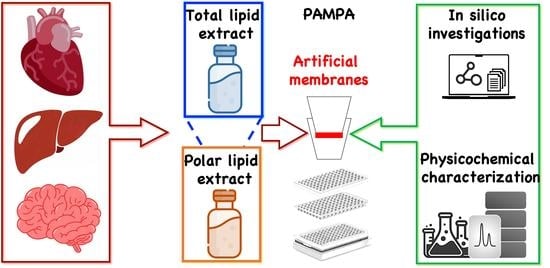
Natural Lipid Extracts as an Artificial Membrane for Drug Permeability Assay: In Vitro and In Silico Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Compound Set Selection and Cheminformatic Tools for Physicochemical and Structural Characterization and Structure–Property Interpretation
2.3. Preparation of Liposomes
2.4. Zeta Potential Measurement
2.5. PAMPA Measurements
2.6. HPLC Analysis
3. Results and Discussion
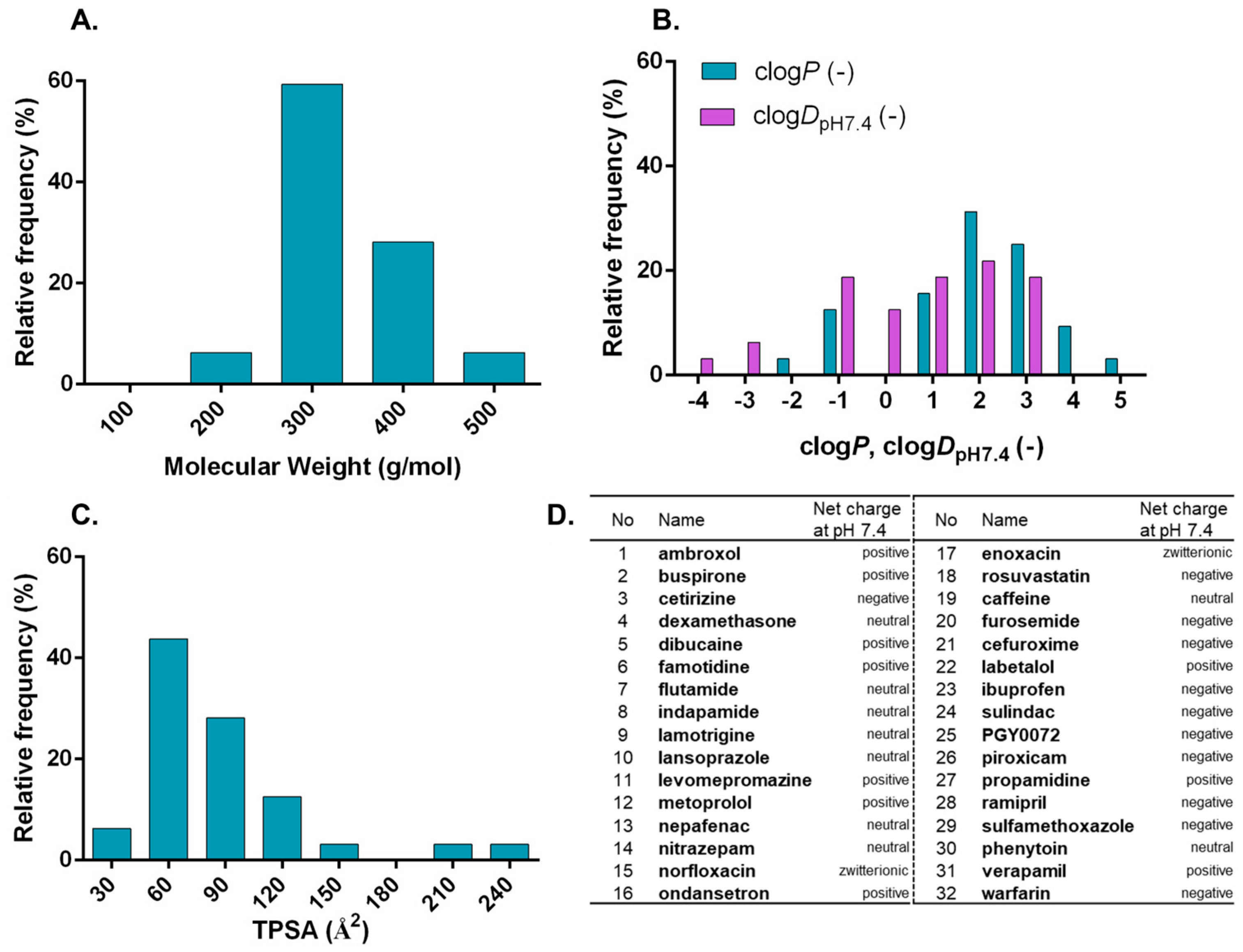
3.1. Characterization of Investigated Drug Compounds
3.2. Characterization of Natural Lipid Extracts
3.2.1. Chemical and Basic Physicochemical Characterization
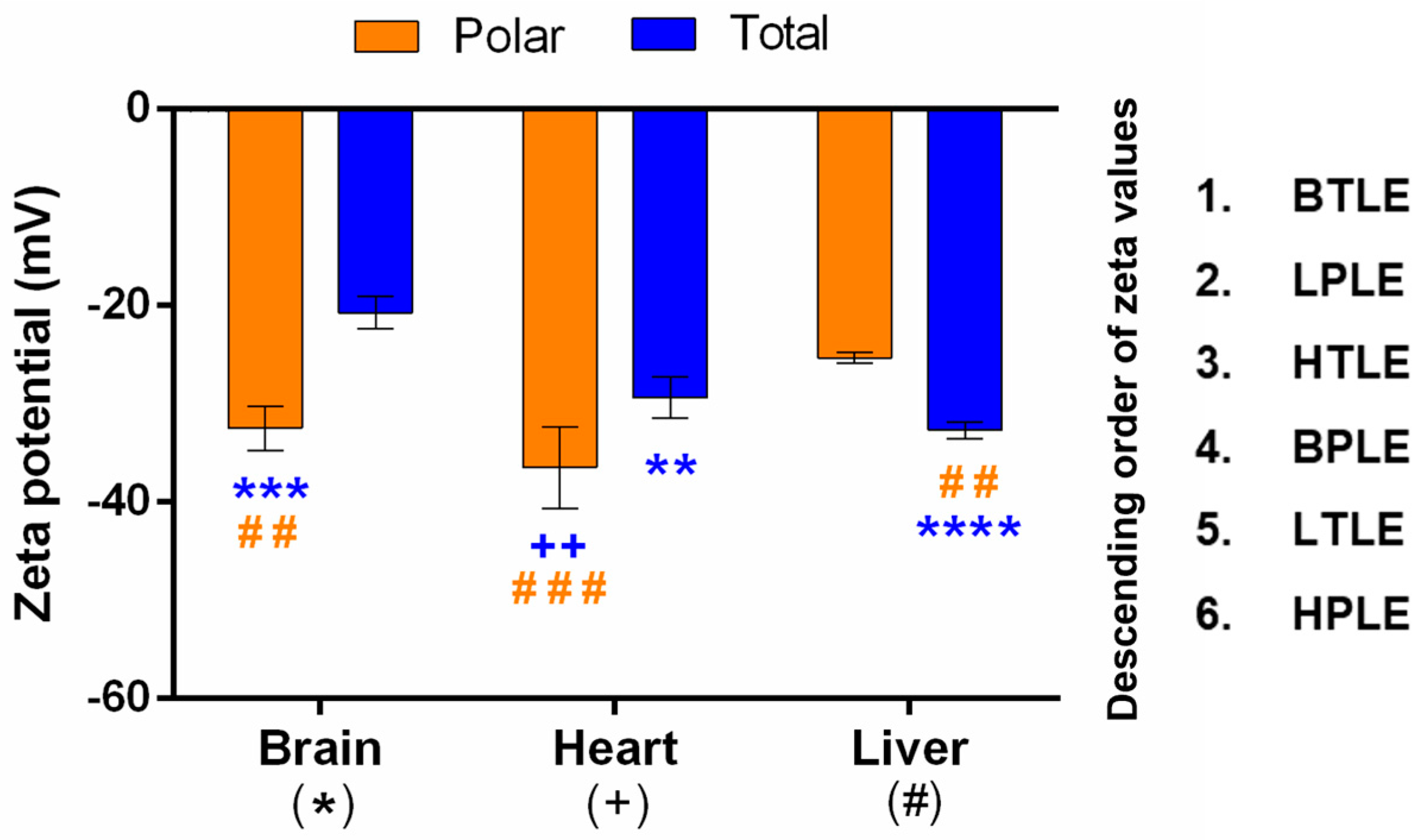
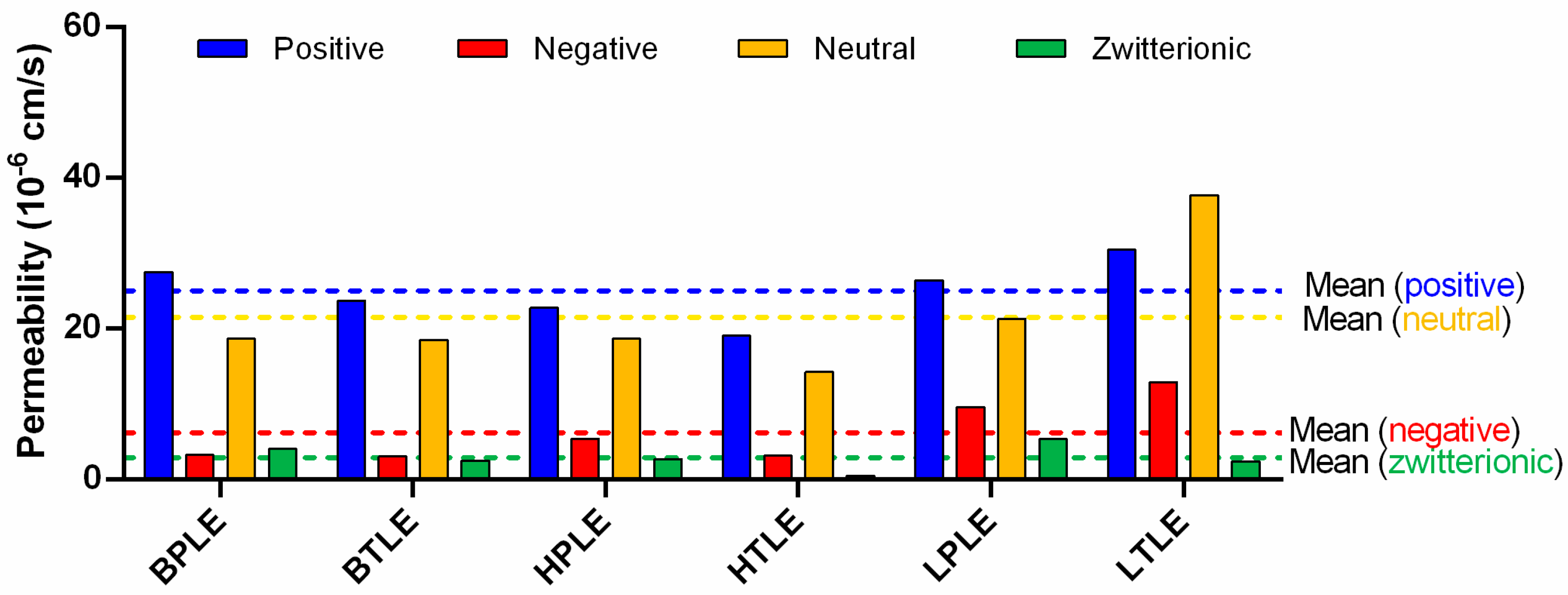
3.2.2. Zeta Potential
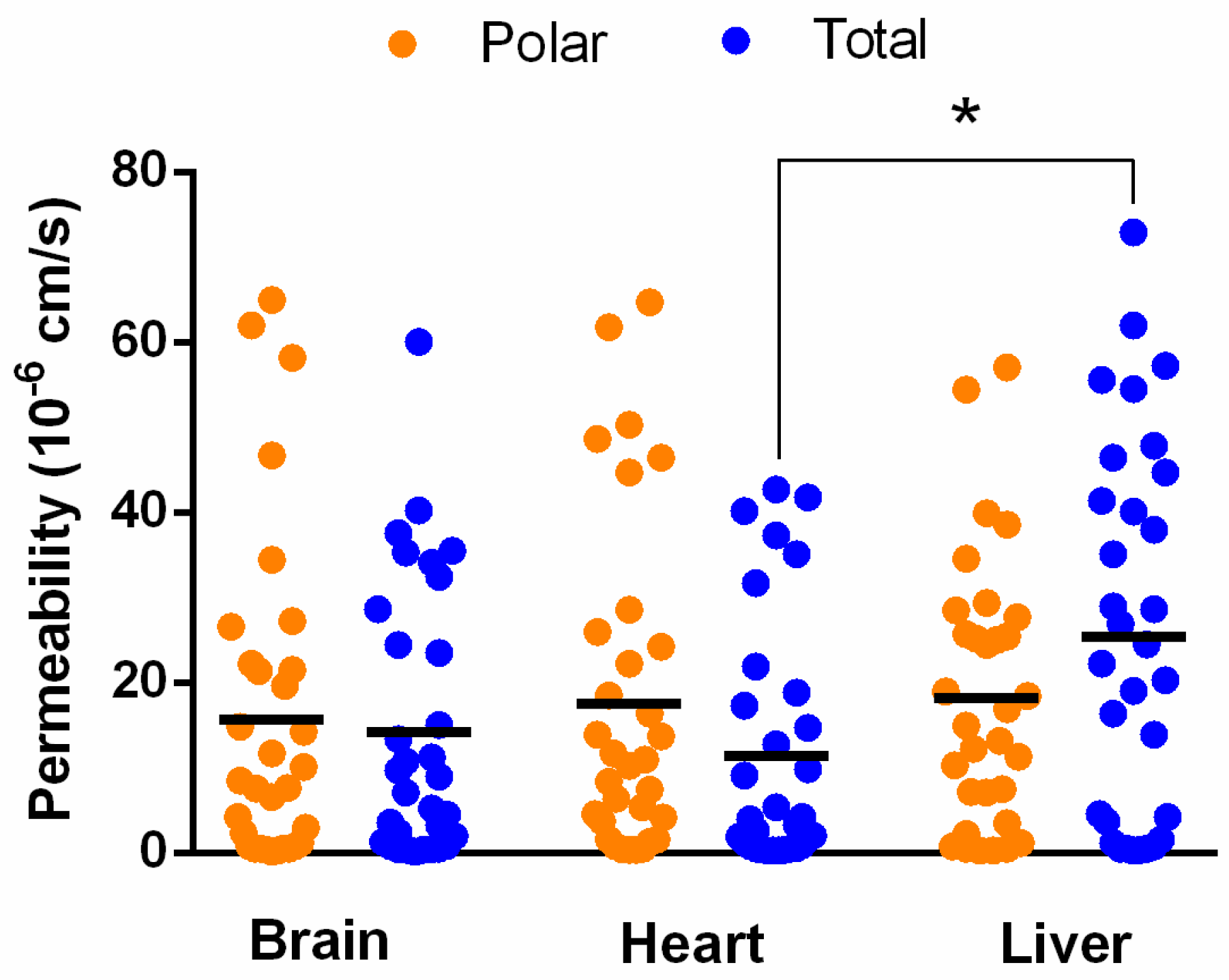
3.3. In Vitro Tissue-Specific Permeability
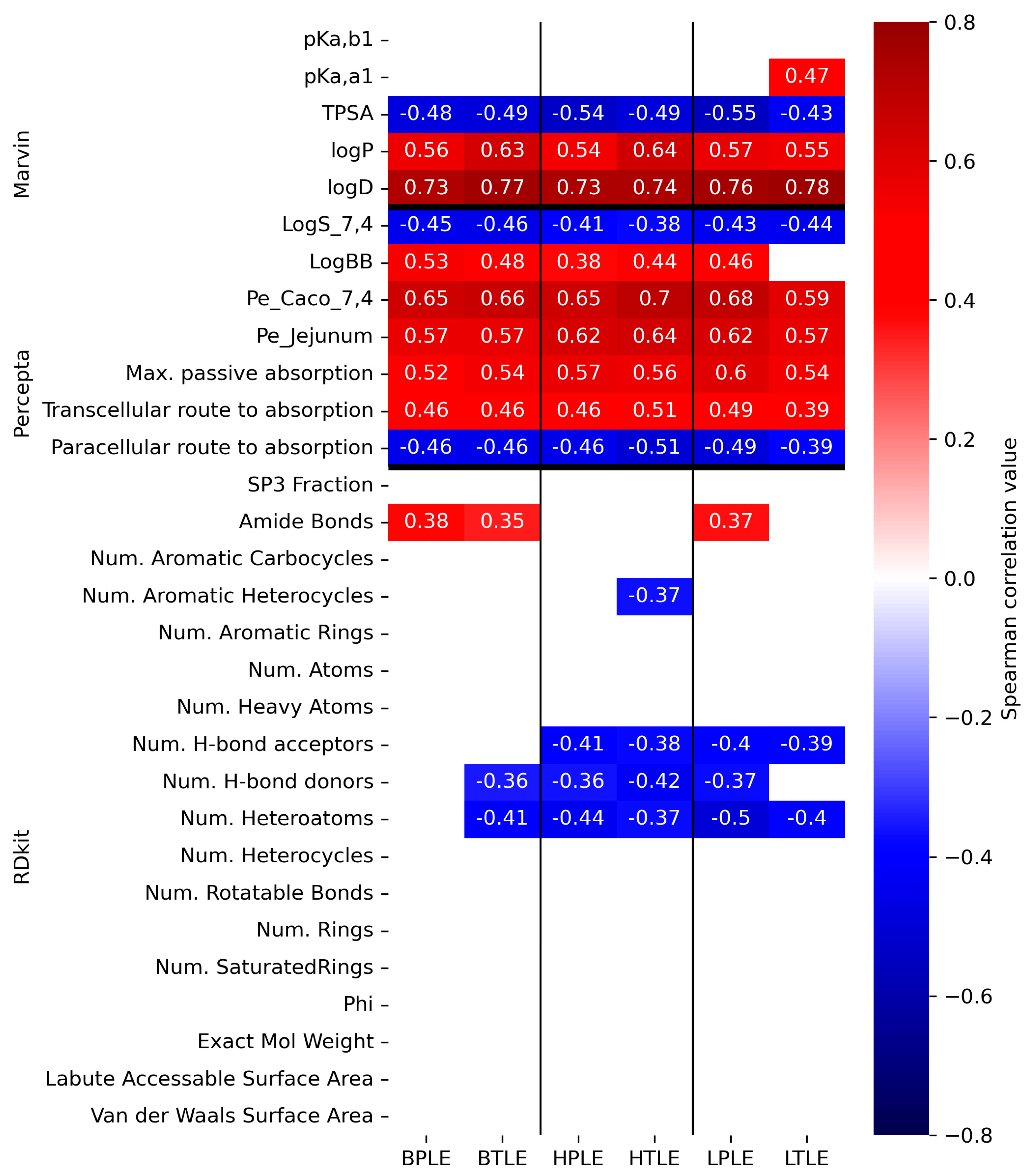
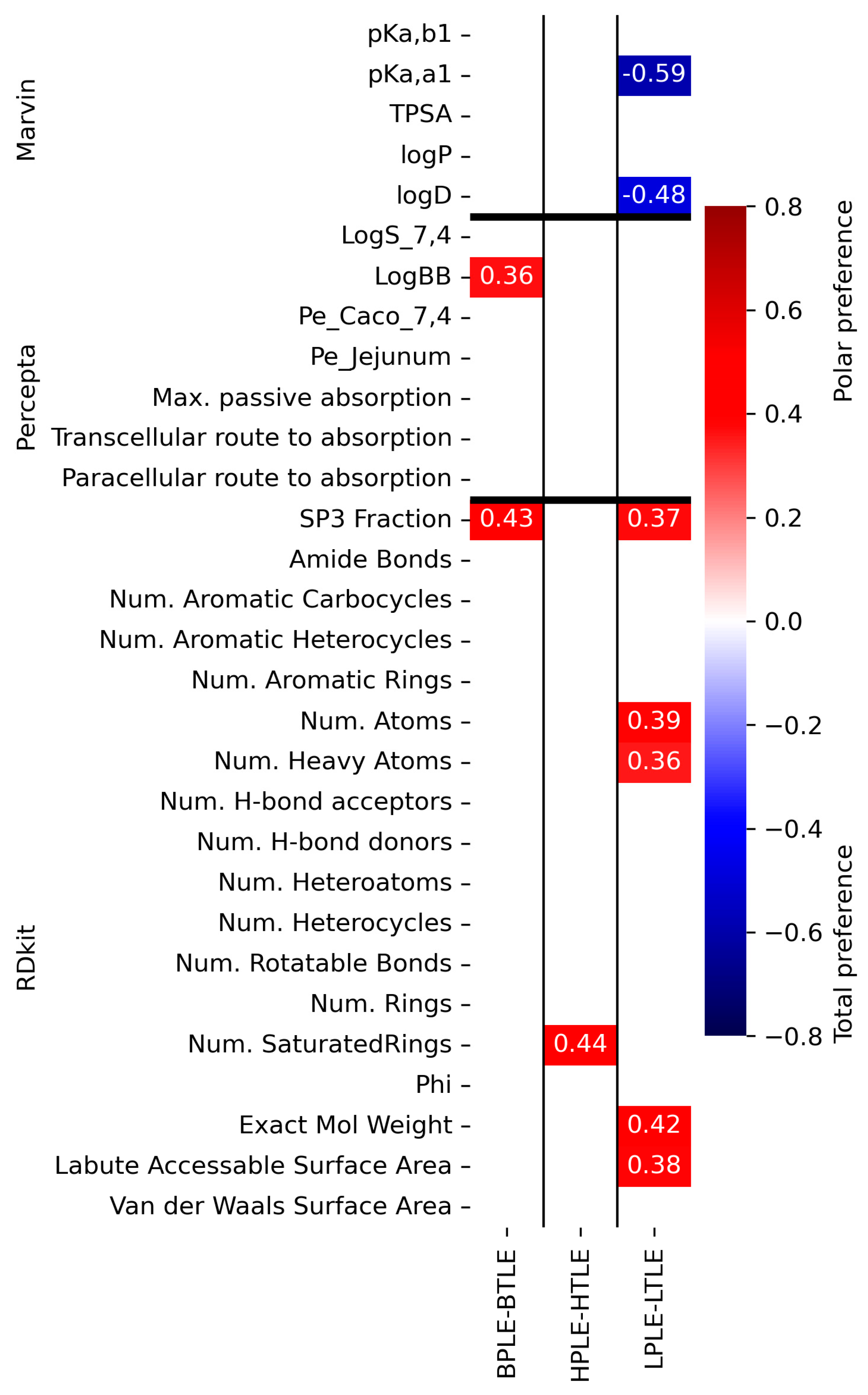
3.4. Cheminformatic Analysis of Tissue-Specific Permeability Data
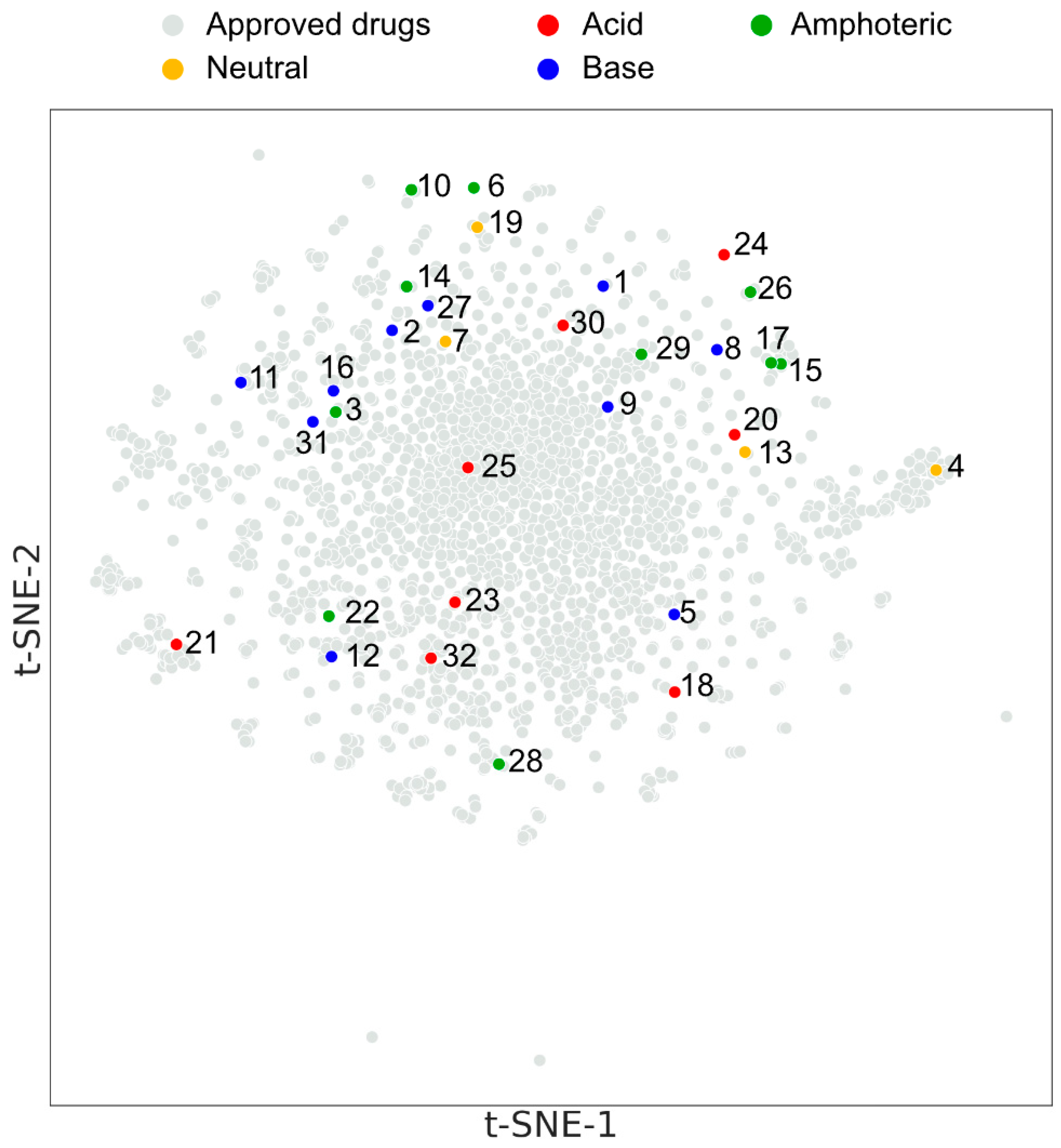
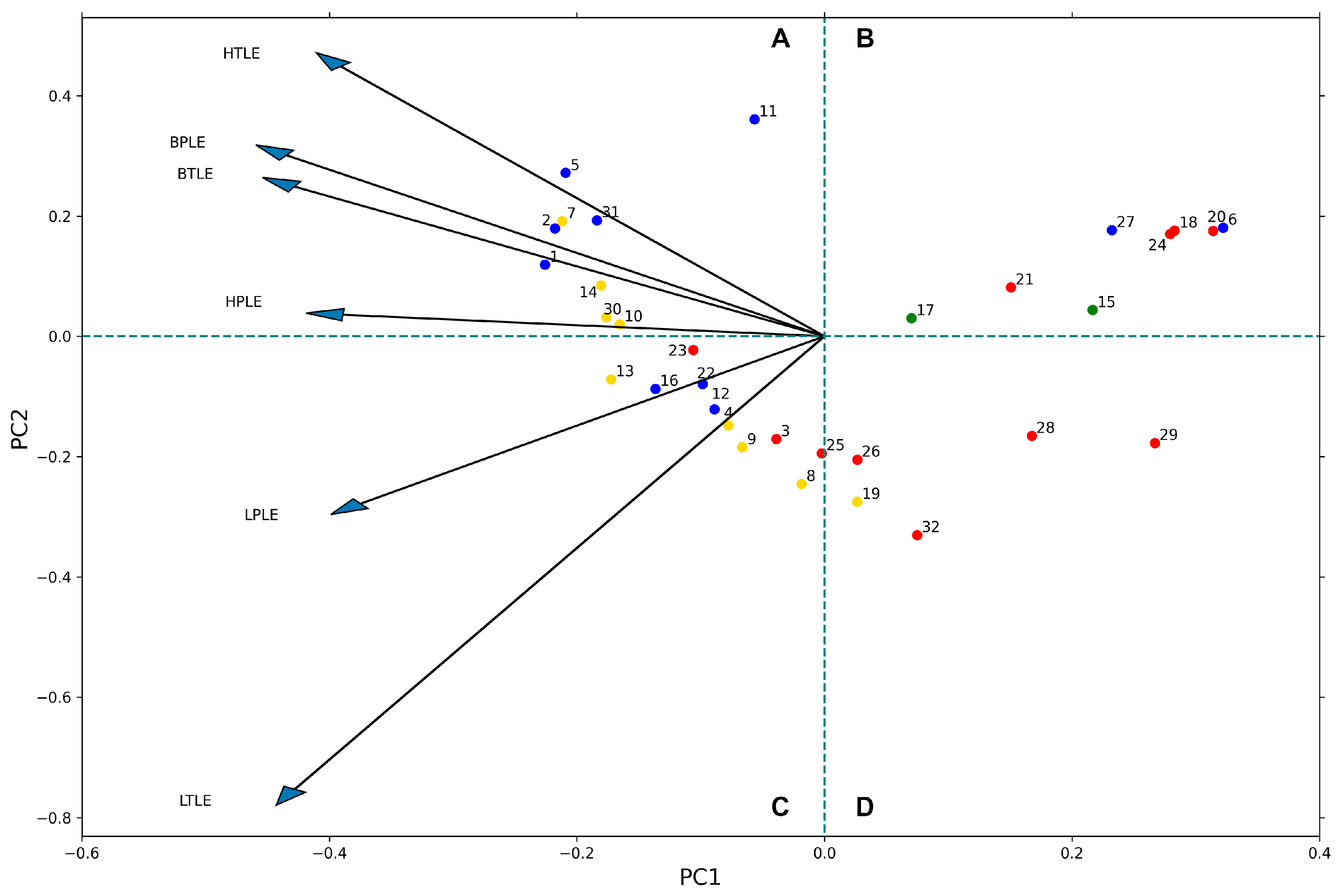
3.4.1. Principal Component Analysis
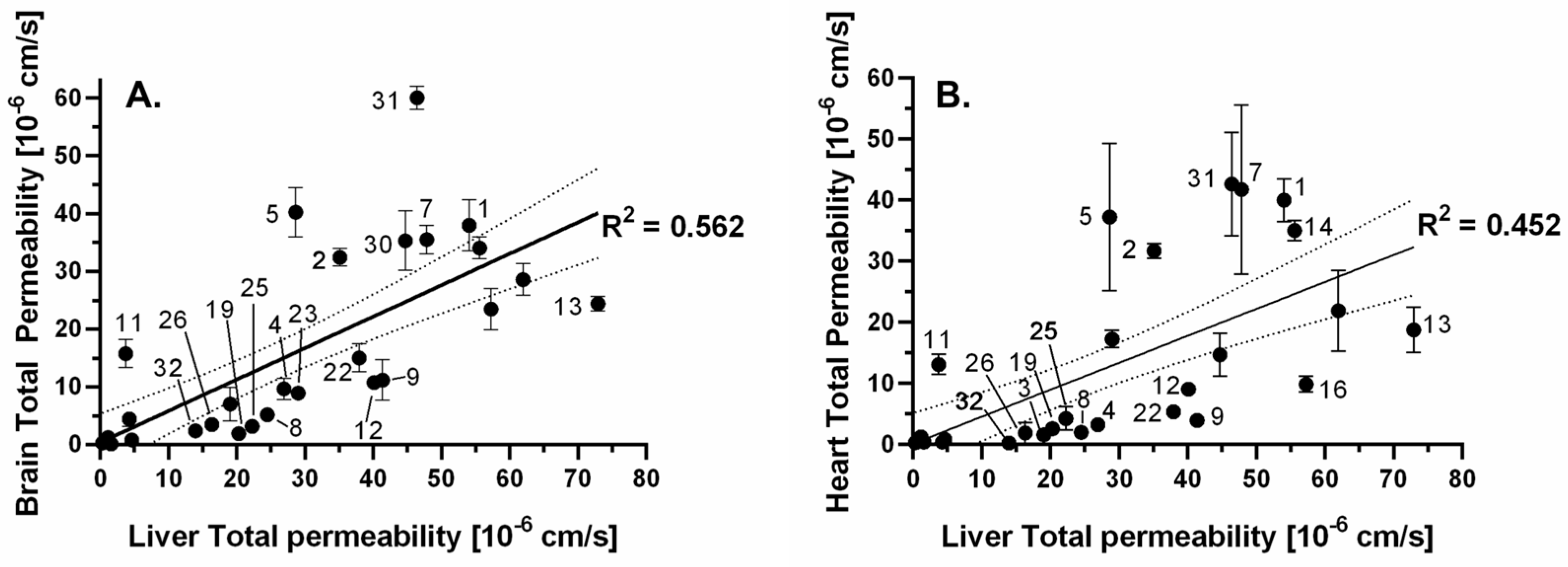
3.4.2. Correlation Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kansy, M.; Senner, F.; Gubernator, K. Physicochemical High Throughput Screening: Parallel Artificial Membrane Permeation Assay in the Description of Passive Absorption Processes. J. Med. Chem. 1998, 41, 1007–1010. [Google Scholar] [CrossRef]
- Wohnsland, F.; Faller, B. High-Throughput Permeability pH Profile and High-Throughput Alkane/Water log P with Artificial Membranes. J. Med. Chem. 2001, 44, 923–930. [Google Scholar] [CrossRef]
- Borbás, E.; Balogh, A.; Bocz, K.; Müller, J.; Kiserdei, É.; Vigh, T.; Sinkó, B.; Marosi, A.; Halász, A.; Dohányos, Z.; et al. In vitro dissolution-permeation evaluation of an electrospun cyclodextrin-based formulation of aripiprazole using μFlux™. Int. J. Pharm. 2015, 491, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Dargó, G.; Vincze, A.; Müller, J.; Kiss, H.J.; Nagy, Z.Z.; Balogh, G.T. Corneal-PAMPA: A novel, non-cell-based assay for prediction of corneal T drug permeability. Eur. J. Pharm. Sci. 2018, 128, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Esső, K.; Dargó, G.; Könczöl, Á.; Balogh, G.T. Tuning the predictive capacity of the PAMPA-BBB model. Eur. J. Pharm. Sci. 2015, 79, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Otasevic, S.; Vojinovic, T. The Lecithin and anionic lipids as an imitation of the lipid membrane in Parallel Artificial Membrane Permeation Assay (PAMPA) blood–brain barrier Models. Prog. Nutr. 2020, 22, e2020035. [Google Scholar] [CrossRef]
- Sugano, K.; Hamada, H.; Machida, M.; Ushio, H.; Saitoh, K.; Terada, K. Optimized conditions of bio-mimetic artificial membrane permeation assay. Int. J. Pharm. 2001, 228, 181–188. [Google Scholar] [CrossRef]
- Di, L.; Kerns, E.H.; Fan, K.; McConnell, O.J.; Carter, G.T. High throughput artificial membrane permeability assay for blood–brain barrier. Eur. J. Med. Chem. 2003, 38, 223–232. [Google Scholar] [CrossRef]
- Bicker, J.; Alves, G.; Fortuna, A.; Soares-Da-Silvad, P.; Falcão, A. A new PAMPA model using an in-house brain lipid extract for screening the blood–brain barrier permeability of drug candidates. Int. J. Pharm. 2016, 501, 102–111. [Google Scholar] [CrossRef]
- Balogh, G.T.; Müller, J.; Könczöl, Á. pH-gradient PAMPA-based in vitro model assay for drug-induced phospholipidosis in early stage of drug discovery. Eur. J. Pharm. Sci. 2013, 49, 81–89. [Google Scholar] [CrossRef]
- Sinkó, B.; Garrigues, T.M.; Balogh, G.T.; Nagy, Z.K.; Tsinman, O.; Avdeef, A.; Takács-Novák, K. Skin–PAMPA: A new method for fast prediction of skin penetration. Eur. J. Pharm. Sci. 2012, 45, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Carrara, S.; Reali, V.; Misiano, P.; Dondio, G.; Bigogno, C. Evaluation of in vitro brain penetration: Optimized PAMPA and MDCKII-MDR1 assay comparison. Int. J. Pharm. 2007, 345, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Lennox, R.B.; McClelland, R.A. Structure and electrochemical properties of microfiltration filter-lipid membrane systems. Anal. Chem. 1982, 54, 76–81. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Solvent Fractionation of Lipid Extracts. Available online: https://cyberlipid.gerli.com/techniques-of-analysis/fractionation-complex-extracts/solvent-fractionation-2/ (accessed on 24 January 2023).
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. Available online: https://lvdmaaten.github.io/publications/papers/JMLR_2008.pdf (accessed on 29 January 2023).
- Sorkun, M.C.; Mullaj, D.; Koelman, J.M.V.A.; Süleyman, S. ChemPlot, a Python library for chemical space visualization. Biol. Med. Chem. 2021. [Google Scholar] [CrossRef]
- Avdeef, A. Permeability—PAMPA, Absorption and Drug Development; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 319–498. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Tinworth, C.P.; Young, R.J. Facts, Patterns, and Principles in Drug Discovery: Appraising the Rule of 5 with Measured Physicochemical Data. J. Med. Chem. 2020, 63, 10091–10108. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Manallack, D.; Prankerd, R.J.; Nassta, G.C.; Ursu, O.; Oprea, T.; Chalmers, D. A Chemogenomic Analysis of Ionization Constants-Implications for Drug Discovery. Chem. Med. Chem. 2013, 8, 242–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://avantilipids.com/product-category/natural-lipids/extracts (accessed on 29 January 2023).
- Avdeef, A. Physicochemical profiling (solubility, permeability and charge state). Curr. Top. Med. Chem. 2001, 1, 277–351. [Google Scholar] [CrossRef] [PubMed]
- Sohlenius-Sternbeck, A.-K.; Terelius, Y. Evaluation of ADMET Predictor in Early Discovery Drug Metabolism and Pharmacokinetics Project Work. Drug Metab. Dispos. 2021, 50, 95–104. [Google Scholar] [CrossRef]
- Talevi, A.; Goodarzi, M.; Ortiz, E.V.; Duchowicz, P.R.; Bellera, C.L.; Pesce, G.; Castro, E.A.; Bruno-Blanch, L.E. Prediction of drug intestinal absorption by new linear and non-linear QSPR. Eur. J. Med. Chem. 2011, 46, 218–228. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Vallianatou, T.; Tsopelas, F.; Tsantili-Kakoulidou, A. Prediction Models for Brain Distribution of Drugs Based on Biomimetic Chromatographic Data. Molecules 2022, 27, 3668. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, T.; Naven, R. Utility of Physicochemical Properties for the Prediction of Toxicological Outcomes: Takeda Perspective. ACS Med. Chem. Lett. 2020, 11, 203–209. [Google Scholar] [CrossRef]
- Lanevskij, K.; Didziapetris, R.; Sazonovas, A. Physicochemical QSAR analysis of hERG inhibition revisited: Towards a quantitative potency prediction. J. Comput. Mol. Des. 2022, 36, 837–849. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Singh, P.K. Analyzing FDA-approved drugs for compliance of pharmacokinetic principles: Should there be a critical screening parameter in drug designing protocols? Expert Opin. Drug Metab. Toxicol. 2021, 17, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Austin, R.P.; Barton, P.; Mohmed, S.; Riley, R.J. The binding of drugs to hepatocytes and its relationship to physicochemical properties. Drug Metab. Dispos. 2005, 33, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Carmona, A.; Tiwari, A.K.; Trippier, P.C. Amide Bond Bioisosteres: Strategies, Synthesis, and Successes. J. Med. Chem. 2020, 63, 12290–12358. [Google Scholar] [CrossRef] [PubMed]
- Hosono, Y.; Morimoto, J.; Sando, S. A comprehensive study on the effect of backbone stereochemistry of a cyclic hexapeptide on membrane permeability and microsomal stability. Org. Biomol. Chem. 2021, 19, 10326–10331. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Naylor, M.R.; Townsend, C.E.; Okumura, C.; Okada, O.; Lokey, R.S. Conformation and Permeability: Cyclic Hexapeptide Diastereomers. J. Chem. Inf. Model. 2019, 59, 2952–2963. [Google Scholar] [CrossRef]
- Ritchie, T.J.; Macdonald, S.J.; Young, R.J.; Pickett, S.D. The impact of aromatic ring count on compound developability: Further insights by examining carbo- and hetero-aromatic and -aliphatic ring types. Drug Discov. Today 2011, 16, 164–171. [Google Scholar] [CrossRef]
- Birch, A.M.; Kenny, P.W.; Simpson, I.; Whittamore, P.R. Matched molecular pair analysis of activity and properties of glycogen phosphorylase inhibitors. Bioorganic Med. Chem. Lett. 2009, 19, 850–853. [Google Scholar] [CrossRef]
- Seiler, P. Interconversion of lipophilicities from hydrocarbon- water systems into octanol-water system. Eur. J. Med. Chem. 1974, 9, 473–479. [Google Scholar]
- Hitchcock, S.A. Structural Modifications that Alter the P-Glycoprotein Efflux Properties of Compounds. J. Med. Chem. 2012, 55, 4877–4895. [Google Scholar] [CrossRef]
- Hunter, C.A. Quantifying Intermolecular Interactions: Guidelines for the Molecular Recognition Toolbox. Angew. Chem. Int. Ed. 2004, 43, 5310–5324. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.H.; Grellier, P.L.; Prior, D.V.; Morris, J.J.; Taylor, P.J.; Doherty, R.M. Hydrogen-bonding. Part 11. A quantitative evaluation of the hydrogen-bond acidity of imides as solutes. J. Org. Chem. 1990, 55, 2227–2229. [Google Scholar] [CrossRef]
- Kenny, P.W. Hydrogen Bonding, Electrostatic Potential, and Molecular Design. J. Chem. Inf. Model. 2009, 49, 1234–1244. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.W. Hydrogen-Bond Donors in Drug Design. J. Med. Chem. 2022, 65, 14261–14275. [Google Scholar] [CrossRef] [PubMed]
- Labute, P. A widely applicable set of descriptors. J. Mol. Graph. Model. 2000, 18, 464–477. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincze, A.; Dékány, G.; Bicsak, R.; Formanek, A.; Moreau, Y.; Koplányi, G.; Takács, G.; Katona, G.; Balogh-Weiser, D.; Arany, Á.; et al. Natural Lipid Extracts as an Artificial Membrane for Drug Permeability Assay: In Vitro and In Silico Characterization. Pharmaceutics 2023, 15, 899. https://doi.org/10.3390/pharmaceutics15030899
Vincze A, Dékány G, Bicsak R, Formanek A, Moreau Y, Koplányi G, Takács G, Katona G, Balogh-Weiser D, Arany Á, et al. Natural Lipid Extracts as an Artificial Membrane for Drug Permeability Assay: In Vitro and In Silico Characterization. Pharmaceutics. 2023; 15(3):899. https://doi.org/10.3390/pharmaceutics15030899
Chicago/Turabian StyleVincze, Anna, Gergely Dékány, Richárd Bicsak, András Formanek, Yves Moreau, Gábor Koplányi, Gergely Takács, Gábor Katona, Diána Balogh-Weiser, Ádám Arany, and et al. 2023. "Natural Lipid Extracts as an Artificial Membrane for Drug Permeability Assay: In Vitro and In Silico Characterization" Pharmaceutics 15, no. 3: 899. https://doi.org/10.3390/pharmaceutics15030899
APA StyleVincze, A., Dékány, G., Bicsak, R., Formanek, A., Moreau, Y., Koplányi, G., Takács, G., Katona, G., Balogh-Weiser, D., Arany, Á., & Balogh, G. T. (2023). Natural Lipid Extracts as an Artificial Membrane for Drug Permeability Assay: In Vitro and In Silico Characterization. Pharmaceutics, 15(3), 899. https://doi.org/10.3390/pharmaceutics15030899