Abstract
Amlodipine is an antihypertensive drug with unknown pharmacogenetic biomarkers. This research is a candidate gene study that looked for associations between amlodipine pharmacokinetics and safety and pharmacogenes. Pharmacokinetic and safety data were taken from 160 volunteers from eight bioequivalence trials. In the exploratory step, 70 volunteers were genotyped for 44 polymorphisms in different pharmacogenes. CYP2D6 poor metabolizers (PMs) showed higher half-life (t1/2) (univariate p-value (puv) = 0.039, multivariate p-value (pmv) = 0.013, β = −5.31, R2 = 0.176) compared to ultrarapid (UMs), normal (NMs) and intermediate metabolizers (IMs). SLC22A1 rs34059508 G/A genotype was associated with higher dose/weight-corrected area under the curve (AUC72/DW) (puv = 0.025; pmv = 0.026, β = 578.90, R2 = 0.060) compared to the G/G genotype. In the confirmatory step, the cohort was increased to 160 volunteers, who were genotyped for CYP2D6, SLC22A1 and CYP3A4. In addition to the previous associations, CYP2D6 UMs showed a lower AUC72/DW (puv = 0.046, pmv = 0.049, β = −68.80, R2 = 0.073) compared to NMs, IMs and PMs and the SLC22A1 rs34059508 G/A genotype was associated with thoracic pain (puv = 0.038) and dizziness (puv = 0.038, pmv = 0.014, log OR = 10.975). To our knowledge, this is the first work to report a strong relationship between amlodipine and CYP2D6 and SLC22A1. Further research is needed to gather more evidence before its application in clinical practice.
1. Introduction
High blood pressure (HBP) is an important risk factor for cardiovascular morbidity and mortality [1]. HBP shows a high prevalence in the population: it affected 45.4% of adults aged 18 or over in 2017–2018 in the US, and up to 74.5% of people aged 60 or over [2]. Patients with HBP are often asymptomatic [1]. However, it is a chronic disease that increases the risk of cardiovascular, cerebrovascular and renal problems. Therefore, the European Society of Hypertension and the European Society of Cardiology (ESH/ESC) guidelines (2018) recommend patients to have their BP controlled and reduced to less than 140/90 mmHg [3]. To achieve this, an optimal control and adherence to the treatment is necessary.
Amlodipine is a calcium channel blocker (CCB) used for the treatment of HBP [4]. Its antihypertensive effect is achieved through the vasodilatation produced by the inhibition of the transmembrane calcium influx into vascular smooth muscle cells and cardiac muscle cells [5]. Amlodipine presents an oral bioavailability of 64%. The plasma peak concentration (Cmax) is reached 6–9 h after administration (tmax). It binds extensively to plasma proteins (98%) and shows a volume of distribution (Vd) of 21 L/kg [6]. It is mainly metabolized by CYP3A4 and none of its metabolites are active [7]. Its elimination half-life (t1/2) ranges between 31 and 37 h after the administration of a single dose, with a clearance (Cl) of 0.42 L/kg*h [6]. The most frequent adverse drug reactions (ADRs) caused by amlodipine are headache, flushing, dizziness and peripheral edema, among others that are less frequent, such as hypotension and thoracic pain [6,8,9]. However, it seems to present a more favorable side effect profile and to be more cardioprotective than other antihypertensive drugs, such as metoprolol, lisinopril or irbesartan [6,10].
When pharmacological treatment is indicated for HBP management, the selection of the drug or drugs is usually based on individual efficacy and tolerability. However, no good response predictors are available, and it is estimated that, in Europe, only 19–40% of patients on treatment achieve target BP [3,11,12]. This lack of efficacy could be partially explained by pharmacogenetics. Genetic variation in genes coding for metabolic enzymes and transporters can alter their activity, which could modify drug exposure, therefore conditioning its efficacy and safety. To date, no clinically relevant pharmacogenetic biomarkers for amlodipine or any other antihypertensive drug have been described. Thus, our objective was to evaluate the impact of 44 single nucleotide polymorphisms (SNPs) in 15 pharmacogenes, including metabolizing enzymes (CYP3A4, CYP3A5, CYP2D6, CYP2B6, CYP2C9, CYP2C19, CYP2C8, CYP2A6, CYP1A2 and UGT1A1) and transporters (SLCO1B1, ABCB1, ABCC2, ABCG2 and SLC22A1) on the pharmacokinetic variability of amlodipine and ADR incidence. The present work is part of the La Princesa Multidisciplinary Initiative for the Implementation of Pharmacogenetics (PriME-PGx) [13].
2. Materials and Methods
2.1. Study Population
The participants of the present pharmacogenetic study were healthy volunteers enrolled in eight amlodipine bioequivalence clinical trials (named from A to H) conducted at the Clinical Trials Unit of Hospital Universitario de La Princesa (UECHUP), Madrid (Spain) from 2012 to 2018. The inclusion criteria were: males or females aged from 18 to 55, free from organic or psychic conditions, with normal medical records, vital signs, electrocardiogram and physical examination and without significant abnormalities in hematology, coagulation, biochemistry, serological and urine analysis. The exclusion criteria were: having received medication two days prior to the start of the study, having a body mass index (BMI) outside the 18.5–30.0 range, being pregnant or breastfeeding women, having history of sensitivity to any drug, having a positive drug screening, smoking or alcoholism, blood donation in the last month and participation in another study with investigational drugs in the three previous months.
The EUDRA-CT numbers of the clinical trials were as follows: 2012-001846-16 (A), 2013-004147-23 (B), 2017-000547-40 (C), 2017-001716-10 (D), 2017-001757-14 (E), 2017-005024-25 (F), 2018-001378-11 (G) and 2018-002075-18 (H). All of them were approved by the Spanish Drugs Agency (AEMPS) and the Research Ethics Committee (CEIm) of the Hospital Universitario de La Princesa. Both the development of the trials and the handling of data were conducted in compliance with Spanish Legislation, the International Council on Harmonization (ICH) guidelines on Good Clinical Practice [14] and the Revised Declaration of Helsinki [15]. A total of 216 volunteers gave written informed consent to participate in the clinical trials and 160 healthy volunteers gave it for the pharmacogenetic study.
2.2. Study Design and Procedures
Data were obtained from eight bioequivalence trials that compared a test and a reference formulation of amlodipine combined with other antihypertensive or hypocholesterolemic agents. A single oral dose was administered under fasting conditions in all trials, which were phase I, open-label, single-center, crossover and randomized with two sequences and two periods for amlodipine. In clinical trial A, amlodipine/atorvastatin (10/10 mg) film-coated tablets were compared with Caduet® film-coated tablets (Pfizer S.A.) at the same dose. Amlodipine/valsartan (10 mg/160 mg) film-coated tablets were compared with Exforge® film-coated tablets (Novartis) at the same dose in clinical trial B. In clinical trials C, D and E, amlodipine/valsartan/hydrochlorothiazide (HTZ) (10/320/25 mg) film-coated tablets were compared with Exforge HCT® film-coated tablets (Novartis Europharm Limited) at the same dose. In clinical trials F, G and H, the test formulations were olmesartan/amlodipine/HTZ (40/10/12.5 mg) film-coated tablets, which were compared with Sevikar HCT® (Daiichi Sankyo Spain, S.A) or with Olmetec Plus® (Daiichi Sankyo Spain, S.A) and Norvas® (Pfizer S.A.) tablets, both 40/10/12.5 mg. The wash-out time between the two periods was 14 days for clinical trials A to E and 21 days for clinical trials F to H. In both periods in all clinical trials, volunteers were confined at UECHUP since 10 p.m. of the day before drug administration (day 0). Drug administration took place at 9:00 a.m. (day 1), and blood samples were extracted at different times for amlodipine plasmatic quantification and for genotyping. Volunteers from clinical trials C to H stayed at UECHUP until 9:30 p.m. (day 1) and volunteers from clinical trials A and B stayed until 9:30 a.m. (day 2). All the volunteers visited UECHUP for additional blood extractions at 24, 48 and 72 h after drug administration. The studies were blinded only for the plasma concentration determination of both formulations. In all of them, the test formulation was demonstrated to be bioequivalent to the reference for amlodipine. Therefore, the arithmetic mean of the pharmacokinetic parameters of both formulations was calculated for each volunteer.
2.3. Pharmacokinetic Analysis
At least 16 blood samples were extracted from pre-dose to 72 h after drug intake in each period of the eight clinical trials. Amlodipine plasma concentration was measured by an external laboratory by high performance liquid chromatography with mass spectrometry (HPLC-MS/MS). The lower limit of quantification was established at 50.40 pg/mL (A), 50.25 pg/mL (B and F), 49.50 pg/mL (C, D, E and H) and 49.90 pg/mL (G). The concentration values obtained were used to calculate pharmacokinetic parameters using WinNonLin Professional Software version 2.0 (Scientific Consulting, Inc., Cary, NC, USA) for clinical trials A and B and version 7 (Scientific Consulting, Inc., Cary, NC, USA) for clinical trials C to H. The area under the curve from pre-dose to 72 h (AUC72) was calculated using the plasmatic concentrations according to the linear trapezoidal rule. The Cmax and tmax were obtained directly from the concentration-time curve and the t1/2 was calculated as −ln2/Ke.
2.4. Safety
Serological, biochemical and hematological tests were performed at different times during the eight clinical trials to assess safety. In addition, vital signs and BP measurements, physical examination and electrocardiograms (ECG) were scheduled at different times after drug administration. Volunteers were asked about the occurrence of adverse events (AEs) and were able to report them spontaneously. AEs causality was analyzed according to the algorithm of the Spanish pharmacovigilance system [16]. Those AEs with a possible, probable or definitive relationship with drug intake were defined as ADRs and were included in the statistical analysis.
2.5. Genotyping
Blood samples collected in EDTA-K2 tubes during the clinical trials were used to extract DNA using a MagNA Pure instrument (Roche Applied Science, USA) or a Maxwell® RSC Automated DNA extractor (Promega Biotech Iberica S.L). In an initial exploratory step, 70 individuals were genotyped for 44 polymorphisms in 15 pharmacogenes. For the initial exploratory phase, genotyping was carried out in a QuantStudio 12 K Flex qPCR instrument with an OpenArray thermal block (Applied Biosystems, Thermofisher, USA) using a custom array (Table 1).

Table 1.
Genotyped SNPs and alleles in which those SNPs are present.
To determine the deletion (*5) or duplication of CYP2D6, a TaqMan copy number variation (CNV) assay (Applied Biosystems, Foster City, CA, USA) was performed with probes for exon 9 (Assay ID: Hs00010001_cn) and intron 2 (Assay ID: Hs04083572_cn) of this gene, as previously described in a published work [17].
Additionally, 90 healthy volunteers were genotyped for the variants in genes with significant associations in the exploratory step and for the main candidates (CYP3A4 rs67666821 and rs35599367). For this confirmatory step, CYP2D6 rs35742686 and rs3892097 were genotyped with qPCR using TaqMan probes (ThermoFisher IDs: C__32407232_50 and C__27102431_D0) in the pharmacogenetics unit of the Clinical Pharmacology Department of the Hospital Universitario de La Princesa in a QuantStudio 12k Flex coupled to a 96-Fast thermal block. The genotyping of CYP2D6 rs5030656, rs1065852, rs28371706, rs28371725, CYP3A4 rs67666821, rs355993678 and SLC22A1 rs34059508 was conducted by MassArray (iPLEX® Gold technology) at the Spanish National Genotyping Center (CEGEN- FPGMX P22-FPGMX-038) [18] and CNV genotyping was performed as previously mentioned.
2.6. Phenotyping and Haplotyping
Enzyme or transporter phenotypes were inferred according to the Clinical Pharmacogenetics Implementation Consortium (CPIC) or the Dutch Pharmacogenetic Working Group (DPWG) guidelines for the following genes: CYP2B6 [19], CYP2C19 [20], CYP2C9 [21], CYP2D6 [22], CYP3A4 [23], CYP3A5 [24], SLCO1B1 [25] and UGT1A1 [26]. For CYP2C8, *1/*1 individuals were classified as normal metabolizers (NM), *1/*2 and *1/*4 as intermediate metabolizers (IM), *4/*4 as poor metabolizers (PM), *1/*3 as rapid metabolizers (RM) and *3/*3 as ultrarapid metabolizers (UM), as previously described [27]. The SNPs genotyped in the remaining genes were analyzed individually.
2.7. Statistical Analysis
SPSS software (version 23, SPSS Inc., Chicago, IL, USA) was used to perform the statistical analysis. AUC72 and Cmax were divided by the dose/weight ratio (DW) to correct the effect of dose and weight. Pharmacokinetic parameters were analyzed according to sex, race, clinical trial, co-administered drugs, genotypes and phenotypes. Variable distributions were checked for normality with a Shapiro–Wilks test. Variables not distributed normally were logarithmically transformed and normality was re-evaluated. For the pharmacokinetic variables following a normal distribution with two categories, a t-test was performed, whereas an ANOVA test followed by the Bonferroni post hoc was applied for those with three or more categories. For those not following a normal distribution, non-parametric tests were used. A Mann–Whitney test was used for variables with two categories and a Kruskal–Wallis test for those with three or more categories. The univariate p-value for statistically significant associations is shown (puv). Those biomarkers that presented a significant association in the univariate analysis (puv < 0.05) were included in the multivariate analysis by means of linear regression. Significant associations are shown as the multivariate p-value (pmv), the non-standardized β coefficient (β) and R2. Additionally, a Chi2 or Fisher exact test was performed in the confirmatory step to evaluate the incidence of ADRs according to sex, race, clinical trial, co-administered drugs, genotypes and phenotypes. Equally, the biomarkers with a significant association in the univariate analysis (puv < 0.05) and the pharmacokinetic parameters AUC and Cmax were included in the multivariate analysis, which involved logistic regression. The pmv and the odds ratio (OR) were shown for statistically significant associations.
3. Results
A total of 160 volunteers participated in this study (Table 2). Women presented lower weight, height and BMI compared to men (p < 0.001, p < 0.001 and p = 0.002, respectively). Age was higher in self-reported Latin-Americans (p = 0.005) compared to self-reported Caucasians and healthy volunteers with other races (individuals who self-reported as Blacks, Asians or Arabs).

Table 2.
Demographic characteristics regarding sex, race and clinical trial.
3.1. Pharmacokinetics
Women showed higher AUC72 and Cmax compared to men (350.09 ± 811.11 h*ng/mL versus 275.88 ± 545.94 h*ng/mL, p < 0.001 and 10.52 ± 2.26 ng/mL versus 8.53 ± 1.59 ng/mL, p < 0.001, respectively), but the differences disappeared after DW correction (Table 3). Women also presented higher tmax (puv = 0.002; pmv = 0.013, β = 0.55, R2 = 0.164) compared to men (Table 3). AUC72/DW, Cmax/DW, tmax and t1/2 were higher when an amlodipine/valsartan formulation was administered (pmv = 0.002, β = 368.25, R2 = 0.073; pmv = 0.01, β = 7.97, R2 = 0.036; pmv = 0.036, β = 0.72, R2 = 0.164 and puv = 0.004, pmv < 0.001, β = 6.04, R2 = 0.077, respectively) compared to the other formulations (Table 3). AUC72/DW was lower when atorvastatin was co-administered (puv = 0.018) compared to the co-administration of other drugs and tmax was lower in clinical trial G (puv = 0.007, pmv < 0.001, β = −0.03, R2 = 0.164) compared to the other clinical trials. In addition, AUC72/DW and tmax were higher in Latin-Americans and healthy volunteers with other races (pmv = 0.029, β = 195.05, R2 = 0.073 and puv = 0.022, pmv = 0.004, β = 0.74, R2 = 0.164, respectively) compared to Caucasians (Table 3).

Table 3.
Pharmacokinetic parameters regarding sex, race, clinical trial and co-administered drug.
In the exploratory step, t1/2 was higher in CYP2D6 PMs compared to UMs + NMs + IMs (puv = 0.039, pmv = 0.013, β = −5.31, R2 = 0.176) (Table 4). Individuals who presented the SLC22A1 rs34059508 G/A genotype showed a higher AUC72/DW (puv = 0.025; pmv = 0.026, β = 578.90, R2 = 0.060) compared to those with G/G genotype (Table 4). No pharmacokinetic differences were found for the remaining pharmacogenetic variables (Table S1).

Table 4.
Pharmacokinetic parameters according to CYP3A4 phenotype and genotypes or phenotypes showing statistically significant associations in the exploratory step.
In the confirmatory phase, 90 additional volunteers were included, reaching a final sample size of 160. Consistently, a significantly lower AUC72/DW was observed in CYP2D6 UMs compared to NMs + IMs + PMs (puv = 0.046, pmv = 0.049, β = −68.80, R2 = 0.073) and t1/2 was higher in PMs compared to IMs + NMs + UMs (puv = 0.006) (Table 5, Figure 1). Volunteers with the SLC22A1 rs34059508 G/A genotype showed a significantly higher AUC72/DW (puv = 0.046) compared to individuals with the G/G genotype, yet no significant differences in t1/2 were observed (Table 5, Figure 1). A tendency of higher exposure could be observed for the CYP3A4 IM phenotype. However, consistently with the exploratory step, the tendency was not significant (Table 5).

Table 5.
Pharmacokinetic parameters regarding the genes analyzed in the confirmatory step.
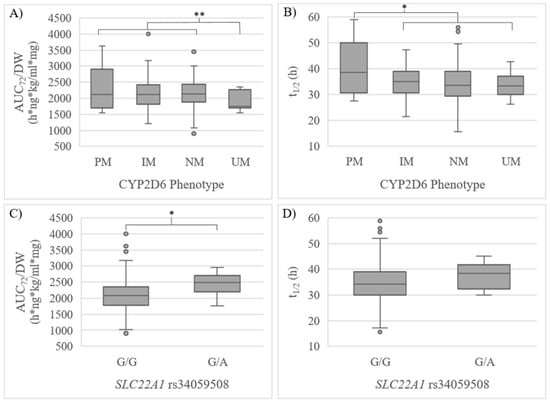
Figure 1.
(A) AUC72/DW and (B) t1/2 regarding CYP2D6 phenotype; (C) AUC72/DW and (D) t1/2 regarding SLC22A1 rs34059508 in the confirmatory step. UM: ultrarapid metabolizer, NM: normal metabolizer, IM: intermediate metabolizer, PM: poor metabolizer. * p < 0.05 in the univariate analysis. ** p < 0.05 in the univariate and multivariate analysis.
3.2. Safety
A total of 66 volunteers suffered from at least one ADR. The most common ADRs were headache (it affected 31.8% of volunteers), dizziness (5.63%), asthenia and eczema (3.12% each), nausea, hypotension and presyncope (2.50% each), diaphoresis (1.88%), diarrhea, cold and vomiting (1.25% each). ADR incidence was lower in men and Caucasians compared to women and to Latin-Americans and healthy volunteers with other races (puv = 0.024, pmv = 0.012, log OR = 2.327; puv = 0.006, pmv = 0.031, log OR = 2.306, respectively). In addition, ADR incidence was higher when a triple combination therapy was administered (puv = 0.011) compared to the administration of a dual therapy, and it was lower in clinical trial B (puv = 0.014) compared to clinical trials A + C–H (Table 6). Latin-Americans and healthy volunteers with other races also suffered from headaches with higher frequency compared to Caucasians (puv = 0.002, pmv = 0.001, log OR = 3.300) (Table 6). The only individual with thoracic pain showed the SLC22A1 rs34059508 G/A genotype (puv = 0.038). This genotype was also associated with a higher frequency of dizziness when compared to the G/G genotype (puv = 0.038, pmv = 0.014, log OR = 10.975) (Table 6). Additionally, those individuals who suffered from dizziness showed higher AUC72/DW compared to those without this ADR (2406.31 ± 475.78 h*ng*kg/mL*mg versus 2107.55 ± 490.26 h*ng*kg/mL*mg, pmv = 0.033, log OR = 1).

Table 6.
Significant differences in the incidence of ADRs according to clinical trial design, demographic characteristics and pharmacogenetic variables.
4. Discussion
Four first-line classes of drugs are used for HBP treatment, usually administered in a combination therapy: angiotensin-converting enzyme (ACE) inhibitors, angiotensin II receptor subtype 1 (AT1) blockers, long-acting CCBs of the dihydropyridine type and thiazide-like diuretics [3]. However, the high and growing incidence of HBP and the low control rates worldwide require that new strategies be developed to reach higher effectiveness of the pharmacological treatments. Pharmacogenetic biomarkers could be a useful tool to achieve the desired response. Pharmacogenetic information regarding amlodipine is scarce and no pharmacogenetic clinical guidelines are available nowadays. Hence, the present work is of great utility as it provides evidence for pharmacogenetic biomarkers and pharmacokinetic and safety variability in amlodipine treatment.
Here, amlodipine pharmacokinetics was consistent with previous works. For instance, the mean tmax (6.24 h) and t1/2 (34.89 h) fall within the ranges specified in the drug label (6–9 h for tmax and 31–37 h for t1/2) [8]. The mean AUC72 in this research was 312.52 ng*h/mL for a dose of 10 mg, compared to the mean AUCt reported in the literature for the same dose, 238.05 ng*h/mL [28]. The concomitant use of drugs could affect the absorption and metabolism of the drug, which could explain the reportedly 24% higher AUC value.
To our knowledge, this is the first work to describe a higher amlodipine exposure and tmax in Latin-Americans and healthy volunteers with other races compared to Caucasians. However, pharmacokinetic differences are well known for several drugs according to race [29], which are believed to be due to the different allele frequency in the CYP P450 genes and other genes involved in drug transport and metabolism among races [30,31]. These genetic differences among races, along with the high incidence of diseases that increases the risk of pharmacological interactions and ADRs, such as diabetes or chronic kidney disease, might lead to different management of antihypertensive treatment in the Latin-American population [32,33]. Further research is needed to evaluate whether these pharmacokinetic differences have an impact on drug response and should be considered in clinical practice.
A higher amlodipine exposure was found in volunteers that were treated with amlodipine and valsartan, which corresponds to clinical trial B. The co-administration of valsartan does not seem to be the cause of the higher exposure, as it was not observed when valsartan and HTZ were co-administered in clinical trials C, D and E and no interaction between amlodipine and valsartan was described in the literature. Additionally, the higher exposure does not correlate with a higher incidence of ADRs, which suggests that these results are futile. However, it is of interest to consider them in the analysis to better understand the interactions among those variables that were considered more relevant.
In this study, individuals with higher CYP2D6 activity showed lower amlodipine exposure and needed less time to eliminate it from the blood. To our knowledge, amlodipine is not known to be a CYP2D6 substrate, unlike diltiazem, another CCB [34]. Previous studies suggested that a fraction of amlodipine is metabolized by CYP enzymes [35], and, according to our data, one of these enzymes could be CYP2D6. The presence of a primary amine group in amlodipine supports this hypothesis, as CYP2D6 shows preference for metabolizing amines [36,37]. Additional in vitro works are required to demonstrate this enzyme–drug interaction, as well as in vivo studies to determine the clinical relevance of CYP2D6 phenotype.
The organic cationic transporter (OCT1), encoded by the SLC22A1 gene, is one of the major hepatic transporters that uptakes diverse drugs from blood into hepatic cells. The SLC22A1 rs34059508 A allele was previously defined as a no function allele [38]. In this study, individuals with the SLC22A1 rs34059508 A allele showed higher drug exposure, which is consistent with different studies with other drugs [38]. No pharmacogenetic information regarding SLC22A1 and amlodipine is known, but it was proposed that CCBs are OCT1’s ligands [39]. According to this, amlodipine could be an OCT1 substrate: SLC22A1 rs34059508 A allele carriers would have a reduced amlodipine uptake into hepatic cells, reducing its metabolism and, consequently, increasing drug exposure. If confirmed in further studies, variants in SLC22A1 may become biomarkers of great relevance in amlodipine treatment.
CYP3A4 is the main enzyme involved in amlodipine metabolism. However, only a non-significant trend of higher exposure in IMs could be observed, without significant differences in safety. To our knowledge, the only study in the literature that analyzed the same CYP3A4 variants found similar results [40]. Other studies with amlodipine focused on CYP3A4*1B, *1G and rs2246709, and the results found were contradictory [41,42,43,44,45]. The lack of correlation between the CYP3A4 phenotype and drug exposure or response could be explained by the high inducibility and inhibition of this enzyme, which might limit its value as a pharmacogenetic biomarker for amlodipine and other drugs [46]. However, the lack of correlation might also be explained by the absence of CYP3A4 PMs; thus, it would be of great interest to analyze whether these results are confirmed when including individuals with this phenotype. CYP3A4 PMs might show higher amlodipine concentrations, as it happens when it is administered in the presence of CYP3A4 inhibitors, such as azole antifungal agents or ritonavir [47].
Regarding safety, the incidence of ADRs was higher in women compared to men, which is consistent with previous works, where a higher response in women was also observed [35,45,48]. The higher exposure found in women before DW correction, along with the different body mass composition and the different hormone profile, may be responsible for the safety differences between men and women [48,49]. The incidence of ADRs was also higher when volunteers were given a triple combination therapy instead of a dual therapy, as was expected, since the administration of a higher number of drugs increases the risk of suffering from an ADR. According to our data, a previous study suggested that dual therapy is safer than triple therapy [50], but these results were not replicated in other works [51,52]. Lastly, Latin-Americans and healthy volunteers with other races suffered from ADRs with higher frequency compared to Caucasians, which is consistent with the higher amlodipine exposure observed in these individuals and also with the results of previous research [53]. Additional studies are required to clarify the clinical relevance of these associations and their utility in clinical practice. The only SNP significantly related to the higher incidence of ADRs was SLC22A1 rs3459508. Individuals who showed the rs3459508 G/A genotype suffered from dizziness and thoracic pain with higher frequency and also showed higher amlodipine exposure. Therefore, safety results reinforce the hypothesis that amlodipine is an OCT1 substrate, as those individuals with lower transporter activity show higher amlodipine exposure and have higher risk of suffering from ADRs.
The main limitation of this research was the administration of a single dose to healthy volunteers, which prevented us from analyzing amlodipine effectiveness or long-term safety. In addition, CYP3A4 could not be properly assessed, as no PMs were identified. By contrast, it should be noticed that the controlled environment in which the study was conducted allowed us to avoid cofounding factors, such as concomitant medications or smoking. Furthermore, additional research, such as more candidate gene studies and genome-wide association studies (GWAS), could provide unknown pharmacogenetic biomarkers involved in amlodipine pharmacokinetics or pharmacodynamics, which could also play an important role in predicting the response to this drug.
5. Conclusions
The CYP2D6 phenotype conditioned amlodipine exposure, which suggests it plays an important role in its metabolism. Furthermore, SLC22A1 affected amlodipine exposure and safety, which suggests it is involved in its transport. To the best of our knowledge, this is the first work to find these associations and to propose CYP2D6 and SLC22A1 as potential pharmacogenetic biomarkers in amlodipine treatment. Even though both associations are robust, their novelty and the absence of information in the literature make it necessary to gather more evidence before implementing their genotyping in routine clinical practice.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pharmaceutics15020404/s1, Table S1: Pharmacokinetic characteristics regarding genotypes or phenotypes in the exploratory step.
Author Contributions
Conceptualization, P.S.-C., P.Z. and F.A.-S.; Data curation, P.S.-C. and P.Z.; Formal analysis, P.S.-C. and P.Z.; Investigation, P.S.-C., P.Z., D.O., G.V.-G., M.R., M.M., L.F.-T., G.M.-A., S.C., A.d.M., M.N.-G., S.M.-V. and F.A.-S.; Methodology, P.S.-C. and P.Z.; Resources, D.O. and F.A.-S.; Writing—original draft, P.S.-C. and P.Z.; Writing—review and editing, D.O., G.V.-G., M.R., M.M., L.F.-T., G.M.-A., S.C., A.d.M., M.N.-G., S.M.-V. and F.A.-S. All authors have read and agreed to the published version of the manuscript.
Funding
P.S.-C. is financed by the FPI-UAM-2021 predoctoral fellowship. G.V.-G. is co-financed by Instituto de Salud Carlos III (ISCIII) and the European Social Fund (PFIS predoctoral grant, number FI20/00090). M.N.-G. is financed by the ICI20/00131 Grant, Acción Estratégica en Salud 2017–2020, ISCIII. P.Z. is financed by a “Contrato Margarita Salas de la convocatoria para la Recualificación del Sistema Universitario Español” (UAM).
Institutional Review Board Statement
The data used in this research were obtained from eight bioequivalence clinical trials conducted at the Clinical Trials Unit of the Hospital Universitario de La Princesa (UECHUP) (EUDRA-CT: 2012-001846-16, 2013-004147-23, 2017-000547-40, 2017-001716-10, 2017-001757-14, 2017-005024-25, 2018-001378-11 and 2018-002075-18). All of them were approved by the Spanish Drugs Agency (AEMPS) and the Research Ethics Committee (CEIm) of the Hospital Universitario de La Princesa. Both the development of the trials and the handling of data were conducted in compliance with Spanish legislation, the International Council on Harmonization (ICH) guidelines on good clinical practice [14] and the Revised Declaration of Helsinki [15].
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data belong to the clinical trials’ sponsors and may be accessible upon reasonable request to the corresponding authors.
Conflicts of Interest
F.A.-S. and D.O. have been consultants or investigators in clinical trials sponsored by the following pharmaceutical companies: Abbott, Alter, Aptatargets, Chemo, Cinfa, FAES, Farmalíder, Ferrer, GlaxoSmithKline, Galenicum, Gilead, Italfarmaco, Janssen-Cilag, Kern, Moderna, MSD, Normon, Novartis, Servier, Silver Pharma, Teva, and Zambon. The remaining authors declare no conflict of interest.
References
- Robles, N.; Macias, J. Hypertension in the Elderly. Cardiovasc. Hematol. Agents Med. Chem. 2015, 12, 136–145. [Google Scholar] [CrossRef]
- Ostchega, Y.; Fryar, C.D.; Nwankwo, T.; Nguyen, D.T. Hypertension Prevalence Among Adults Aged 18 and Over: United States, 2017–2018. NCHS Data Brief. 2020, 364, 1–8. [Google Scholar]
- Jordan, J.; Kurschat, C.; Reuter, H. Arterial Hypertension. Dtsch. Ärztebl. Int. 2018, 115, 557–568. [Google Scholar] [CrossRef]
- Ananchenko, G.; Novakovic, J.; Lewis, J. Amlodipine Besylate. In Profiles of Drug Substances, Excipients and Related Methodology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 37, pp. 31–77. ISBN 978-0-12-397220-0. [Google Scholar]
- Alvarez, J.C.; Mayer-Duverneuil, C.; Cappy, J.; Lorin de la Grandamison, G.; Knapp-Gisclon, A. Postmortem Fatal and Non-Fatal Concentrations of Amlodipine. Forensic Sci. Int. 2020, 316, 110555. [Google Scholar] [CrossRef] [PubMed]
- Meredith, P.A.; Elliott, H.L. Clinical Pharmacokinetics of Amlodipine. Clin. Pharmacokinet. 1992, 22, 22–31. [Google Scholar] [CrossRef] [PubMed]
- FDA Norvasc (Amlodipine Besylate) Label. Highlights of Prescribing Information. 2011. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/019787s047lbl.pdf (accessed on 10 November 2022).
- AEMPS Ficha Técnica AMLODIPINO CINFA 10 Mg Comprimidos EFG. 2022. Available online: https://cima.aemps.es/cima/dochtml/ft/65461/FichaTecnica_65461# (accessed on 20 November 2022).
- Rabah, F.; El-Naggari, M.; Al-Nabhani, D. Amlodipine: The Double Edged Sword: Amlodipine: The Double Edged Sword. J. Paediatr. Child Health 2017, 53, 540–542. [Google Scholar] [CrossRef] [PubMed]
- Park, C.G. Is Amlodipine More Cardioprotective than Other Antihypertensive Drug Classes? Korean J. Intern. Med. 2014, 29, 301–304. [Google Scholar] [CrossRef]
- NCD Risk Factor Collaboration (NCD-RisC). Worldwide Trends in Hypertension Prevalence and Progress in Treatment and Control from 1990 to 2019: A Pooled Analysis of 1201 Population-Representative Studies with 104 Million Participants. Lancet Lond. Engl. 2021, 398, 957–980. [Google Scholar] [CrossRef]
- Suárez, C. Single-Pill Telmisartan and Amlodipine: A Rational Combination for the Treatment of Hypertension. Drugs 2011, 71, 2295–2305. [Google Scholar] [CrossRef]
- Zubiaur, P.; Mejía-Abril, G.; Navares-Gómez, M.; Villapalos-García, G.; Soria-Chacartegui, P.; Saiz-Rodríguez, M.; Ochoa, D.; Abad-Santos, F. PriME-PGx: La Princesa University Hospital Multidisciplinary Initiative for the Implementation of Pharmacogenetics. J. Clin. Med. 2021, 10, 3772. [Google Scholar] [CrossRef]
- Vijayananthan, A.; Nawawi, O. The Importance of Good Clinical Practice Guidelines and Its Role in Clinical Trials. Biomed. Imaging Interv. J. 2008, 4, e5. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki Ethical Principles for Medical Research Involving Human Subjects. Available online: https://jamanetwork.com/journals/jama/fullarticle/1760318 (accessed on 20 November 2022).
- Aguirre, C.; García, M. Evaluación de la causalidad en las comunicaciones de reacciones adversas a medicamentos. Algoritmo del Sistema Español de Farmacovigilancia. Med. Clínica 2016, 147, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Belmonte, C.; Ochoa, D.; Román, M.; Saiz-Rodríguez, M.; Wojnicz, A.; Gómez-Sánchez, C.I.; Martín-Vílchez, S.; Abad-Santos, F. Influence of CYP2D6, CYP3A4, CYP3A5 and ABCB1 Polymorphisms on Pharmacokinetics and Safety of Aripiprazole in Healthy Volunteers. Basic Clin. Pharmacol. Toxicol. 2018, 122, 596–605. [Google Scholar] [CrossRef] [PubMed]
- CeGen-FPGMX Centro Nacional de Genotipado (CeGen-ISCIII). 2022.
- Desta, Z.; Gammal, R.S.; Gong, L.; Whirl-Carrillo, M.; Gaur, A.H.; Sukasem, C.; Hockings, J.; Myers, A.; Swart, M.; Tyndale, R.F.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-Containing Antiretroviral Therapy. Clin. Pharmacol. Ther. 2019, 106, 726–733. [Google Scholar] [CrossRef]
- Scott, S.A.; Sangkuhl, K.; Stein, C.M.; Hulot, J.-S.; Mega, J.L.; Roden, D.M.; Klein, T.E.; Sabatine, M.S.; Johnson, J.A.; Shuldiner, A.R. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 Genotype and Clopidogrel Therapy: 2013 Update. Clin. Pharmacol. Ther. 2013, 94, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Theken, K.N.; Lee, C.R.; Gong, L.; Caudle, K.E.; Formea, C.M.; Gaedigk, A.; Klein, T.E.; Agúndez, J.A.G.; Grosser, T. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin. Pharmacol. Ther. 2020, 108, 191–200. [Google Scholar] [CrossRef]
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.-J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777. [Google Scholar] [CrossRef]
- DPWG. Annotation of DPWG Guideline for Quetiapine and CYP3A4. 2021. Available online: https://www.pharmgkb.org/guidelineAnnotation/PA166265421 (accessed on 1 November 2022).
- Birdwell, K.; Decker, B.; Barbarino, J.; Peterson, J.; Stein, C.; Sadee, W.; Wang, D.; Vinks, A.; He, Y.; Swen, J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Johnson, S.G.; Caudle, K.E.; Haidar, C.E.; Voora, D.; Wilke, R.A.; Maxwell, W.D.; McLeod, H.L.; Krauss, R.M.; Roden, D.M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1 and Simvastatin-Induced Myopathy: 2014 Update. Clin. Pharmacol. Ther. 2014, 96, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Gammal, R.; Court, M.; Haidar, C.; Iwuchukwu, O.; Gaur, A.; Alvarellos, M.; Guillemette, C.; Lennox, J.; Whirl-Carrillo, M.; Brummel, S.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing. Clin. Pharmacol. Ther. 2016, 99, 363–369. [Google Scholar] [CrossRef]
- Campodónico, D.M.; Zubiaur, P.; Soria-Chacartegui, P.; Casajús, A.; Villapalos-García, G.; Navares-Gómez, M.; Gómez-Fernández, A.; Parra-Garcés, R.; Mejía-Abril, G.; Román, M.; et al. CYP2C8 *3 and *4 Define CYP2C8 Phenotype: An Approach with the Substrate Cinitapride. Clin. Transl. Sci. 2022, 15, 2613–2624. [Google Scholar] [CrossRef]
- Abad-Santos, F.; Novalbos, J.; Gálvez-Múgica, M.-A.; Gallego-Sandín, S.; Almeida, S.; Vallée, F.; García, A.G. Assessment of Sex Differences in Pharmacokinetics and Pharmacodynamics of Amlodipine in a Bioequivalence Study. Pharmacol. Res. 2005, 51, 445–452. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Pacanowski, M.; Bull, J.; Zhang, L. Racial/Ethnic Differences in Drug Disposition and Response: Review of Recently Approved Drugs. Clin. Pharmacol. Ther. 2015, 97, 263–273. [Google Scholar] [CrossRef]
- Zhou, Y.; Ingelman-Sundberg, M.; Lauschke, V. Worldwide Distribution of Cytochrome P450 Alleles: A Meta-Analysis of Population-Scale Sequencing Projects. Clin. Pharmacol. Ther. 2017, 102, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Bell, E.C.; Zhang, Y.; Liang, D. Racial Disparity in Drug Disposition in the Digestive Tract. Int. J. Mol. Sci. 2021, 22, 1038. [Google Scholar] [CrossRef]
- Correa, R.; Harsha Tella, S.; Elshimy, G.; Davidson, J.A. The Status of Diabetes and Its Complications in Latin-American Population: A Review Article. Diabetes Res. Clin. Pract. 2020, 168, 108274. [Google Scholar] [CrossRef]
- Gonzalez-Bedat, M.; Rosa-Diez, G.; Pecoits-Filho, R.; Ferreiro, A.; García-García, G.; Cusumano, A.; Fernandez-Cean, J.; Noboa, O.; Douthat, W. Burden of Disease: Prevalence and Incidence of ESRD in Latin America. Clin. Nephrol. 2015, 83, 3–6. [Google Scholar] [CrossRef]
- Molden, E.; Åsberg, A.; Christensen, H. CYP2D6 Is Involved in O-Demethylation of Diltiazem: An In Vitro Study with Transfected Human Liver Cells. Eur. J. Clin. Pharmacol. 2000, 56, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Seeland, U.; Regitz-Zagrosek, V. Sex and Gender Differences in Cardiovascular Drug Therapy. In Sex and Gender Differences in Pharmacology; Regitz-Zagrosek, V., Ed.; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2013; Volume 214, pp. 211–236. ISBN 978-3-642-30725-6. [Google Scholar]
- Abdoh, A.; Al-Omari, M.M.; Badwan, A.A.; Jaber, A.M.Y. Amlodipine Besylate–Excipients Interaction in Solid Dosage Form. Pharm. Dev. Technol. 2004, 9, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Costache, A.D.; Trawick, D.; Bohl, D.; Sem, D.S. AmineDB: Large Scale Docking of Amines with CYP2D6 and Scoring for Druglike Properties—Towards Defining the Scope of the Chemical Defense against Foreign Amines in Humans. Xenobiotica 2007, 37, 221–245. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Gong, L.; Giacomini, K.; Altman, R.B.; Klein, T.E. PharmGKB Summary: Very Important Pharmacogene Information for SLC22A1. Pharmacogenet. Genom. 2014, 24, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.C.; Khuri, N.; Liang, X.; Stecula, A.; Chien, H.-C.; Yee, S.W.; Huang, Y.; Sali, A.; Giacomini, K.M. Discovery of Competitive and Noncompetitive Ligands of the Organic Cation Transporter 1 (OCT1; SLC22A1). J. Med. Chem. 2017, 60, 2685–2696. [Google Scholar] [CrossRef] [PubMed]
- Saiz-Rodríguez, M.; Almenara, S.; Navares-Gómez, M.; Ochoa, D.; Román, M.; Zubiaur, P.; Koller, D.; Santos, M.; Mejía, G.; Borobia, A.M.; et al. Effect of the Most Relevant CYP3A4 and CYP3A5 Polymorphisms on the Pharmacokinetic Parameters of 10 CYP3A Substrates. Biomedicines 2020, 8, 94. [Google Scholar] [CrossRef]
- Bhatnagar, V.; Garcia, E.P.; O’Connor, D.T.; Brophy, V.H.; Alcaraz, J.; Richard, E.; Bakris, G.L.; Middleton, J.P.; Norris, K.C.; Wright, J.; et al. CYP3A4 and CYP3A5 Polymorphisms and Blood Pressure Response to Amlodipine among African-American Men and Women with Early Hypertensive Renal Disease. Am. J. Nephrol. 2010, 31, 95–103. [Google Scholar] [CrossRef]
- Guo, C.; Pei, Q.; Tan, H.; Huang, Z.; Yuan, H.; Yang, G. Effects of Genetic Factors on the Pharmacokinetics and Pharmacodynamics of Amlodipine in Primary Hypertensive Patients. Biomed. Rep. 2015, 3, 195–200. [Google Scholar] [CrossRef]
- Han, J.M.; Yee, J.; Chung, J.E.; Lee, K.E.; Park, K.; Gwak, H.S. Effects of Cytochrome P450 Oxidoreductase Genotypes on the Pharmacokinetics of Amlodipine in Healthy Korean Subjects. Mol. Genet. Genom. Med. 2020, 8, e1201. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wen, G.; Lu, Y.; Wen, J.; Ji, Y.; Xing, X.; Li, Y.; Wen, J.; Yuan, H. CYP3A4*1G and CYP3A5*3 Genetic Polymorphisms Alter the Antihypertensive Efficacy of Amlodipine in Patients with Hypertension Following Renal Transplantation. Int J. Clin. Pharmacol. Ther. 2017, 55, 109–118. [Google Scholar] [CrossRef]
- Zuo, X.; Zhang, W.; Yuan, H.; Barrett, J.S.; Hua, Y.; Huang, Z.; Zhou, H.; Pei, Q.; Guo, C.; Wang, J.; et al. ABCB1 Polymorphism and Gender Affect the Pharmacokinetics of Amlodipine in Chinese Patients with Essential Hypertension: A Population Analysis. Drug Metab. Pharmacokinet. 2014, 29, 305–311. [Google Scholar] [CrossRef]
- Zhou, S.; Yung Chan, S.; Cher Goh, B.; Chan, E.; Duan, W.; Huang, M.; McLeod, H.L. Mechanism-Based Inhibition of Cytochrome P450 3A4 by Therapeutic Drugs. Clin. Pharmacokinet. 2005, 44, 279–304. [Google Scholar] [CrossRef]
- Zhou, S.-F. Drugs Behave as Substrates, Inhibitors and Inducers of Human Cytochrome P450 3A4. Curr. Drug Metab. 2008, 9, 310–322. [Google Scholar] [CrossRef]
- Kloner, R.A.; Sowers, J.R.; DiBona, G.F.; Gaffney, M.; Marilee, W. Sex- and Age-Related Antihypertensive Effects of Amlodipine. Am. J. Cardiol. 1996, 77, 713–722. [Google Scholar] [CrossRef]
- Soldin, O.P.; Mattison, D.R. Sex Differences in Pharmacokinetics and Pharmacodynamics. Clin. Pharmacokinet. 2009, 48, 143–157. [Google Scholar] [CrossRef]
- Sung, K.-C.; Oh, Y.-S.; Cha, D.-H.; Hong, S.-J.; Won, K.-H.; Yoo, K.-D.; Rha, S.-W.; Ahn, Y.-K.; Ahn, J.-C.; Jang, J.-Y.; et al. Efficacy and Tolerability of Telmisartan/Amlodipine + Hydrochlorothiazide Versus Telmisartan/Amlodipine Combination Therapy for Essential Hypertension Uncontrolled With Telmisartan/Amlodipine: The Phase III, Multicenter, Randomized, Double-Blind TAHYTI Study. Clin. Ther. 2018, 40, 50–63.e3. [Google Scholar] [CrossRef] [PubMed]
- Düsing, R.; Waeber, B.; Destro, M.; Santos Maia, C.; Brunel, P. Triple-Combination Therapy in the Treatment of Hypertension: A Review of the Evidence. J. Hum. Hypertens. 2017, 31, 501–510. [Google Scholar] [CrossRef]
- Salam, A.; Atkins, E.R.; Hsu, B.; Webster, R.; Patel, A.; Rodgers, A. Efficacy and Safety of Triple versus Dual Combination Blood Pressure-Lowering Drug Therapy: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. J. Hypertens. 2019, 37, 1567–1573. [Google Scholar] [CrossRef] [PubMed]
- McDowell, S.E.; Coleman, J.J.; Ferner, R.E. Systematic Review and Meta-Analysis of Ethnic Differences in Risks of Adverse Reactions to Drugs Used in Cardiovascular Medicine. BMJ 2006, 332, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).