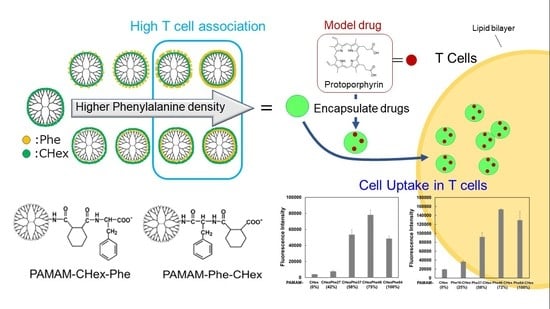
T Cell-Association of Carboxy-Terminal Dendrimers with Different Bound Numbers of Phenylalanine and Their Application to Drug Delivery
Abstract
:1. Introduction
2. Materials and Methods
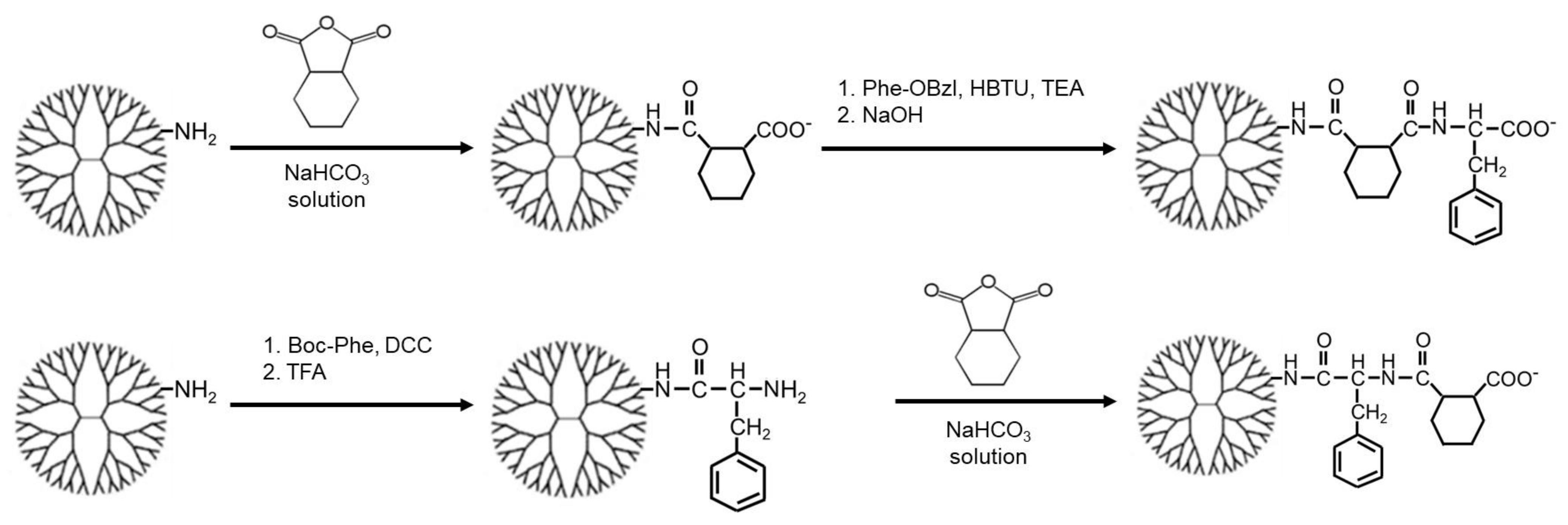
2.1. Synthesis
2.2. Characterization
2.3. Association of Dendrimers with Immune Cells
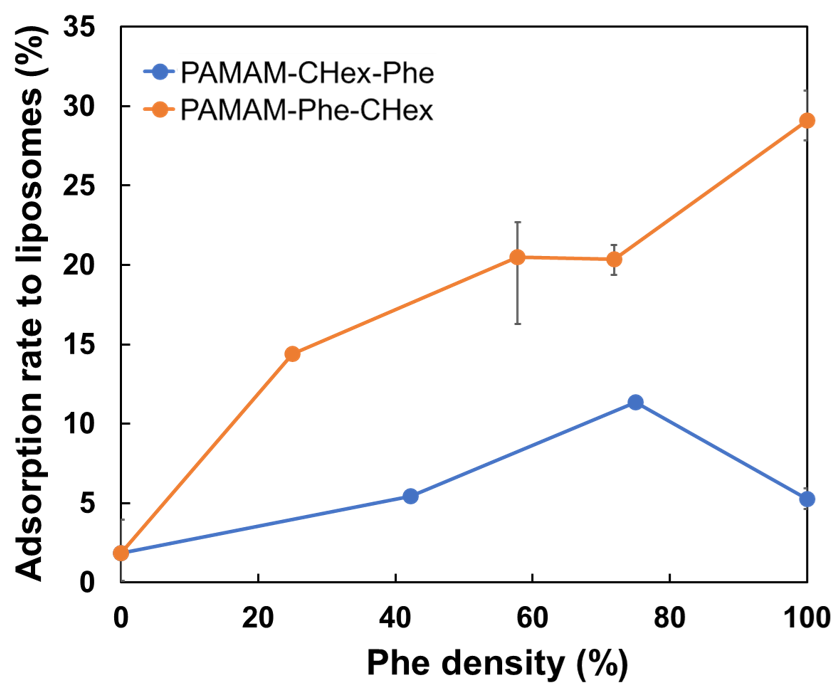
2.4. Adsorption of Dendrimers to Liposomes
2.5. Encapsulation of PpIX in Dendrimers
2.6. Delivery of PpIX-Encapsulating Dendrimers into T Cells
3. Results and Discussion
3.1. Synthesis of Carboxy-Terminal Dendrimers Modified with Different Bound Numbers of Phe
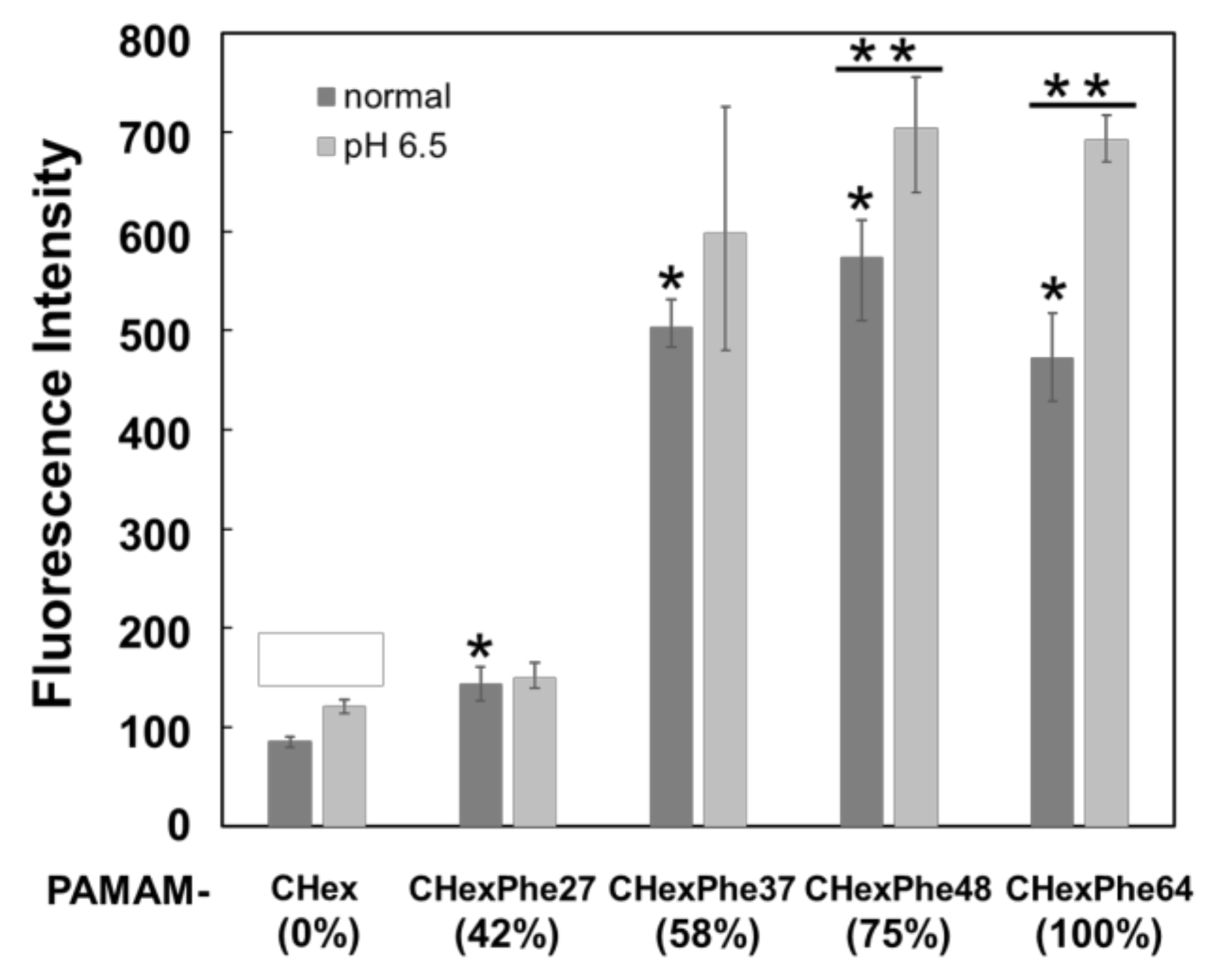
3.2. Cell Association of Carboxy-Terminal Dendrimers Modified with Various Numbers of Phe
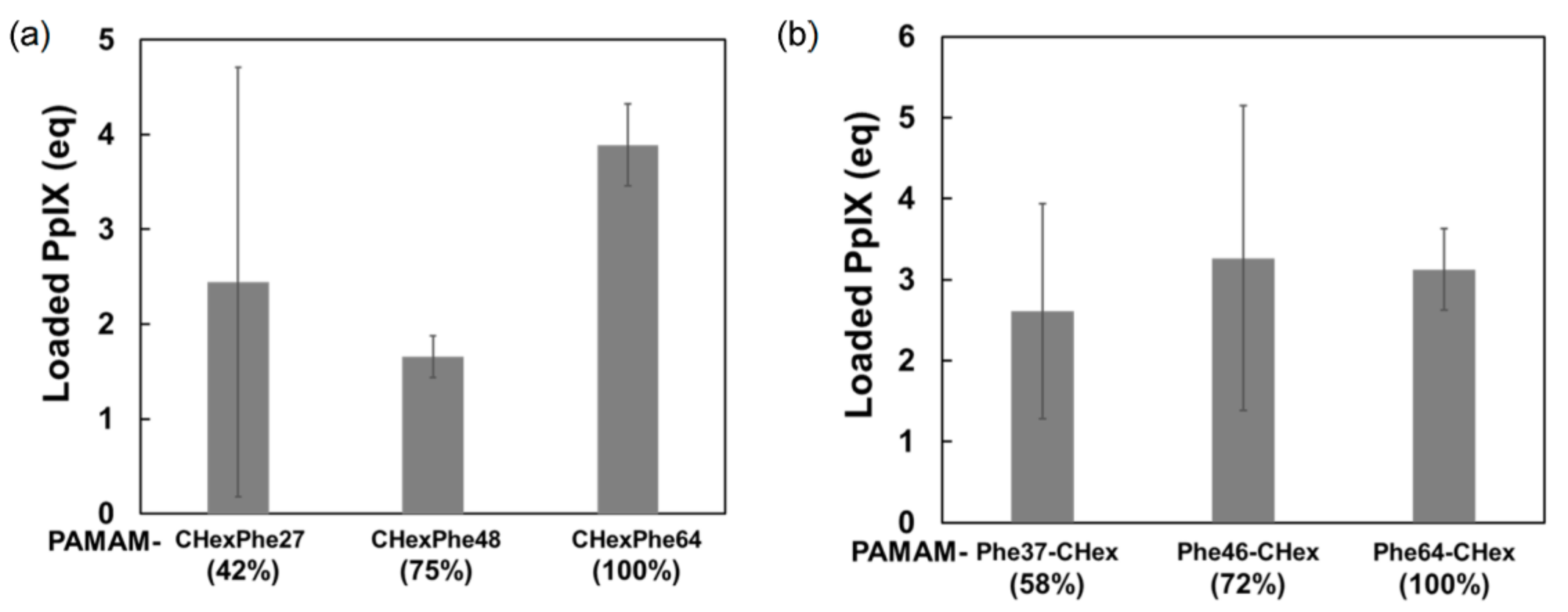
3.3. Model Drug Delivery into T Cells Using PAMAM-CHex-Phe and PAMAM-Phe-CHex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, T.M.; Cullis, P.R. Drug delivery systems: Entering the mainstream. Science 2004, 303, 1818–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riehemann, K.; Schneider, S.W.; Luger, T.A.; Godin, B.; Ferrari, M.; Fuchs, H. Nanomedicine—Challenge and perspectives. Angew. Chem. Int. Ed. 2009, 48, 872–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svenson, S.; Tomalia, D.A. Dendrimers in biomedical applications—Reflections on the field. Adv. Drug Deliv. Rev. 2005, 57, 2106–2129. [Google Scholar] [CrossRef]
- Khandare, J.; Calderón, M.; Dagia, N.M.; Haag, R. Multifunctional dendritic polymers in nanomedicine: Opportunities and challenges. Chem. Soc. Rev. 2012, 41, 2824–2848. [Google Scholar] [CrossRef] [Green Version]
- Mignani, S.; Rodrigues, J.; Tomas, H.; Zablocka, M.; Shi, X.; Caminade, A.M.; Majoral, J.P. Dendrimers in combination with natural products and analogues as anti-cancer agents. Chem. Soc. Rev. 2018, 47, 514–532. [Google Scholar] [CrossRef]
- Sherje, A.P.; Jadhav, M.; Dravyakar, B.R.; Kadam, D. Dendrimers: A versatile nanocarrier for drug delivery and targeting. Int. J. Pharm. 2018, 548, 707–720. [Google Scholar] [CrossRef]
- Kawamura, W.; Miura, Y.; Kokuryo, D.; Toh, K.; Yamada, N.; Nomoto, T.; Matsumoto, Y.; Sueyoshi, D.; Liu, X.; Aoki, I.; et al. Density-tunable conjugation of cyclic RGD ligands with polyion complex vesicles for the neovascular imaging of orthotopic glioblastomas. Sci. Technol. Adv. Mater. 2015, 16, 035004. [Google Scholar] [CrossRef]
- Ishii, T.; Miyata, K.; Anraku, Y.; Naito, M.; Yi, Y.; Jinbo, T.; Takae, S.; Fukusato, Y.; Hori, M.; Osada, K.; et al. Enhanced target recognition of nanoparticles by cocktail PEGylation with chains of varying lengths. Chem. Commun. 2016, 52, 1517–1519. [Google Scholar] [CrossRef]
- Alder-Abramovich, L.; Reches, M.; Sedman, V.L.; Allen, S.; Tendler, S.J.B.; Gazit, E. Thermal and chemical stability of diphenylalanine peptide nanotubes: Implications for nanotechnological applications. Langmuir 2006, 22, 1313–1320. [Google Scholar] [CrossRef]
- Singh, V.; Rai, R.K.; Arora, A.; Sinha, N.; Thakur, A.K. Therapeutic implication of L-phenylalanine aggregation mechanism and its modulation by D-phenylalanine in phenylketonuria. Sci. Rep. 2014, 4, 3875–3882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler-Abramovich, L.; Vaks, L.; Carny, O.; Trudler, D.; Magno, A.; Caflisch, A.; Frenkel, D.; Gazit, E. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat. Chem. Biol. 2012, 8, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Do, T.D.; Kincannon, W.M.; Bowers, M.T. Phenylalanine oligomers and fibrils: The mechanism of assembly and the importance of tetramers and counterions. J. Am. Chem. Soc. 2015, 137, 10080–10083. [Google Scholar] [CrossRef] [PubMed]
- Griffith, E.C.; Perkins, R.J.; Telesford, D.M.; Adams, E.M.; Cwiklik, L.; Allen, H.C.; Roeselová, M.; Vaida, V. Interaction of l-phenylalanine with a phospholipid monolayer at the water-air interface. J. Phys. Chem. B 2015, 119, 9038–9048. [Google Scholar] [CrossRef] [PubMed]
- Perkins, R.; Vaida, V. Phenylalanine increases membrane permeability. J. Am. Chem. Soc. 2017, 139, 14388–14391. [Google Scholar] [CrossRef] [Green Version]
- Nishita, M.; Park, S.Y.; Nishio, T.; Kamizaki, K.; Wang, Z.; Tamada, K.; Takumi, T.; Hashimoto, R.; Otani, H.; Pazour, G.J.; et al. Ror2 signaling regulates Golgi structure and transport through IFT20 for tumor invasiveness. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef] [Green Version]
- Darvin, P.; Toor, S.M.; Sasidharan, N.V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Joffre, O.P.; Segura, E.; Savina, A.; Amigorena, S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012, 12, 557–569. [Google Scholar] [CrossRef]
- Joshi, M.D.; Unger, W.J.; Storm, G.; van Kooyk, Y.; Mastrobattista, E. Targeting tumor antigens to dendritic cells using particulate carriers. J. Control. Release 2012, 161, 25–37. [Google Scholar] [CrossRef]
- Schmid, D.; Park, C.G.; Hartl, C.A.; Subedi, N.; Cartwright, A.N.; Puerto, R.B.; Zheng, Y.; Maiarana, J.; Freeman, G.J.; Wucherpfennig, K.W.; et al. T cell-targeting nanoparticles focus delivery of immunotherapy to improve antitumor immunity. Nat. Commun. 2017, 8, 1747–1758. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Slack, S.D.; Levy, C.N.; Hughes, S.M.; Jiang, Y.; Yogodzinski, C.; Roychoudhury, P.; Jerome, K.R.; Schiffer, J.T.; Hladik, F.; et al. Hybrid nanocarriers incorporating mechanistically distinct drugs for lymphatic CD4+ T cell activation and HIV-1 latency reversal. Sci. Adv. 2019, 5, eaav6322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, R.; Sakakibara, M.; Nagashima, T.; Sangai, T.; Arai, M.; Fujimori, T.; Takano, S.; Shida, T.; Nakatani, Y.; Miyazaki, M. Accumulation of regulatory T cells in sentinel lymph nodes is a prognostic predictor in patients with node-negative breast cancer. Eur. J. Cancer 2009, 45, 2123–2131. [Google Scholar] [CrossRef] [PubMed]
- Cochran, A.J.; Huang, R.R.; Lee, J.; Itakura, E.; Leong, S.P.; Essner, R. Tumour-induced immune modulation of sentinel lymph nodes. Nat. Rev. Immunol. 2006, 6, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Swartz, M.A.; Lund, A.W. Lymphatic and interstitial flow in the tumour microenvironment: Linking mechanobiology with immunity. Nat. Rev. Cancer 2012, 12, 210–219. [Google Scholar] [CrossRef]
- Niki, Y.; Ogawa, M.; Makiura, R.; Magata, Y.; Kojima, C. Optimization of dendrimer structure for sentinel lymph node imaging: Effects of generation and terminal group. Nanomedicine 2015, 11, 2119–2127. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Nagashima, S.; Nakajima, K.; Ohira, T.; Izawa, T.; Yamate, J.; Higashikawa, K.; Kuge, Y.; Ogawa, M.; Kojima, C. Carboxyl-, sulfonyl-, and phosphate-terminal dendrimers as a nanoplatform with lymph node targeting. Int. J. Pharm. 2020, 576, 119021. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Nishio, M.; Nagashima, S.; Nakajima, K.; Ohira, T.; Nakai, S.; Nakase, I.; Higashikawa, K.; Kuge, Y.; Matsumoto, A.; et al. Association of hydrophobic carboxyl-terminal dendrimers with lymph node-resident lymphocytes. Polymers 2020, 12, 1474. [Google Scholar] [CrossRef]
- Shiba, H.; Nishio, M.; Sawada, M.; Tamaki, M.; Michigami, M.; Nakai, S.; Nakase, I.; Fujii, I.; Matsumoto, A.; Kojima, C. Carboxy-terminal dendrimers with phenylalanine for a pH-sensitive delivery system into immune cells including T cells. J. Mater. Chem. B 2022, 10, 2463–2470. [Google Scholar] [CrossRef]
- Tamaki, M.; Fukushima, D.; Kojima, C. Dual pH-sensitive and UCST-type thermosensitive dendrimers: Phenylalanine-modified polyamidoamine dendrimers with carboxyl termini. RSC Adv. 2018, 8, 28147–28151. [Google Scholar] [CrossRef] [Green Version]
- Kojima, C.; Fu, Y.; Tamaki, M. Control of stimuli sensitivity in pH-switchable LCST/UCST-type thermosensitive dendrimers by changing the dendrimer structure. Polymers 2022, 14, 2426. [Google Scholar] [CrossRef] [PubMed]
- Hubera, V.; Camisaschi, C.; Berzi, A.; Ferro, S.; Lugini, L.; Triulzi, T.; Tuccitto, A.; Tagliabue, E.; Castelli, C.; Rivoltini, L. Cancer acidity: An ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin. Cancer Biol. 2017, 43, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Crippen, G.M. Atomic physicochemical parameters for three-dimensional-structure-directed quantitative structure-activity relationships. 2. Modeling dispersive and hydrophobic interactions. J. Chem. Inf. Comput. Sci. 1987, 27, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Shima, F.; Akagi, T.; Uto, T.; Akashi, M. Manipulating the antigen-specific immune response by the hydrophobicity of amphiphilic poly(γ-glutamic acid) nanoparticles. Biomaterials 2013, 34, 9709–9716. [Google Scholar] [CrossRef]
- Ke, X.; Howard, G.P.; Tang, H.; Cheng, B.; Saung, M.T.; Santos, J.L.; Mao, H.Q. Physical and chemical profiles of nanoparticles for lymphatic targeting. Adv. Drug Deliv. Rev. 2019, 151–152, 72–93. [Google Scholar] [CrossRef] [PubMed]
- Yuba, E. Design of pH-sensitive polymer-modified liposomes for antigen delivery and their application in cancer immunotherapy. Polym. J. 2016, 48, 761–771. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, L.V.; Rolf, J.; Emslie, E.; Shi, Y.B.; Taylor, P.M.; Cantrell, D.A. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013, 14, 500–508. [Google Scholar] [CrossRef] [Green Version]
- Kühne, A.; Tzvetkov, M.V.; Hagos, Y.; Lage, H.; Burckhardt, G.; Brockmöller, J. Influx and efflux transport as determinants of melphalan cytotoxicity: Resistance to melphalan in MDR1 overexpressing tumor cell lines. Biochem. Pharmacol. 2009, 78, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, C.; Toi, Y.; Harada, A.; Kono, K. Preparation of poly(ethylene glycol)-attached dendrimers encapsulating photosensitizers for application to photodynamic therapy. Bioconjugate Chem. 2007, 18, 663–670. [Google Scholar] [CrossRef]




| Dendrimer 1 | Bound Number | ||
|---|---|---|---|
| Phe (Terminal Density 3) | CHex | FITC | |
| PAMAM-CHex-Phe64 2 | 64 (100%) | 64 | 7.0 |
| PAMAM-CHex-Phe48 | 48 (75%) | 64 | 4.0 |
| PAMAM-CHex-Phe37 | 37 (58%) | 64 | 3.2 |
| PAMAM-CHex-Phe27 | 27 (42%) | 64 | 4.1 |
| PAMAM-Phe64-CHex 2 | 64 (100%) | 62 | 4.0 |
| PAMAM-Phe46-CHex 2 | 46 (72%) | ~64 | 4.7 |
| PAMAM-Phe37-CHex 2 | 37 (58%) | ~64 | 3.3 |
| PAMAM-Phe16-CHex 2 | 16 (25%) | ~64 | 2.5 |
| PAMAM-CHex | 0 (0%) | 64 | 4.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiba, H.; Hirose, T.; Fu, Y.; Michigami, M.; Fujii, I.; Nakase, I.; Matsumoto, A.; Kojima, C. T Cell-Association of Carboxy-Terminal Dendrimers with Different Bound Numbers of Phenylalanine and Their Application to Drug Delivery. Pharmaceutics 2023, 15, 888. https://doi.org/10.3390/pharmaceutics15030888
Shiba H, Hirose T, Fu Y, Michigami M, Fujii I, Nakase I, Matsumoto A, Kojima C. T Cell-Association of Carboxy-Terminal Dendrimers with Different Bound Numbers of Phenylalanine and Their Application to Drug Delivery. Pharmaceutics. 2023; 15(3):888. https://doi.org/10.3390/pharmaceutics15030888
Chicago/Turabian StyleShiba, Hiroya, Tomoka Hirose, Yunshen Fu, Masataka Michigami, Ikuo Fujii, Ikuhiko Nakase, Akikazu Matsumoto, and Chie Kojima. 2023. "T Cell-Association of Carboxy-Terminal Dendrimers with Different Bound Numbers of Phenylalanine and Their Application to Drug Delivery" Pharmaceutics 15, no. 3: 888. https://doi.org/10.3390/pharmaceutics15030888