Ginsenoside Rg3 Reduces the Toxicity of Graphene Oxide Used for pH-Responsive Delivery of Doxorubicin to Liver and Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of GO–Rg3 and GO–Rg3–DOX Conjugates
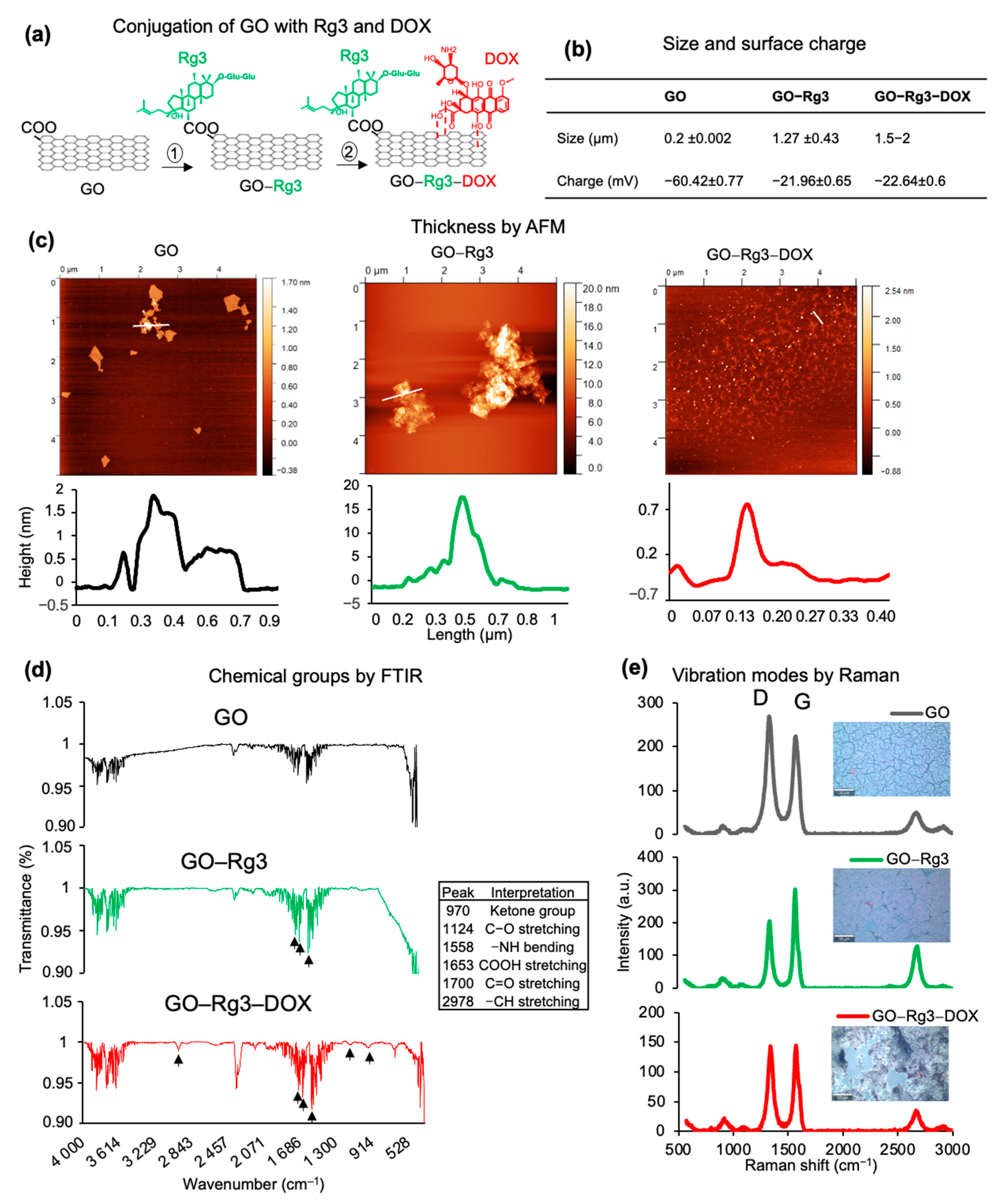
2.3. Characterization of GO, GO–Rg3, and GO–Rg3–DOX Conjugates
2.4. Viability Assay Using Alamarblue Assay
2.5. Drug Release Analysis
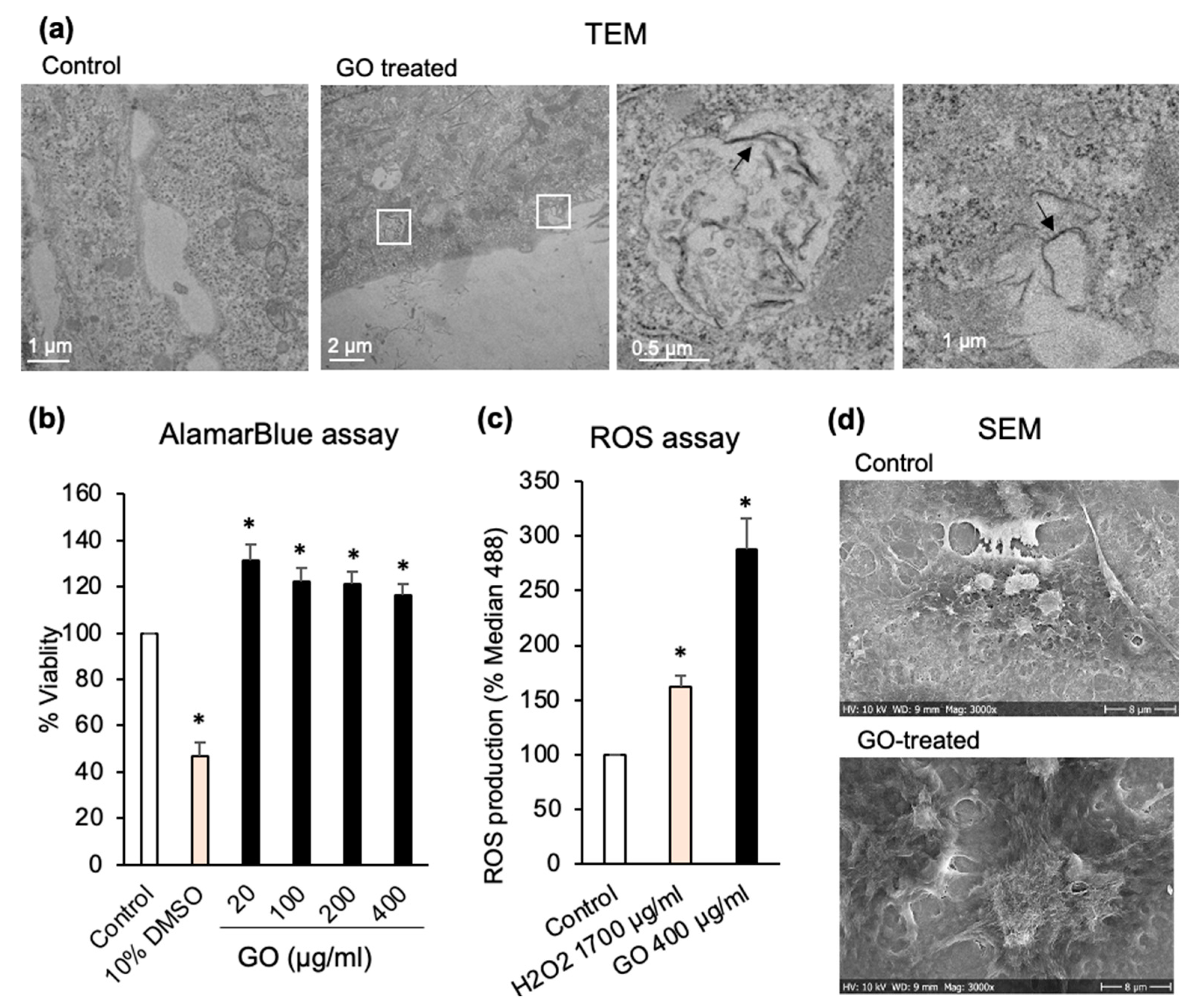
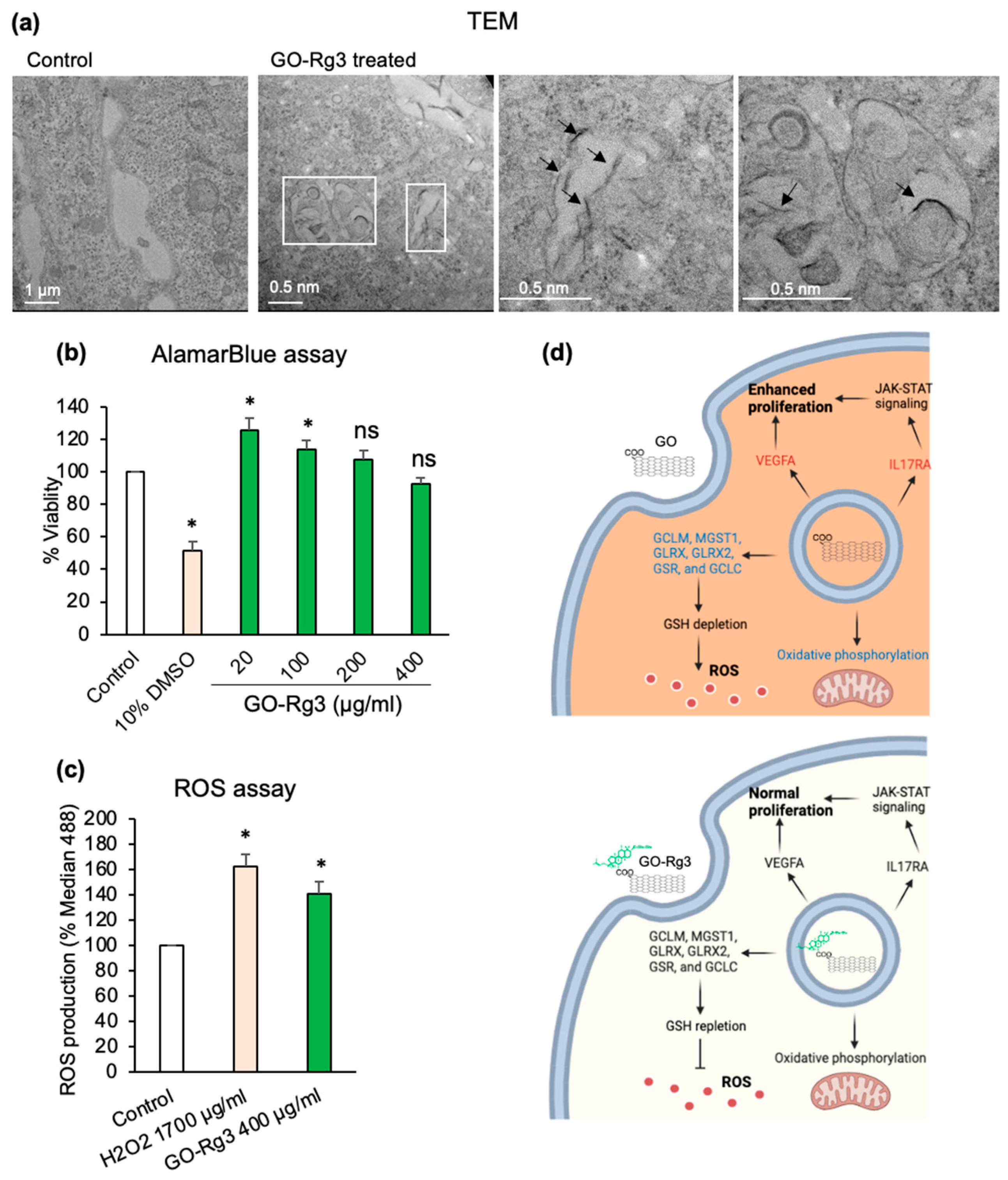
2.6. ROS Measurement
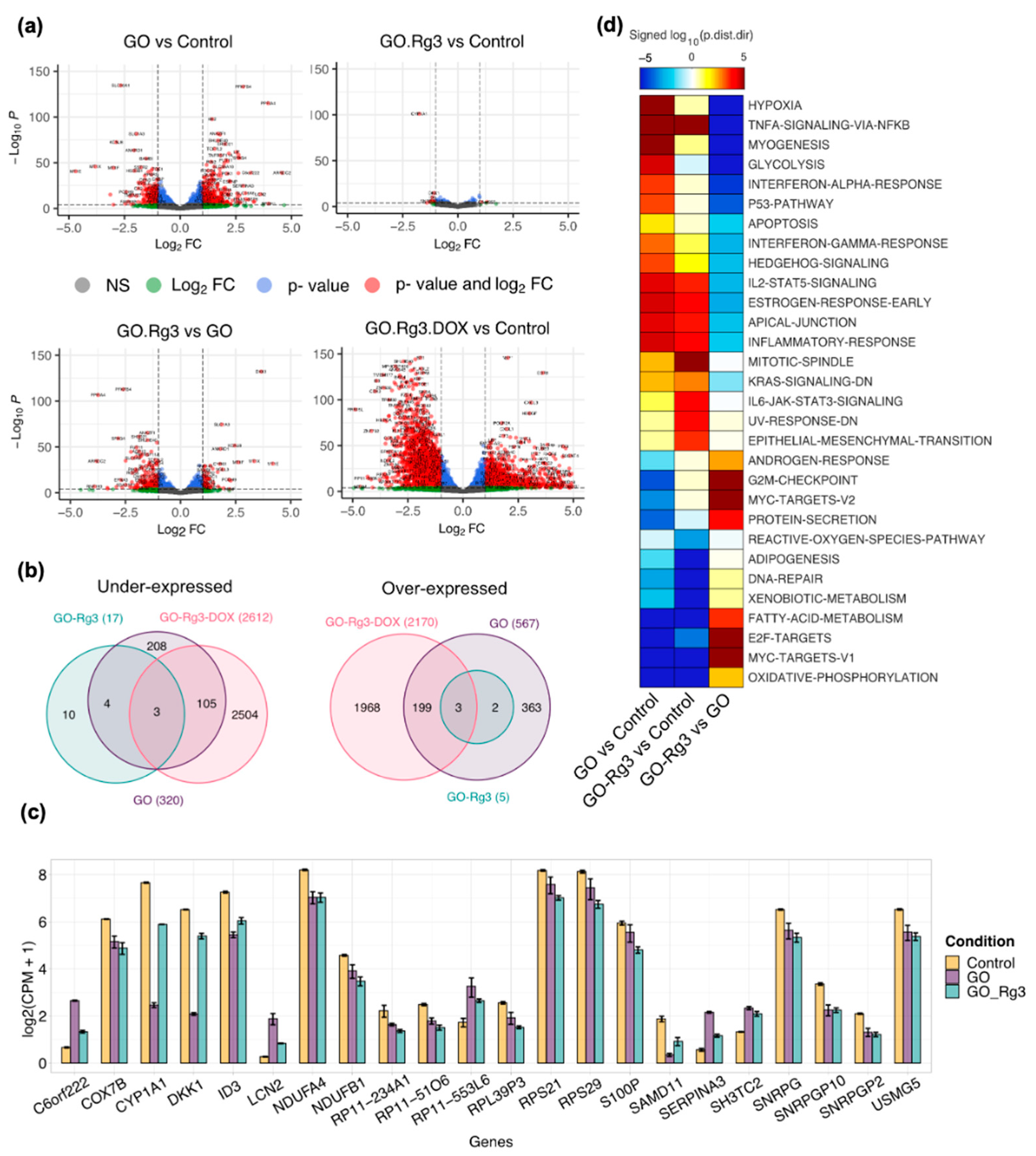
2.7. RNA Extraction and Sequencing
2.8. Differential Expression (DE) Analysis
2.9. Gene Set Analysis
2.10. Scanning Electron Microscopy (SEM)
2.11. Transmission Electron Microscopy (TEM)
2.12. Confocal Microscopy
3. Results
3.1. Rg3 and DOX Were Loaded onto GO to Create GO-Rg3 and GO–Rg3–DOX Conjugates
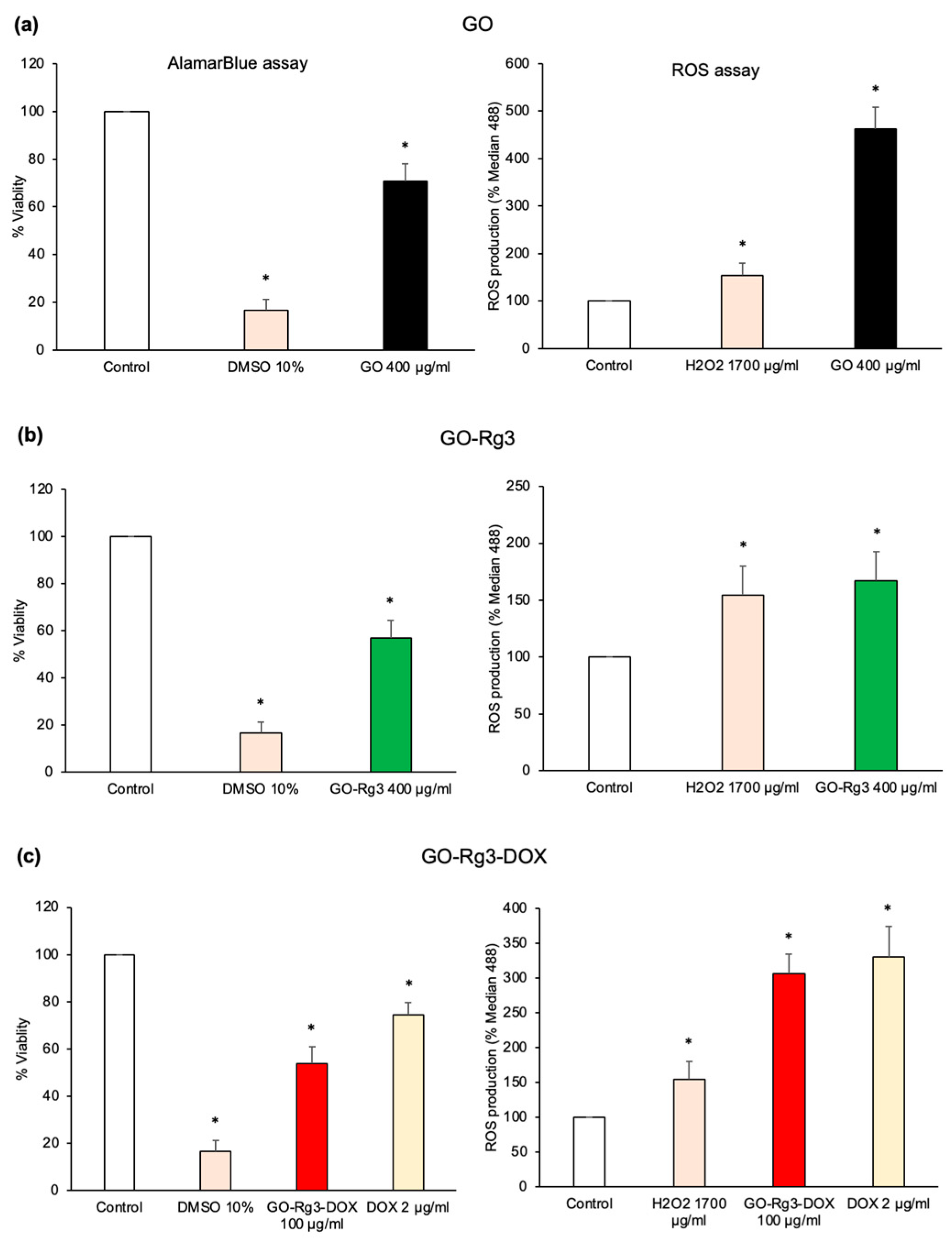
3.2. GO Increases ROS Production in Huh7 Cancer Cells
3.3. Conjugation of GO with Rg3 Mitigates ROS Production Induced by the Nanocarrier
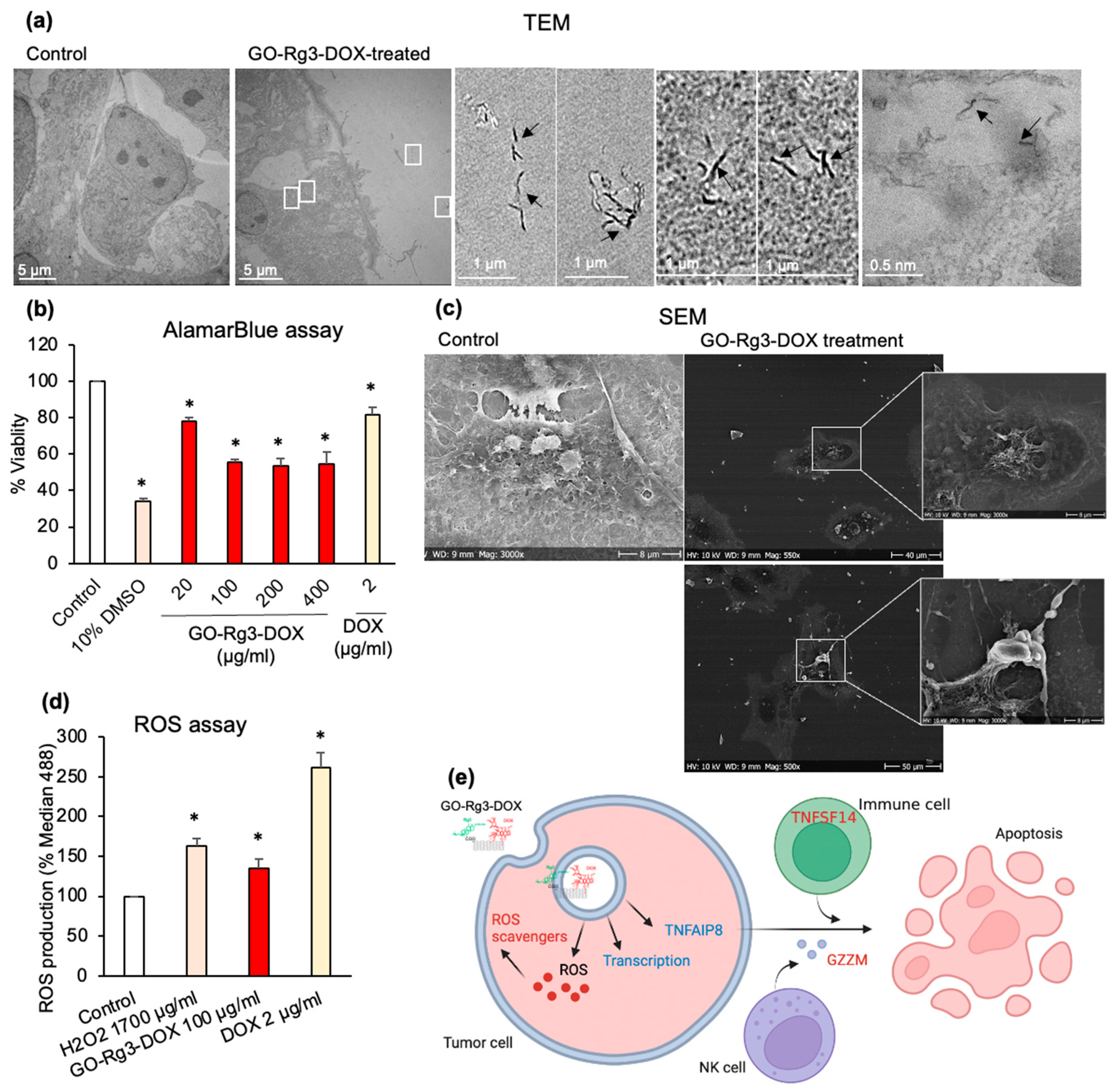
3.4. Effect of GO–Rg3–DOX on Hepatocarcinoma and Breast Cancer Cells
3.4.1. pH Dependent Release of DOX from GO–Rg3–DOX
3.4.2. DOX Delivered in the Form of GO–Rg3–DOX Is More Effective Than Free DOX
3.4.3. GO–Rg3–DOX Is Cytotoxic against Human MDA-MB-231 Breast Cancer Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bernstein, W.B. Cancer in the Tropics. In Hunter’s Tropical Medicine and Emerging Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 178–188. [Google Scholar]
- Hu, S.W.; Wang, J.; Zhang, T.T.; Li, X.L.; Chen, H.Y.; Xu, J.J. Targeted Transmembrane Delivery of Ca 2+ via FA-Nanogel for Synergistically Enhanced Chemotherapy. ACS Appl. Mater. Interfaces 2019, 11, 16412–16420. [Google Scholar] [CrossRef]
- Liu, G.; Tsai, H.; Zeng, X.; Zuo, Y.; Tao, W.; Han, J.; Mei, L. Phosphorylcholine-Based Stealthy Nanocapsules Enabling Tumor Microenvironment-Responsive Doxorubicin Release for Tumor Suppression. Theranostics 2017, 7, 1192–1203. [Google Scholar] [CrossRef]
- Chen, C.-T.; Wang, Z.-H.; Hsu, C.-C.; Lin, H.-H.; Chen, J.-H. In Vivo Protective Effects of Diosgenin against Doxorubicin-Induced Cardiotoxicity. Nutrients 2015, 7, 4938–4954. [Google Scholar] [CrossRef]
- Su, Z.; Ye, J.; Qin, Z.; Ding, X. Protective Effects of Madecassoside against Doxorubicin Induced Nephrotoxicity in Vivo and in Vitro. Sci. Rep. 2016, 5, 18314. [Google Scholar] [CrossRef]
- Vyas, M.; Simbo, D.A.; Mursalin, M.; Mishra, V.; Bashary, R.; Khatik, G.L. Drug Delivery Approaches for Doxorubicin in the Management of Cancers. Curr. Cancer Rev. 2019, 16, 320–331. [Google Scholar] [CrossRef]
- Pandit, S.; Rahimi, S.; Derouiche, A.; Boulaoued, A.; Mijakovic, I. Sustained Release of Usnic Acid from Graphene Coatings Ensures Long Term Antibiofilm Protection. Sci. Rep. 2021, 11, 9956. [Google Scholar] [CrossRef]
- Rahimi, S.; Chen, Y.; Zareian, M.; Pandit, S.; Mijakovic, I. Cellular and Subcellular Interactions of Graphene-Based Materials with Cancerous and Non-Cancerous Cells. Adv. Drug Deliv. Rev. 2022, 189, 114467. [Google Scholar] [CrossRef]
- Pattnaik, S.; Swain, K.; Lin, Z. Graphene and Graphene-Based Nanocomposites: Biomedical Applications and Biosafety. J. Mater Chem. B 2016, 4, 7813–7831. [Google Scholar] [CrossRef]
- Pei, X.; Zhu, Z.; Gan, Z.; Chen, J.; Zhang, X.; Cheng, X.; Wan, Q.; Wang, J. PEGylated Nano-Graphene Oxide as a Nanocarrier for Delivering Mixed Anticancer Drugs to Improve Anticancer Activity. Sci. Rep. 2020, 10, 2717. [Google Scholar] [CrossRef]
- Lei, H.; Xie, M.; Zhao, Y.; Zhang, F.; Xu, Y.; Xie, J. Chitosan/Sodium Alginate Modificated Graphene Oxide-Based Nanocomposite as a Carrier for Drug Delivery. Ceram Int. 2016, 42, 17798–17805. [Google Scholar] [CrossRef]
- Zhou, T.; Zhou, X.; Xing, D. Controlled Release of Doxorubicin from Graphene Oxide Based Charge-Reversal Nanocarrier. Biomaterials 2014, 35, 4185–4194. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, C.; Wang, Y.; Li, Y.; Du, L.; Cui, Z.; Wu, S. Cytotoxicity of Graphene Oxide and Graphene Oxide Loaded with Doxorubicin on Human Multiple Myeloma Cells. Int. J. Nanomed. 2014, 9, 1413. [Google Scholar] [CrossRef] [PubMed]
- Zainal-Abidin, M.H.; Hayyan, M.; Ngoh, G.C.; Wong, W.F. Doxorubicin Loading on Functional Graphene as a Promising Nanocarrier Using Ternary Deep Eutectic Solvent Systems. ACS Omega 2020, 5, 1656–1668. [Google Scholar] [CrossRef]
- Lv, Y.; Tao, L.; Annie Bligh, S.W.; Yang, H.; Pan, Q.; Zhu, L. Targeted Delivery and Controlled Release of Doxorubicin into Cancer Cells Using a Multifunctional Graphene Oxide. Mater. Sci. Eng. C 2016, 59, 652–660. [Google Scholar] [CrossRef]
- Wang, L.; Yu, D.; Dai, R.; Fu, D.; Li, W.; Guo, Z.; Cui, C.; Xu, J.; Shen, S.; Ma, K. PEGylated Doxorubicin Cloaked Nano-Graphene Oxide for Dual-Responsive Photochemical Therapy. Int. J. Pharm 2019, 557, 66–73. [Google Scholar] [CrossRef]
- Duan, G.; Kang, S.; Tian, X.; Garate, J.A.; Zhao, L.; Ge, C.; Zhou, R. Protein Corona Mitigates the Cytotoxicity of Graphene Oxide by Reducing Its Physical Interaction with Cell Membrane. Nanoscale 2015, 7, 15214–15224. [Google Scholar] [CrossRef] [PubMed]
- Gulzar, A.; Xu, J.; Xu, L.; Yang, P.; He, F.; Yang, D.; An, G.; Ansari, M.B. Redox-Responsive UCNPs-DPA Conjugated NGO-PEG-BPEI-DOX for Imaging-Guided PTT and Chemotherapy for Cancer Treatment. Dalton. Trans. 2018, 47, 3921–3930. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, H.; Shi, J. Inorganic Nanoparticle-Based Drug Codelivery Nanosystems to Overcome the Multidrug Resistance of Cancer Cells. Mol. Pharm. 2014, 11, 2495–2510. [Google Scholar] [CrossRef]
- CHOI, K. Botanical Characteristics, Pharmacological Effects and Medicinal Components of Korean Panax Ginseng C A Meyer. Acta Pharm. Sin. 2008, 29, 1109–1118. [Google Scholar] [CrossRef]
- Rhim, H.; Kim, H.; Lee, D.Y.; Oh, T.H.; Nah, S.-Y. Ginseng and Ginsenoside Rg3, a Newly Identified Active Ingredient of Ginseng, Modulate Ca2+ Channel Currents in Rat Sensory Neurons. Eur. J. Pharm. 2002, 436, 151–158. [Google Scholar] [CrossRef]
- Kim, Y.C.; Kim, S.R.; Markelonis, G.J.; Oh, T.H. Ginsenosides Rb1and Rg3protect Cultured Rat Cortical Cells from Glutamate-Induced Neurodegeneration. J. Neurosci. Res. 1998, 53, 426–432. [Google Scholar] [CrossRef]
- Shinkai, K.; Akedo, H.; Mukai, M.; Imamura, F.; Isoai, A.; Kobayashi, M.; Kitagawa, I. Inhibition of in Vitro Tumor Cell Invasion by Ginsenoside Rg 3. Jpn. J. Cancer Res. 1996, 87, 357–362. [Google Scholar] [CrossRef]
- Mochizuki, M.; Yoo, Y.; Matsuzawa, K.; Sato, K.; Saiki, I.; Tonooka, S.; Samukawa, K.; Azuma, I. Inhibitory Effect of Tumor Metastasis in Mice by Saponins, Ginsenoside-Rb2, 20(R)- and 20(S)-Ginsenoside-Rg3, of Red Ginseng. Biol. Pharm. Bull. 1995, 18, 1197–1202. [Google Scholar] [CrossRef]
- Tian-min, X.U.; Ying, X.I.N.; Man-hua, C.U.I.; Xin, J.; Li-ping, G.U.; Rg, G.; Drugs, M.; Rg, G.; Chemistry, M.; Medicine, P.; et al. Inhibitory effect of ginsenoside Rg3 combined with cyclophosphamide on growth and angiogenesis of ovarian cancer. Chin. Med. J. 2007, 120, 584–588. [Google Scholar]
- Yue, P.Y.K.; Wong, D.Y.L.; Wu, P.K.; Leung, P.Y.; Mak, N.K.; Yeung, H.W.; Liu, L.; Cai, Z.; Jiang, Z.-H.; Fan, T.P.D.; et al. The Angiosuppressive Effects of 20(R)- Ginsenoside Rg3. Biochem. Pharm. 2006, 72, 437–445. [Google Scholar] [CrossRef]
- Xu, T.M.; Cui, M.H.; Xin, Y.; Gu, L.P.; Jiang, X.; Su, M.M.; Wang, D.D.; Wang, W.J. Inhibitory Effect of Ginsenoside Rg3 on Ovarian Cancer Metastasis. Chin. Med. J. 2008, 121, 1394–1397. [Google Scholar] [CrossRef]
- Tang, H.; Ye, Z.; Ren, Y.; Zhu, Z.; Wu, Y. Investigation on the Mechanism of Ginsenoside Rg3 in Treating Murine Primary Mammary Tumor. Front. Med. China 2009, 3, 421–425. [Google Scholar] [CrossRef]
- Lu, P.; Su, W.; Miao, Z.; Niu, H.; Liu, J.; Hua, Q. Effect and Mechanism of Ginsenoside Rg3 on Postoperative Life Span of Patients with Non-Small Cell Lung Cancer. Chin. J. Integr. Med. 2008, 14, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Cheng, J.; Huang, Y.P.; Han, S.L.; Liu, N.X.; Zhu, G.B.; Yao, J.G. Effect of Adjuvant Chemotherapy of Ginsenoside Rg3 Combined with Mitomycin C and Tegafur in Advanced Gastric Cancer. Chin. J. Gastrointest. Surg. 2007, 10, 64–66. [Google Scholar]
- Huang, J. Efficacy of Shenyi Capsule Combined with Gemcitabine plus Cisplatin in Treatment of Advanced Esophageal Cancer: A Randomized Controlled Trial. J. Chin. Integr. Med. 2009, 7, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhu, H.; Zhu, Y.; Feng, J.; Chen, Z.; Li, G.; Zhang, X.; Zhang, Z.; Tang, J.; Shi, M.; et al. A Randomized, Prospective, Multi-Centre Clinical Trial of NP Regimen (Vinorelbine+cisplatin) plus Gensing Rg3 in the Treatment of Advanced Non-Small Cell Lung Cancer Patients. Chin. J. Lung Cancer 2006, 9, 254–258. [Google Scholar] [CrossRef]
- Wang, X.; Chen, L.; Wang, T.; Jiang, X.; Zhang, H.; Li, P.; Lv, B.; Gao, X. Ginsenoside Rg3 Antagonizes Adriamycin-Induced Cardiotoxicity by Improving Endothelial Dysfunction from Oxidative Stress via Upregulating the Nrf2-ARE Pathway through the Activation of Akt. Phytomedicine 2015, 22, 875–884. [Google Scholar] [CrossRef]
- Li, L.; Ni, J.; Li, M.; Chen, J.; Han, L.; Zhu, Y.; Kong, D.; Mao, J.; Wang, Y.; Zhang, B.; et al. Ginsenoside Rg3 Micelles Mitigate Doxorubicin-Induced Cardiotoxicity and Enhance Its Anticancer Efficacy. Drug Deliv. 2017, 24, 1617–1630. [Google Scholar] [CrossRef]
- Zare-Zardini, H.; Taheri-Kafrani, A.; Amiri, A.; Bordbar, A.K. New Generation of Drug Delivery Systems Based on Ginsenoside Rh2-, Lysine- and Arginine-Treated Highly Porous Graphene for Improving Anticancer Activity. Sci. Rep. 2018, 8, 586. [Google Scholar] [CrossRef]
- Kazempour, M.; Namazi, H.; Akbarzadeh, A.; Kabiri, R. Synthesis and Characterization of PEG-Functionalized Graphene Oxide as an Effective PH-Sensitive Drug Carrier. Artif. Cells Nanomed. Biotechnol. 2019, 47, 90–94. [Google Scholar] [CrossRef]
- Zhang, W.; Guo, Z.; Huang, D.; Liu, Z.; Guo, X.; Zhong, H. Synergistic Effect of Chemo-Photothermal Therapy Using PEGylated Graphene Oxide. Biomaterials 2011, 32, 8555–8561. [Google Scholar] [CrossRef]
- Zhao, Q.; Zheng, X.; Jiang, J.; Zhou, H.; Hu, P. Determination of Ginsenoside Rg3 in Human Plasma and Urine by High Performance Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B Anal. Technol. Biomed Life Sci. 2010, 878, 2266–2273. [Google Scholar] [CrossRef]
- Mistiran, A.F.; Dzarr, A.A.; Gan, S.H. HPLC Method Development and Validation for Simultaneous Detection of Arabinoside-C and Doxorubicin. Toxicol. Mech. Methods 2010, 20, 472–481. [Google Scholar] [CrossRef]
- Fernández-Merino, M.J.; Guardia, L.; Paredes, J.I.; Villar-Rodil, S.; Solís-Fernández, P.; Martínez-Alonso, A.; Tascón, J.M.D. Vitamin C Is an Ideal Substitute for Hydrazine in the Reduction of Graphene Oxide Suspensions. J. Phys. Chem. C 2010, 114, 6426–6432. [Google Scholar] [CrossRef]
- Pfister, D.; Núñez, N.G.; Pinyol, R.; Govaere, O.; Pinter, M.; Szydlowska, M.; Gupta, R.; Qiu, M.; Deczkowska, A.; Weiner, A.; et al. NASH Limits Anti-Tumour Surveillance in Immunotherapy-Treated HCC. Nature 2021, 592, 450–456. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Github 2018. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 26 September 2022).
- Väremo, L.; Nielsen, J.; Nookaew, I. Enriching the Gene Set Analysis of Genome-Wide Data by Incorporating Directionality of Gene Expression and Combining Statistical Hypotheses and Methods. Nucleic. Acids Res. 2013, 41, 4378–4391. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Konkena, B.; Vasudevan, S. Understanding Aqueous Dispersibility of Graphene Oxide and Reduced Graphene Oxide through p K a Measurements. J. Phys. Chem. Lett. 2012, 3, 867–872. [Google Scholar] [CrossRef]
- Tang, H.; Zhao, Y.; Shan, S.; Yang, X.; Liu, D.; Cui, F.; Xing, B. Wrinkle- and Edge-Adsorption of Aromatic Compounds on Graphene Oxide as Revealed by Atomic Force Microscopy, Molecular Dynamics Simulation, and Density Functional Theory. Env. Sci. Technol. 2018, 52, 7689–7697. [Google Scholar] [CrossRef]
- Kudin, K.N.; Ozbas, B.; Schniepp, H.C.; Prud’homme, R.K.; Aksay, I.A.; Car, R. Raman Spectra of Graphite Oxide and Functionalized Graphene Sheets. Nano. Lett. 2008, 8, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Beams, R.; Gustavo Cançado, L.; Novotny, L. Raman Characterization of Defects and Dopants in Graphene. J. Phys. Condens. Matter. 2015, 27, 083002. [Google Scholar] [CrossRef]
- Kim, D.-G.; Jung, K.H.; Lee, D.-G.; Yoon, J.-H.; Choi, K.S.; Kwon, S.W.; Shen, H.-M.; Morgan, M.J.; Hong, S.-S.; Kim, Y.-S. 20(S)-Ginsenoside Rg3 Is a Novel Inhibitor of Autophagy and Sensitizes Hepatocellular Carcinoma to Doxorubicin. Oncotarget 2014, 5, 4438–4451. [Google Scholar] [CrossRef]
- Huang, J.; Tang, X.; Ikejima, T.; Sun, X.; Wang, X.; Xi, R.; Wu, L. A New Triterpenoid from Panax Ginseng Exhibits Cytotoxicity through P53 and the Caspase Signaling Pathway in the HepG2 Cell Line. Arch. Pharm. Res. 2008, 31, 323–329. [Google Scholar] [CrossRef]
- Chatterjee, N.; Eom, H.J.; Choi, J. A Systems Toxicology Approach to the Surface Functionality Control of Graphene-Cell Interactions. Biomaterials 2014, 35, 1109–1127. [Google Scholar] [CrossRef] [PubMed]
- Lammel, T.; Boisseaux, P.; Fernández-Cruz, M.L.; Navas, J.M. Internalization and Cytotoxicity of Graphene Oxide and Carboxyl Graphene Nanoplatelets in the Human Hepatocellular Carcinoma Cell Line Hep G2. Part Fibre Toxicol. 2013, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Fu, Y.; Wei, T.; Le Guyader, L.; Gao, G.; Liu, R.-S.; Chang, Y.-Z.; Chen, C. The Triggering of Apoptosis in Macrophages by Pristine Graphene through the MAPK and TGF-Beta Signaling Pathways. Biomaterials 2012, 33, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Guo, S.; Nishina, Y.; Bianco, A. Reaction between Graphene Oxide and Intracellular Glutathione Affects Cell Viability and Proliferation. ACS Appl. Mater Interfaces 2021, 13, 3528–3535. [Google Scholar] [CrossRef]
- Postma, B.; Poppelier, M.J.; van Galen, J.C.; Prossnitz, E.R.; van Strijp, J.A.G.; de Haas, C.J.C.; van Kessel, K.P.M. Chemotaxis Inhibitory Protein of Staphylococcus Aureus Binds Specifically to the C5a and Formylated Peptide Receptor. J. Immunol. 2004, 172, 6994–7001. [Google Scholar] [CrossRef]
- Szmidt, M.; Stankiewicz, A.; Urbańska, K.; Jaworski, S.; Kutwin, M.; Wierzbicki, M.; Grodzik, M.; Burzyńska, B.; Góra, M.; Chwalibog, A.; et al. Graphene Oxide Down-Regulates Genes of the Oxidative Phosphorylation Complexes in a Glioblastoma. BMC Mol. Biol. 2019, 20, 2. [Google Scholar] [CrossRef]
- Pramanik, K.C.; Boreddy, S.R.; Srivastava, S.K. Role of Mitochondrial Electron Transport Chain Complexes in Capsaicin Mediated Oxidative Stress Leading to Apoptosis in Pancreatic Cancer Cells. PLoS ONE 2011, 6, e20151. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, L.; Li, M.; Zhao, R.; Yang, X.; Ji, T.; Gu, Z.; Yin, J.-J.; Gao, X.; Nie, G. Deciphering the Underlying Mechanisms of Oxidation-State Dependent Cytotoxicity of Graphene Oxide on Mammalian Cells. Toxicol. Lett. 2015, 237, 61–71. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 Signalling Axis in Cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Lahiani, M.H.; Gokulan, K.; Williams, K.; Khare, S. Ex Vivo Human Colon Tissue Exposure to Pristine Graphene Activates Genes Involved in the Binding, Adhesion and Proliferation of Epithelial Cells. Int. J. Mol. Sci. 2021, 22, 11443. [Google Scholar] [CrossRef]
- Numasaki, M.; Watanabe, M.; Suzuki, T.; Takahashi, H.; Nakamura, A.; McAllister, F.; Hishinuma, T.; Goto, J.; Lotze, M.T.; Kolls, J.K.; et al. IL-17 Enhances the Net Angiogenic Activity and In Vivo Growth of Human Non-Small Cell Lung Cancer in SCID Mice through Promoting CXCR-2-Dependent Angiogenesis. J. Immunol. 2005, 175, 6177–6189. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Sun, C.; Liao, C.; Cui, L.; Li, H.; Qu, G.; Yu, W.; Song, N.; Cui, Y.; Wang, Z.; et al. Graphene Enhances Cellular Proliferation through Activating the Epidermal Growth Factor Receptor. J. Agric. Food Chem. 2016, 64, 5909–5918. [Google Scholar] [CrossRef] [PubMed]
- Romaldini, A.; Spanò, R.; Catalano, F.; Villa, F.; Poggi, A.; Sabella, S. Sub-Lethal Concentrations of Graphene Oxide Trigger Acute-Phase Response and Impairment of Phase-I Xenobiotic Metabolism in Upcyte® Hepatocytes. Front Bioeng. Biotechnol. 2022, 10, 808. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.-M.; Jung, H.-J.; Choi, W.-Y.; Lim, C.-J. Antioxidative, Anti-Inflammatory, and Matrix Metalloproteinase Inhibitory Activities of 20(S)-Ginsenoside Rg3 in Cultured Mammalian Cell Lines. Mol. Biol. Rep. 2013, 40, 269–279. [Google Scholar] [CrossRef]
- Gum, S.I.; Cho, M.K. The Amelioration of N-Acetyl-p-Benzoquinone Imine Toxicity by Ginsenoside Rg3: The Role of Nrf2-Mediated Detoxification and Mrp1/Mrp3 Transports. Oxid. Med. Cell Longev. 2013, 2013, 957947. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Kallinowski, F.; Okunieff, P. Review Blood Flow, Oxygen and Nutrient Supply, and Metabolic Microenvironment of Human Tumors: A Review. Cancer Res. 1989, 49, 6449–6465. [Google Scholar]
- Gatenby, R.A.; Gillies, R.J. Why Do Cancers Have High Aerobic Glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Liu, Z.; Ma, Y.; Huang, Y.; Chen, Y. High-Efficiency Loading and Controlled Release of Doxorubicin Hydrochloride on Graphene Oxide. J. Phys. Chem. C 2008, 112, 17554–17558. [Google Scholar] [CrossRef]
- Kataoka, K.; Matsumoto, T.; Yokoyama, M.; Okano, T.; Sakurai, Y.; Fukushima, S.; Okamoto, K.; Kwon, G.S. Doxorubicin-Loaded Poly(Ethylene Glycol)–Poly(β-Benzyl-l-Aspartate) Copolymer Micelles: Their Pharmaceutical Characteristics and Biological Significance. J. Control. Release 2000, 64, 143–153. [Google Scholar] [CrossRef]
- Zheng, Y.; Yan, Y.; Lin, L.; He, Q.; Hu, H.; Luo, R.; Xian, D.; Wu, J.; Shi, Y.; Zeng, F.; et al. Titanium Carbide MXene-Based Hybrid Hydrogel for Chemo-Photothermal Combinational Treatment of Localized Bacterial Infection. Acta Biomater. 2022, 142, 113–123. [Google Scholar] [CrossRef]
- Mizutani, H.; Tada-Oikawa, S.; Hiraku, Y.; Kojima, M.; Kawanishi, S. Mechanism of Apoptosis Induced by Doxorubicin through the Generation of Hydrogen Peroxide. Life Sci. 2005, 76, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Hou, Q.; Zhao, T.; Zhang, H.; Zhang, Q.; Wu, L.; Fan, Z. Granzyme M Directly Cleaves Inhibitor of Caspase-Activated DNase (CAD) to Unleash CAD Leading to DNA Fragmentation. J. Immunol. 2006, 177, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Gara, S.; Jia, L.; Merino, M.; Agarwal, S.; Zhang, L.; Cam, M.; Patel, D.; Kebebew, E. Germline HABP2 Mutation Causing Familial Nonmedullary Thyroid Cancer. N. Engl. J. Med. 2015, 373, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Kischkel, F.; Lawrence, D.; Tinel, A.; LeBlanc, H.; Virmani, A.; Schow, P.; Gazdar, A.; Blenis, J.; Arnott, D.; Ashkenazi, A. Death Receptor Recruitment of Endogenous Caspase-10 and Apoptosis Initiation in the Absence of Caspase-8. J. Biol. Chem. 2001, 276, 46639–46646. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahimi, S.; van Leeuwen, D.; Roshanzamir, F.; Pandit, S.; Shi, L.; Sasanian, N.; Nielsen, J.; Esbjörner, E.K.; Mijakovic, I. Ginsenoside Rg3 Reduces the Toxicity of Graphene Oxide Used for pH-Responsive Delivery of Doxorubicin to Liver and Breast Cancer Cells. Pharmaceutics 2023, 15, 391. https://doi.org/10.3390/pharmaceutics15020391
Rahimi S, van Leeuwen D, Roshanzamir F, Pandit S, Shi L, Sasanian N, Nielsen J, Esbjörner EK, Mijakovic I. Ginsenoside Rg3 Reduces the Toxicity of Graphene Oxide Used for pH-Responsive Delivery of Doxorubicin to Liver and Breast Cancer Cells. Pharmaceutics. 2023; 15(2):391. https://doi.org/10.3390/pharmaceutics15020391
Chicago/Turabian StyleRahimi, Shadi, Daniel van Leeuwen, Fariba Roshanzamir, Santosh Pandit, Lei Shi, Nima Sasanian, Jens Nielsen, Elin K. Esbjörner, and Ivan Mijakovic. 2023. "Ginsenoside Rg3 Reduces the Toxicity of Graphene Oxide Used for pH-Responsive Delivery of Doxorubicin to Liver and Breast Cancer Cells" Pharmaceutics 15, no. 2: 391. https://doi.org/10.3390/pharmaceutics15020391
APA StyleRahimi, S., van Leeuwen, D., Roshanzamir, F., Pandit, S., Shi, L., Sasanian, N., Nielsen, J., Esbjörner, E. K., & Mijakovic, I. (2023). Ginsenoside Rg3 Reduces the Toxicity of Graphene Oxide Used for pH-Responsive Delivery of Doxorubicin to Liver and Breast Cancer Cells. Pharmaceutics, 15(2), 391. https://doi.org/10.3390/pharmaceutics15020391






