Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis and Characterization of the Compounds
2.2. Geometry Optimizations of Complex 2 Using DFT Calculations
2.3. Single Crystal X-ray Analysis of [Cu(phen)(L)(NO3)]
2.4. Biological Experiments: Cell-Based Studies
2.4.1. In Vitro Cytotoxicity
2.4.2. Cell Culture for Molecular Biology Assays
2.4.3. Cell Cycle Analysis
2.4.4. Apoptosis, Autophagy, and Mitochondrial Membrane Potential (ΔΨ m) Analyses
2.4.5. Reactive Oxygen Species (ROS) Production Determination
2.5. Cellular Uptake of Copper in A2780 and 22Rv1 Cancer Cells
2.6. Interactions with Model Proteins
2.7. Statistical Evaluation
3. Results and Discussion
3.1. Synthesis and Characterization of the Complexes
3.2. Geometry Optimization Using DFT Calculations
3.3. Single-Crystal X-ray Analysis of [Cu(phen)(L)(NO3)]
3.4. In Vitro Cytotoxicity of Complexes 1–6
3.5. Effect of Complex 2 on the Cell Cycle of A2780 Cells
3.6. Induction of Apoptosis and Associated Processes
3.7. Mitochondrial Membrane Potential Disruption
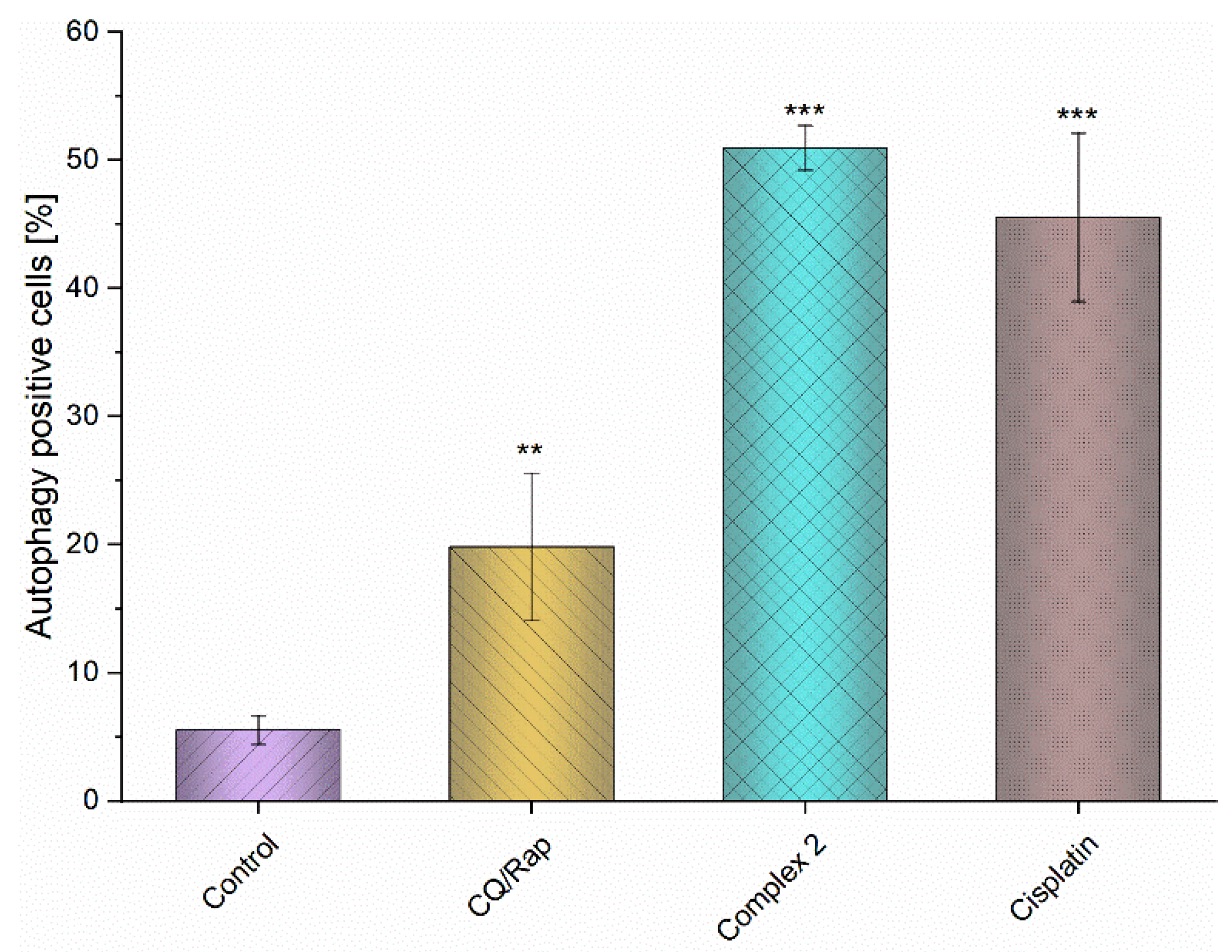
3.8. Intracellular Production of Reactive Oxygen Species (ROS) and Induction of Autophagy
3.9. Accumulation of Copper in A2780 and 22Rv1 Cancer Cells
3.10. Interactions with Model Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rozmer, Z.; Perjesi, P. Naturally occurring chalcones and their biological activities. Phytochem. Rev. 2016, 15, 87–120. [Google Scholar] [CrossRef]
- Sulpizio, C.; Breibeck, J.; Romplel, A. Recent progress in synthesis and characterization of metal chalcone complexes and their potential as bioactive agents. J. Inorg. Biochem. 2018, 374, 497–524. [Google Scholar] [CrossRef]
- Huang, X.; Liu, Z.; Wang, M.; Yin, X.; Wang, Y.; Dai, L.; Wang, H. Platinum(IV) complexes conjugated with chalcone analogs as dual targeting anticancer agents: In vitro and in vivo studies. Bioorg. Chem. 2020, 105, 104430. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Huang, R.; Wang, Z.; Li, L.; Gou, S.; Liao, Z.; Wang, H. Pt(IV) complexes conjugating with chalcone analogues as inhibitors of microtube polymerization exhibited selective inhibition in human cancer cells. Eur. J. Med. Chem. 2018, 146, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Dkhar, L.; Banothu, V.; Pinder, E.; Phillips, R.M.; Kaminsky, W.; Kollipara, M.R. Ru, Rh and Ir metal complexes of pyridyl chalcone derivatives: Their potent antibacterial activity, comparable cytotoxicity potency and selectivity to cisplatin. Polyhedron 2020, 185, 114606. [Google Scholar] [CrossRef]
- Kaushal, R.; Kaur, M. Bio-medical potential of chalcone derivatives and their metal complexes as antidiabetic agents: A review. J. Coord. Chem. 2021, 74, 725–742. [Google Scholar] [CrossRef]
- Johnson, J.; Yardily, A. Co(II), Ni(II), Cu(II), and Zn(II) metal complexes of chalcone: Synthesis, characterization, thermal, antimicrobial, photocatalytic degradation of dye and molecular modeling studies. Appl. Organomet. Chem. 2022, 36, e6465. [Google Scholar] [CrossRef]
- Křikavová, R.; Vančo, J.; Trávníček, Z.; Hutyra, J.; Dvořák, Z. Design and characterization of highly in vitro antitumor active ternary copper(II) complexes containing 2′-hydroxychalcone ligands. J. Inorg. Biochem. 2016, 163, 8–17. [Google Scholar] [CrossRef]
- Shao, Y.; Molnar, L.F.; Jung, Y.; Kussmann, J.; Ochsenfeld, C.; Brown, S.T.; Gilbert, A.T.B.; Slipchenko, L.V.; Levchenko, S.V.; O’Neill, D.P.; et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- APEX3 Software Suite, © 2023 Bruker AXS Inc.: Madison, WI, USA, 2023.
- Sheldrick, G.M. SHELXTL Version 2014/7. Available online: http://shelx.uni-ac.gwdg.de/SHELX/index.php (accessed on 5 April 2020).
- Putz, H.; Brandenburg, K. Diamond-Crystal and Molecular Structure Visualization, Crystal Impact-Dr. H. Putz & Dr. K; Brandenburg GbR: Bonn, Germany, 2006. [Google Scholar]
- Hernychova, L.; Alexandri, E.; Tzakos, A.G.; Zatloukalová, M.; Primykiri, A.; Gerothanassis, I.P.; Uhrik, L.; Šebela, M.; Kopečný, D.; Jedinák, L.; et al. Serum albumin as a primary non-covalent binding protein for nitro-oleic acid. Int. J. Biol. Macromol. 2022, 203, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Arndt, C.; Koristka, S.; Feldmann, A.; Bachmann, M. Native Polyacrylamide Gels. In Electrophoretic Separation of Proteins. Methods in Molecular Biology; Kurien, B., Scofield, R., Eds.; Humana Press: New York, NY, USA, 2019; Volume 1855, pp. 87–91. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- TIBCO Software Inc. Statistica (Data Analysis Software System), Version 13. 2018. Available online: http://tibco.com (accessed on 28 April 2022).
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds: Part B: Applications in Coordination, Organometallic, and Bioinorganic Chemistry, 6th ed.; Wiley Publications: Hoboken, NJ, USA, 2008; pp. 1–273. [Google Scholar] [CrossRef]
- Geary, W.J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Ito, K.; Ito, T. Magnetic moments of copper(II) complexes. Aust. J. Chem. 1958, 11, 406–414. [Google Scholar] [CrossRef]
- Casanova, D.; Cirera, J.; Llunell, M.; Alemany, P.; Avnir, D.; Alvarez, S. Minimal distortion pathways in polyhedral rearrangements. J. Am. Chem. Soc. 2004, 126, 1755–1763. [Google Scholar] [CrossRef]
- Kathiresan, S.; Mugesh, S.; Annaraj, J.; Murugan, M. Mixed-ligand copper(II) Schiff base complexes: The vital role of co-ligands in DNA/protein interactions and cytotoxicity. New J. Chem. 2017, 41, 1267–1283. [Google Scholar] [CrossRef]
- Hirohama, T.; Kuranuki, Y.; Ebina, E.; Sugizaki, T.; Arii, H.; Chikira, M.; Selvi, P.T.; Palaniandavar, M. Copper(II) complexes of 1,10-phenanthroline-derived ligands: Studies on DNA binding properties and nuclease activity. J. Inorg. Biochem. 2005, 99, 1205–1219. [Google Scholar] [CrossRef]
- Vančo, J.; Trávníček, Z.; Hošek, J.; Dvořák, Z. Heteroleptic copper(II) complexes of prenylated flavonoid osajin behave as selective and effective antiproliferative and anti-inflammatory agents. J. Inorg. Biochem. 2022, 226, 111639. [Google Scholar] [CrossRef]
- Vančo, J.; Trávníček, Z.; Hošek, J.; Malina, T.; Dvořák, Z. Copper(II) Complexes Containing Natural Flavonoid Pomiferin Show Considerable In Vitro Cytotoxicity and Anti-inflammatory Effects. Int. J. Mol. Sci. 2021, 22, 7626. [Google Scholar] [CrossRef]
- Ly, J.D.; Grubb, D.R.; Lawen, A. The mitochondrial membrane potential (deltapsi(m)) in apoptosis; an update. Apoptosis 2003, 8, 115–128. [Google Scholar] [CrossRef]
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Anshul, G.; Mumper, R.J. Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat. Rev. 2009, 35, 32–46. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef] [PubMed]
- Poillet-Perez, L.; Despouy, G.; Delage-Mourroux, R.; Boyer-Guittaut, M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015, 4, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Zhu, H.; Sheng, F.; Tian, Y.; Zhou, J.; Chen, Y.; Li, S.; Lin, J. Activation of the MAPK11/12/13/14 (p38 MAPK) pathway regulates the transcription of autophagy genes in response to oxidative stress induced by a novel copper complex in HeLa cells. Autophagy 2014, 10, 1285–1300. [Google Scholar] [CrossRef]










| Molecular formula | C29 H24 Cu N4 O5 |
| Formula weight | 572.06 |
| Temperature | 295(2) K |
| Wavelength | 0.71073 Å |
| Crystal system | Monoclinic |
| Space group | P21/n |
| Unit cell dimensions | a = 10.454(4) Å, α= 90° |
| b = 16.197(5) Å, β= 96.95(2)° | |
| c = 14.748(5) Å, γ = 90° | |
| Volume | 2478.8(15) Å3 |
| Z | 4 |
| Density (calculated) | 1.533 g/cm3 |
| Absorption coefficient | 0.931 mm−1 |
| F(000) | 1180 |
| Crystal size | 0.200 × 0.180 × 0.160 mm3 |
| θ range for data collection | 2.26 to 24.82° |
| Index ranges | −12 ≤ h ≤ 12, −19 ≤ k ≤ 19, −17 ≤ l ≤ 17 |
| Reflections collected | 23,346 |
| Independent reflections | 4255 [R(int) = 0.1006] |
| Completeness to θ = 24.821° | 99.30% |
| Absorption correction | Multi-Scan method (SADABS) |
| Refinement method | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 4255/0/352 |
| Goodness-of-fit on F2 | 1.051 |
| Final R indices [I > 2σ(I)] | R1 = 0.0449, wR2 = 0.0821 |
| R indices (all data) | R1 = 0.0869, wR2 = 0.0957 |
| Largest diff. peak and hole | 0.312 and −0.308 e. Å−3 |
| Selected Interatomic Parameters | In Vacuum | In Water | In Vacuum | In Water | X-ray # | X-ray * |
|---|---|---|---|---|---|---|
| [Cu(bphen)(L)(NO3)] ϖB97X-D/def2-tzvp | [Cu(bphen)(L)]+ ϖB97X-D/def2-tzvp | [Cu(phen)(L)(NO3)] | [Cu(phen)(chal1)(NO3)] | |||
| Cu–O1 | 1.906 | 1.903 | 1.857 | 1.881 | 1.872 (2) | 1.8839 (18) |
| Cu–O2 | 1.959 | 1.949 | 1.907 | 1.926 | 1.924 (2) | 1.9260 (18) |
| Cu–O6 | 2.159 | 2.358 | - | - | 2.341 (3) | 2.429 (2) |
| Cu–N1 | 2.035 | 2.018 | 2.016 | 2.014 | 2.004 (3) | 2.001 (2) |
| Cu–N2 | 2.050 | 2.025 | 2.025 | 2.015 | 2.004 (3) | 1.995 (2) |
| N1–Cu–O2 | 164.14 | 171.82 | 174.25 | 174.76 | 166.40 (11) | 173.51 (8) |
| N2–Cu–O1 | 162.44 | 167.51 | 172.26 | 173.10 | 168.54 (11) | 168.68 (9) |
| N1–Cu–O6 | 103.28 | 96.87 | - | - | 105.85 (12) | 95.29 (8) |
| O1–Cu–O6 | 102.02 | 93.33 | - | - | 96.49 (11) | 91.61 (8) |
| Bond Length [Å] | Angle [°] | ||
|---|---|---|---|
| Cu(1)-O(1) | 1.872 (2) | O(1)-Cu(1)-O(2) | 91.69 (10) |
| Cu(1)-O(2) | 1.924 (2) | O(1)-Cu(1)-N(2) | 168.54 (11) |
| Cu(1)-N(2) | 2.004 (3) | O(2)-Cu(1)-N(2) | 94.67 (11) |
| Cu(1)-N(1) | 2.004 (3) | O(1)-Cu(1)-N(1) | 89.51 (11) |
| Cu(1)-O(6) | 2.341 (3) | O(2)-Cu(1)-N(1) | 166.40 (11) |
| N(3)-O(5) | 1.223 (4) | N(2)-Cu(1)-N(1) | 82.09 (11) |
| N(3)-O(4) | 1.230 (4) | O(1)-Cu(1)-O(6) | 96.49 (11) |
| N(3)-O(6) | 1.239 (4) | O(2)-Cu(1)-O(6) | 87.48 (11) |
| N(2)-Cu(1)-O(6) | 93.30 (11) | ||
| N(1)-Cu(1)-O(6) | 105.85 (12) | ||
| Compound | Human Cancer Cell Lines | |||||||
|---|---|---|---|---|---|---|---|---|
| HeLa | 22Rv1 | MCF7 | PC3 | HepG2 | HOS | A549 | G361 | |
| 1 | 3.3 ± 0.2 | 7.9 ± 0.6 | 13.7 ± 1.8 | >50 | 16.5 ± 0.3 | 5.2 ± 0.8 | 14.9 ± 2.6 | 6.9 ± 0.5 |
| 2 | 2.0 ± 0.1 | 1.0 ± 0.1 | 1.6 ± 0.1 | 2.2 ± 0.2 | 2.3 ± 0.5 | 2.1 ± 0.1 | 2.1 ± 0.2 | 2.0 ± 0.1 |
| 3 | 3.2 ± 0.2 | 3.9 ± 0.4 | 17.7 ± 0.9 | 16.8 ± 2.6 | 6.9 ± 1.3 | 4.6 ± 0.9 | 3.3 ± 0.1 | 3.8 ± 0.7 |
| 4 | 21.8 ± 0.2 | >50 | >50 | >50 | >50 | >50 | >50 | 2.9 ± 0.1 |
| 5 | 23.6 ± 0.8 | 21.5 ± 1.6 | 33.8 ± 2.9 | >50 | >50 | >50 | >50 | 23.8 ± 0.9 |
| 6 | 35.6 ± 0.1 | >50 | >50 | >50 | >50 | 32.8 ± 0.9 | >50 | >50 |
| HL | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| cisplatin | 27.0 ± 5.3 | 18.2 ± 0.8 | 23.0 ± 2.6 | >50 | >50 | >50 | >50 | 7.5 ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trávníček, Z.; Malina, T.; Vančo, J.; Šebela, M.; Dvořák, Z. Heteroleptic Copper(II) Complexes Containing 2′-Hydroxy-4-(Dimethylamino)Chalcone Show Strong Antiproliferative Activity. Pharmaceutics 2023, 15, 307. https://doi.org/10.3390/pharmaceutics15020307
Trávníček Z, Malina T, Vančo J, Šebela M, Dvořák Z. Heteroleptic Copper(II) Complexes Containing 2′-Hydroxy-4-(Dimethylamino)Chalcone Show Strong Antiproliferative Activity. Pharmaceutics. 2023; 15(2):307. https://doi.org/10.3390/pharmaceutics15020307
Chicago/Turabian StyleTrávníček, Zdeněk, Tomáš Malina, Ján Vančo, Marek Šebela, and Zdeněk Dvořák. 2023. "Heteroleptic Copper(II) Complexes Containing 2′-Hydroxy-4-(Dimethylamino)Chalcone Show Strong Antiproliferative Activity" Pharmaceutics 15, no. 2: 307. https://doi.org/10.3390/pharmaceutics15020307
APA StyleTrávníček, Z., Malina, T., Vančo, J., Šebela, M., & Dvořák, Z. (2023). Heteroleptic Copper(II) Complexes Containing 2′-Hydroxy-4-(Dimethylamino)Chalcone Show Strong Antiproliferative Activity. Pharmaceutics, 15(2), 307. https://doi.org/10.3390/pharmaceutics15020307