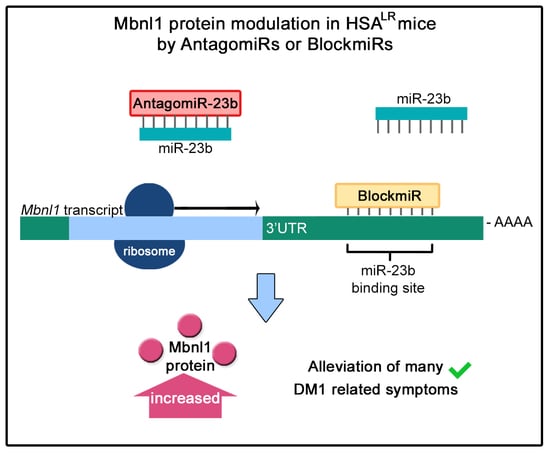
BlockmiR AONs as Site-Specific Therapeutic MBNL Modulation in Myotonic Dystrophy 2D and 3D Muscle Cells and HSALR Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mouse 3D Skeletal Muscle Tissues Biofabrication
2.3. BlockmiR Design
2.4. Cell Transfection
2.5. Toxicity Assay
2.6. MagnetoELISA
2.7. RNA Extraction, RT-qPCR, and RT-PCR
2.8. Immunofluorescence
2.9. Protein Extraction and Analysis
2.10. RNASeq Transcriptome Analysis
2.11. STRING Analysis
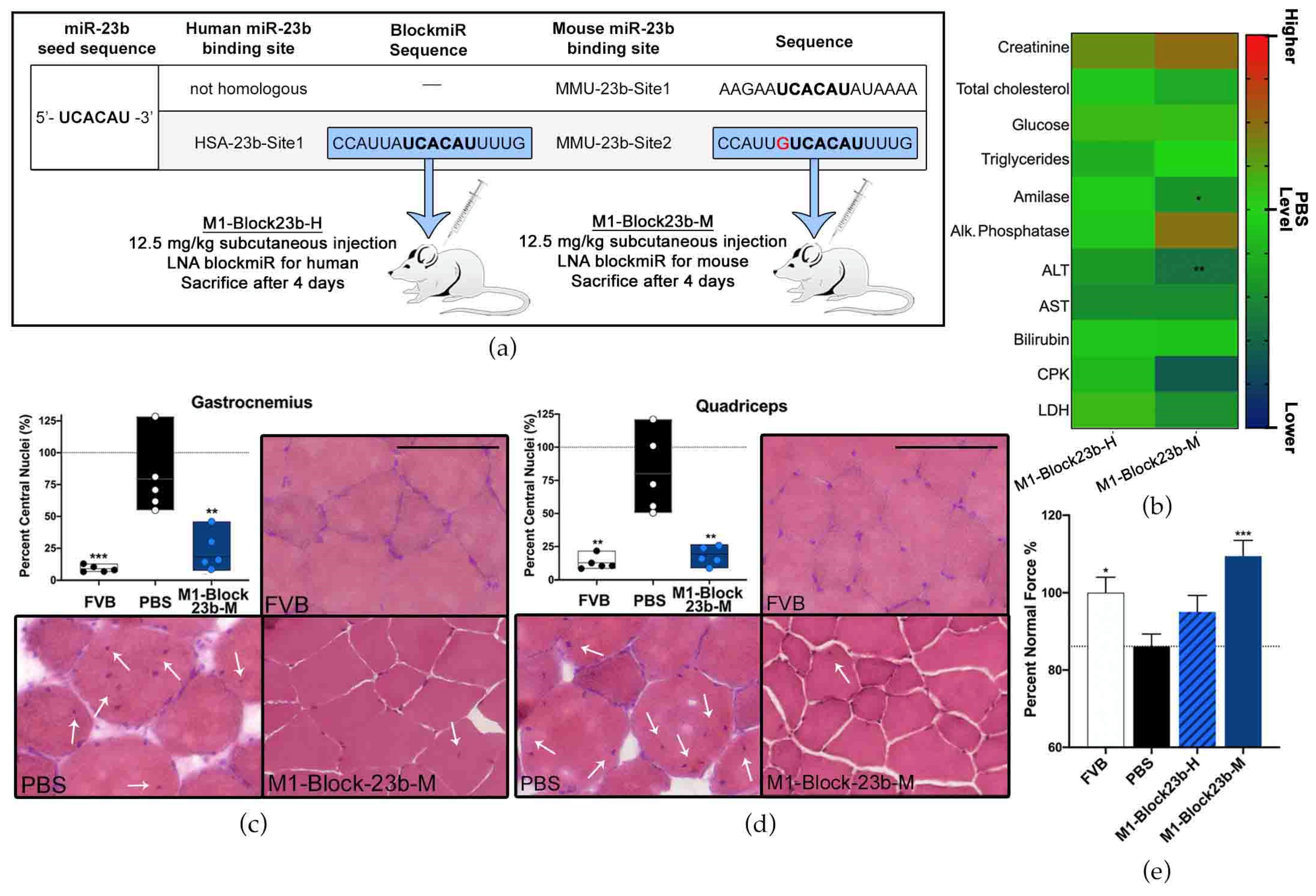
2.12. Transgenic Mice and BlockmiR Administration
2.13. Forelimb Grip Strength Test
2.14. Blood Biochemistry
2.15. Muscle Histology
2.16. Statistical Analysis
3. Results
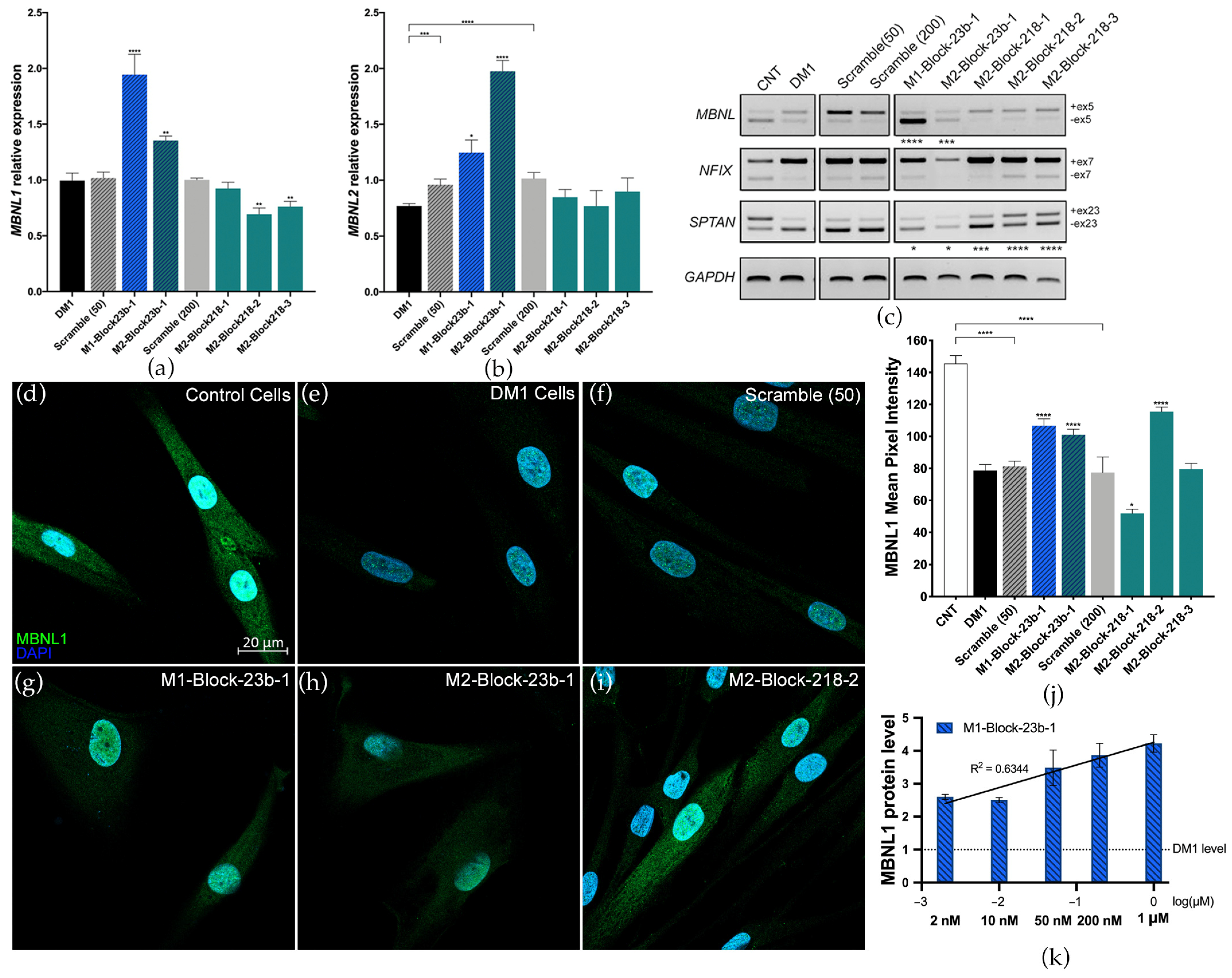
3.1. Preliminary Screen for blockmiRs in DM1 Cells
3.2. Blocking miRNA Binding-Sites Shows Therapeutic Effects at the Transcript and Protein Level
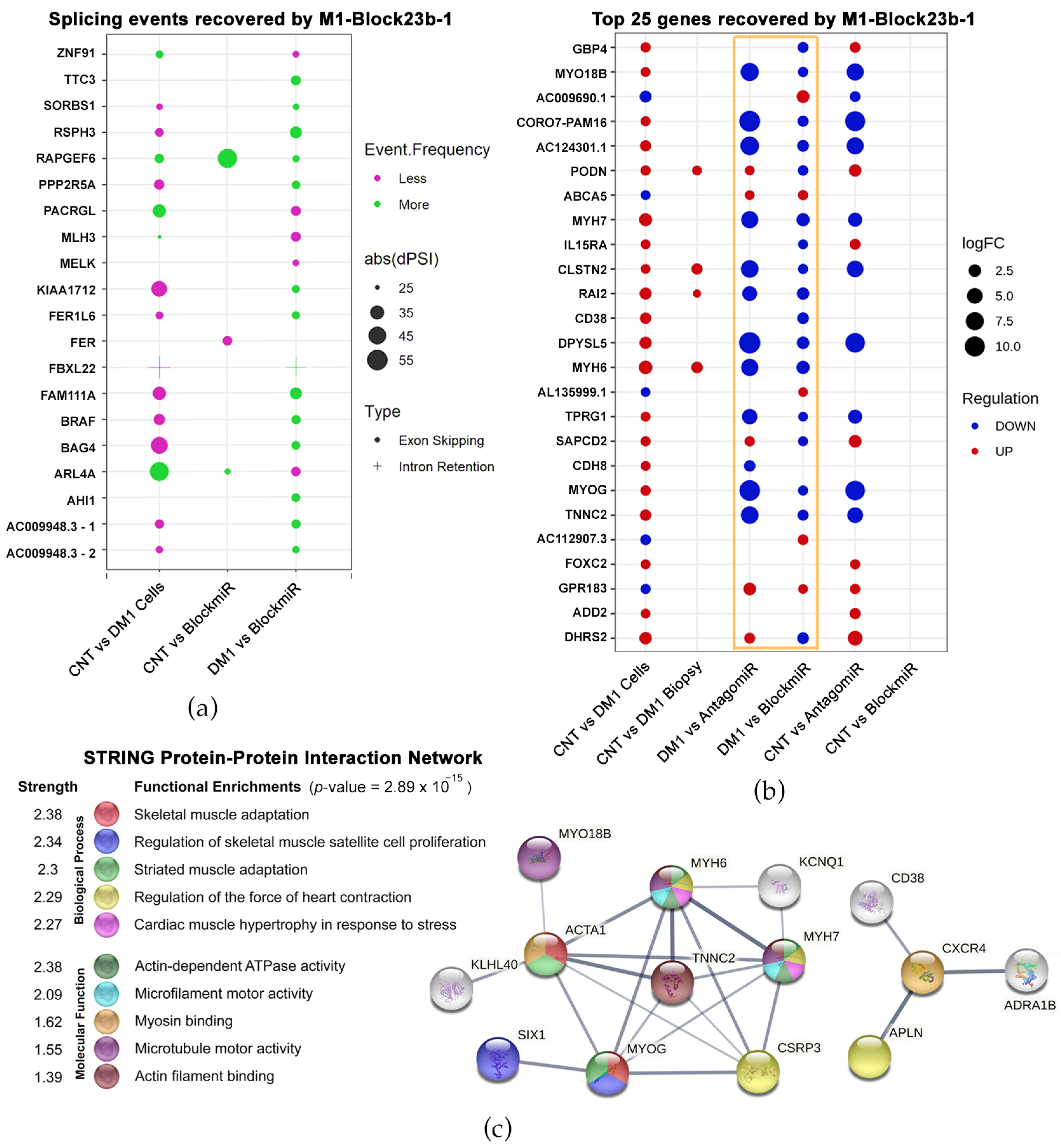
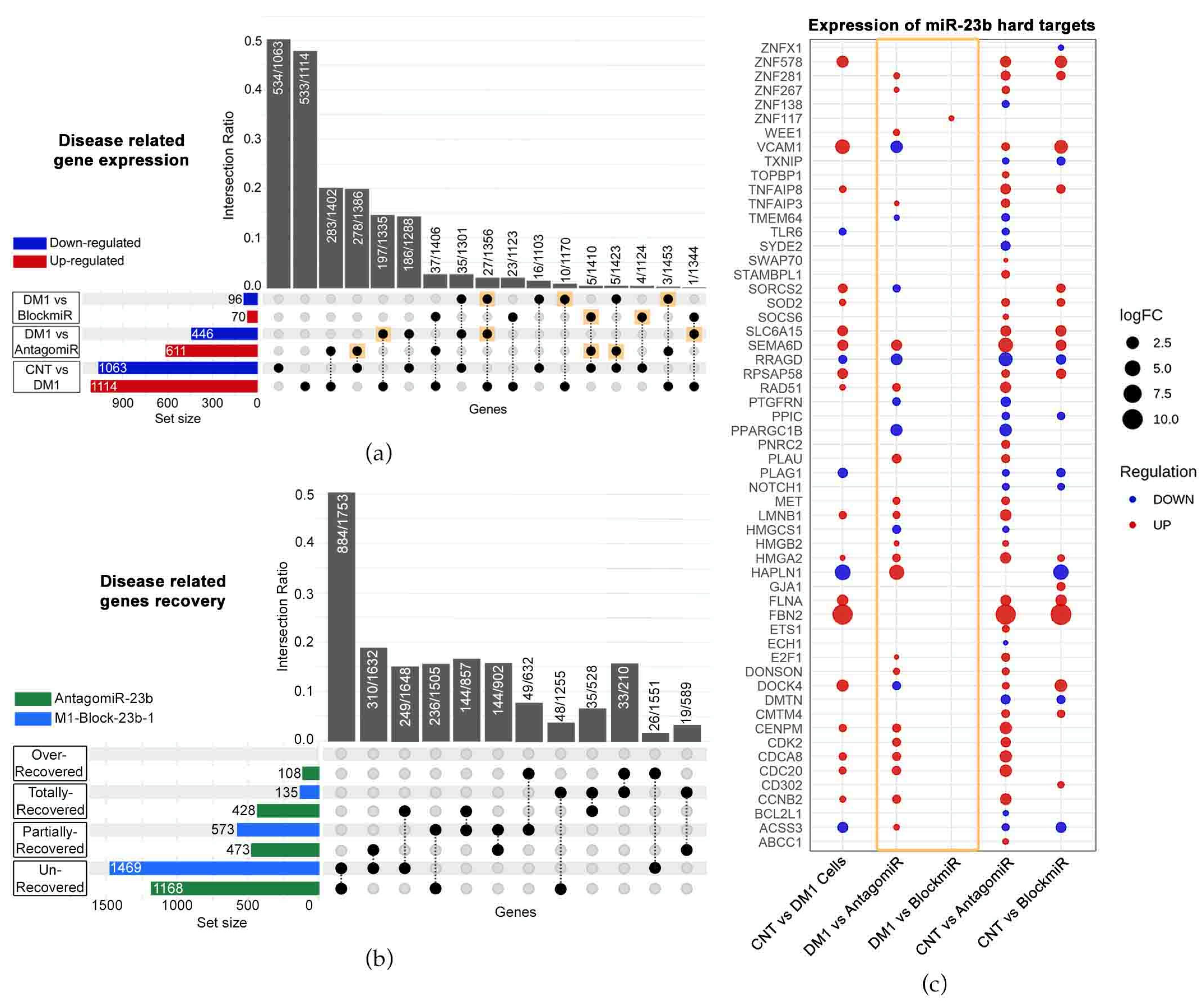
3.3. Transcriptomic Analysis Reveals Improvements by M1-Block-23b-1
3.4. M1-Block-23b-1 Has Highly Specific Effects
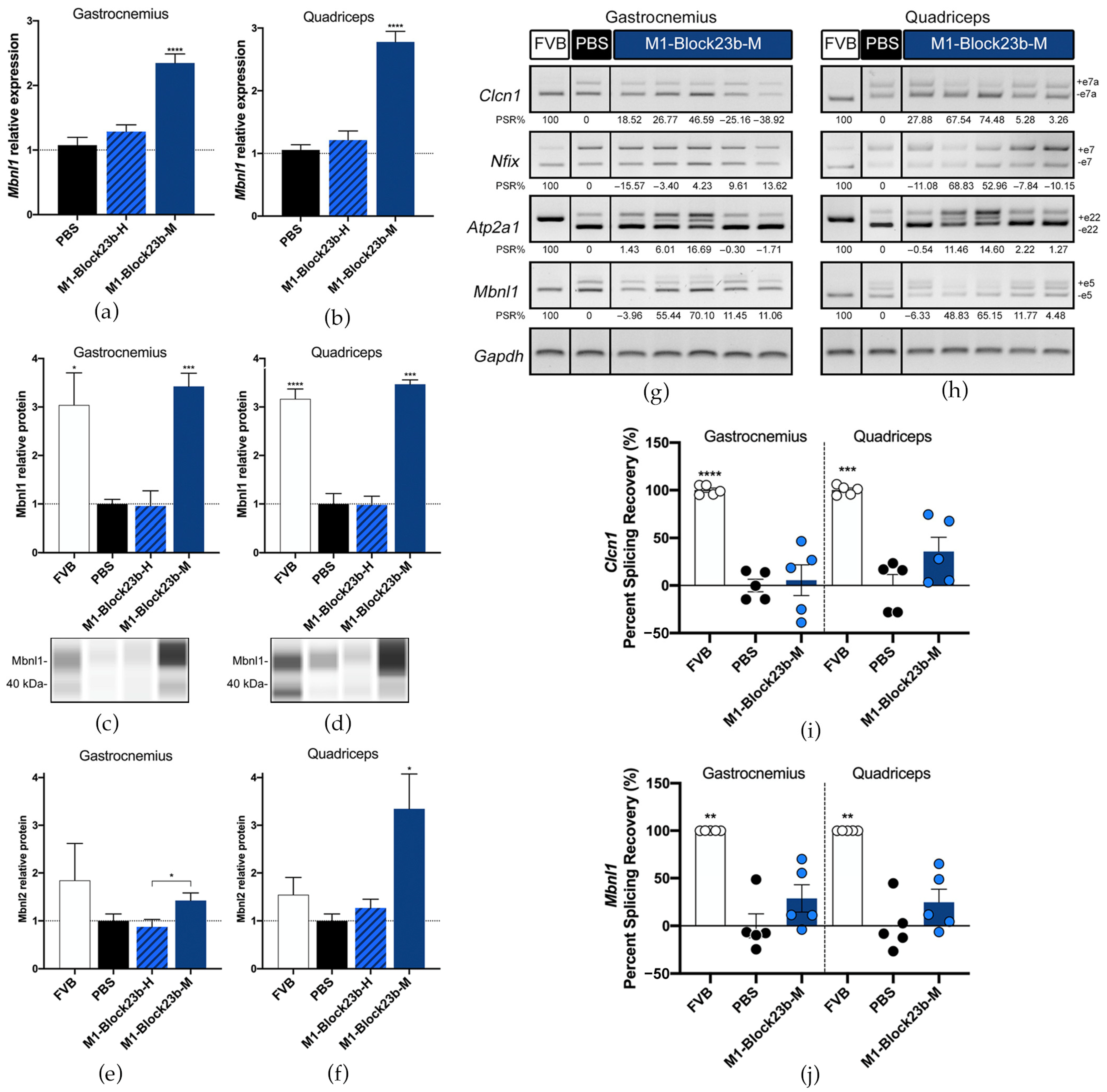
3.5. BlockmiR Administration In Vivo Increases Mbnl1 Protein and Rescues Symptoms
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, N.E.; Butterfield, R.J.; Mayne, K.; Newcomb, T.; Imburgia, C.; Dunn, D.; Duval, B.; Feldkamp, M.L.; Weiss, R.B. Population-based prevalence of myotonic dystrophy type 1 using genetic analysis of statewide blood screening program. Neurology 2021, 96, e1045–e1053. [Google Scholar] [CrossRef] [PubMed]
- Landfeldt, E.; Nikolenko, N.; Jimenez-Moreno, C.; Cumming, S.; Monckton, D.G.; Gorman, G.; Turner, C.; Lochmuller, H. Disease burden of myotonic dystrophy type 1. J. Neurol. 2019, 266, 998–1006. [Google Scholar] [CrossRef] [Green Version]
- Dhont, S.; Callens, R.; Stevens, D.; Bauters, F.; De Bleecker, J.L.; Derom, E.; Van Braeckel, E. Myotonic dystrophy type 1 as a major risk factor for severe COVID-19? Acta Neurol. Belg. 2020, 121, 1761–1765. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Gilabert, M.; Artero, R.; Lopez-Castel, A. The myotonic dystrophy type 1 drug development pipeline: 2022 edition. Drug Discov. Today 2023, 28, 103489. [Google Scholar] [CrossRef] [PubMed]
- Tsilfidis, C.; MacKenzie, A.E.; Mettler, G.; Barcelo, J.; Korneluk, R.G. Correlation between CTG trinucleotide repeat length and frequency of severe congenital myotonic dystrophy. Nat. Genet. 1992, 1, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.W.; Urbinati, C.R.; Teng-Umnuay, P.; Stenberg, M.G.; Byrne, B.J.; Thornton, C.A.; Swanson, M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000, 19, 4439–4448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Mankodi, A.; Swanson, M.S.; Moxley, R.T.; Thornton, C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004, 13, 3079–3088. [Google Scholar] [CrossRef] [Green Version]
- Batra, R.; Manchanda, M.; Swanson, M.S. Global insights into alternative polyadenylation regulation. RNA Biol. 2015, 12, 597–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanadia, R.N.; Urbinati, C.R.; Crusselle, V.J.; Luo, D.; Lee, Y.J.; Harrison, J.K.; Oh, S.P.; Swanson, M.S. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns 2003, 3, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, P.; Stepniak-Konieczna, E.; Sobczak, K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014, 42, 10873–10887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Miller, J.W.; Mankodi, A.; Kanadia, R.N.; Yuan, Y.; Moxley, R.T.; Swanson, M.S.; Thornton, C.A. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006, 15, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Ward, A.J.; Cherone, J.M.; Giudice, J.; Wang, T.T.; Treacy, D.J.; Lambert, N.J.; Freese, P.; Saxena, T.; Cooper, T.A.; et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015, 25, 858–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Costa, J.M.; Llamusi, M.B.; Garcia-Lopez, A.; Artero, R. Alternative splicing regulation by Muscleblind proteins: From development to disease. Biol. Rev. Camb. Philos. Soc. 2011, 86, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Ladd, A.N.; Charlet, N.; Cooper, T.A. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 2001, 21, 1285–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, T.M.; Lueck, J.D.; Swanson, M.S.; Dirksen, R.T.; Thornton, C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Investig. 2007, 117, 3952–3957. [Google Scholar] [CrossRef] [Green Version]
- Savkur, R.S.; Philips, A.V.; Cooper, T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001, 29, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Pang, P.D.; Alsina, K.M.; Cao, S.; Koushik, A.B.; Wehrens, X.H.T.; Cooper, T.A. CRISPR -mediated expression of the fetal Scn5a isoform in adult mice causes conduction defects and arrhythmias. J. Am. Heart Assoc. 2018, 7, e010393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual-Gilabert, M.; Lopez-Castel, A.; Artero, R. Myotonic dystrophy type 1 drug development: A pipeline toward the market. Drug Discov. Today 2021, 26, 1765–1772. [Google Scholar] [CrossRef]
- Mignon, L. IONIS-DMPKRx Clinical Program in Myotonic Dystrophy. Available online: https://www.myotonic.org/sites/default/files/MDF_Mignon_09Sep2017.pdf (accessed on 23 November 2022).
- Rinaldi, C.; Wood, M.J.A. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018, 14, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Wang, D.D.; Hu, W.B.; Zeng, W.Q.; Xu, X.; Li, Q.X.; Bi, F.F.; Yang, H.; Qiu, J. Calcitriol increases MBNL1 expression and alleviates myotonic dystrophy phenotypes in HSA(LR) mouse models. J. Transl. Med. 2022, 20, 588. [Google Scholar] [CrossRef] [PubMed]
- Cerro-Herreros, E.; Gonzalez-Martinez, I.; Moreno, N.; Espinosa-Espinosa, J.; Fernandez-Costa, J.M.; Colom-Rodrigo, A.; Overby, S.J.; Seoane-Miraz, D.; Poyatos-Garcia, J.; Vilchez, J.J.; et al. Preclinical characterization of antagomiR-218 as a potential treatment for Myotonic Dystrophy. Mol. Ther. Nucleic Acids 2021, 26, 174–191. [Google Scholar] [CrossRef] [PubMed]
- Cerro-Herreros, E.; Sabater-Arcis, M.; Fernandez-Costa, J.M.; Moreno, N.; Perez-Alonso, M.; Llamusi, B.; Artero, R. miR-23b and miR-218 silencing increase Muscleblind-like expression and alleviate myotonic dystrophy phenotypes in mammalian models. Nat. Commun. 2018, 9, 2482. [Google Scholar] [CrossRef] [PubMed]
- Cerro-Herreros, E.; Gonzalez-Martinez, I.; Moreno-Cervera, N.; Overby, S.; Perez-Alonso, M.; Llamusi, B.; Artero, R. Therapeutic potential of AntagomiR-23b for treating myotonic dystrophy. Mol. Ther. Nucleic Acids 2020, 21, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Arcis, M.; Bargiela, A.; Furling, D.; Artero, R. miR-7 restores phenotypes in myotonic dystrophy muscle cells by repressing hyperactivated autophagy. Mol. Ther. Nucleic Acids 2020, 19, 278–292. [Google Scholar] [CrossRef] [PubMed]
- Rau, F.; Freyermuth, F.; Fugier, C.; Villemin, J.P.; Fischer, M.C.; Jost, B.; Dembele, D.; Gourdon, G.; Nicole, A.; Duboc, D.; et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011, 18, 840–845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.W.; Cai, H.F.; Wei, X.F.; Sun, J.J.; Lan, X.Y.; Lei, C.Z.; Lin, F.P.; Qi, X.L.; Plath, M.; Chen, H. miR-30-5p regulates muscle differentiation and alternative splicing of muscle-related genes by targeting MBNL. Int. J. Mol. Sci. 2016, 17, 182. [Google Scholar] [CrossRef] [Green Version]
- Overby, S.J.; Cerro-Herreros, E.; Gonzalez-Martinez, I.; Varela, M.A.; Seoane-Miraz, D.; Jad, Y.; Raz, R.; Moller, T.; Perez-Alonso, M.; Wood, M.J.; et al. Proof of concept of peptide-linked blockmiR-induced MBNL functional rescue in myotonic dystrophy type 1 mouse model. Mol. Ther. Nucleic Acids 2022, 27, 1146–1155. [Google Scholar] [CrossRef]
- Overby, S.J. Proof of Concept of Therapeutic Gene Modulation of MBNL1/2 in Myotonic Dystrophy. Ph.D. Thesis, Universitat de València, Valencia, Spain, 2022. [Google Scholar]
- Fernandez-Garibay, X.; Ortega, M.A.; Cerro-Herreros, E.; Comelles, J.; Martinez, E.; Artero, R.; Fernandez-Costa, J.M.; Ramon-Azcon, J. Bioengineeredin vitro3D model of myotonic dystrophy type 1 human skeletal muscle. Biofabrication 2021, 13, 035035. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Muñoz, G.A.; Fernández-Costa, J.M.; Ortega, M.A.; Balaguer-Trias, J.; Martin-Lasierra, E.; Ramón-Azcón, J. Plasmonic nanocrystals on polycarbonate substrates for direct and label-free biodetection of Interleukin-6 in bioengineered 3D skeletal muscles. Nanophotonics 2021, 10, 4477–4488. [Google Scholar] [CrossRef]
- Mankodi, A.; Logigian, E.; Callahan, L.; McClain, C.; White, R.; Henderson, D.; Krym, M.; Thornton, C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 2000, 289, 1769–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arandel, L.; Polay Espinoza, M.; Matloka, M.; Bazinet, A.; De Dea Diniz, D.; Naouar, N.; Rau, F.; Jollet, A.; Edom-Vovard, F.; Mamchaoui, K.; et al. Immortalized human myotonic dystrophy muscle cell lines to assess therapeutic compounds. Dis. Model Mech. 2017, 10, 487–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Albors, A.; Castano, A.G.; Fernandez-Garibay, X.; Ortega, M.A.; Balaguer, J.; Ramon-Azcon, J. Microphysiological sensing platform for an in-situ detection of tissue-secreted cytokines. Biosens. Bioelectron. X 2019, 2, 100025. [Google Scholar] [CrossRef] [PubMed]
- Holt, I.; Mittal, S.; Furling, D.; Butler-Browne, G.S.; Brook, J.D.; Morris, G.E. Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes Cells 2007, 12, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Moreno, N.; Gonzalez-Martinez, I.; Artero, R.; Cerro-Herreros, E. Rapid determination of MBNL1 protein levels by quantitative dot blot for the evaluation of antisense oligonucleotides in myotonic dystrophy myoblasts. Methods Mol. Biol. 2022, 2434, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Tapial, J.; Ha, K.C.H.; Sterne-Weiler, T.; Gohr, A.; Braunschweig, U.; Hermoso-Pulido, A.; Quesnel-Vallieres, M.; Permanyer, J.; Sodaei, R.; Marquez, Y.; et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017, 27, 1759–1768. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Frost, R.A.; Nystrom, G.J.; Lang, C.H. Lipopolysaccharide regulates proinflammatory cytokine expression in mouse myoblasts and skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R698–R709. [Google Scholar] [CrossRef]
- Wagner, S.D.; Struck, A.J.; Gupta, R.; Farnsworth, D.R.; Mahady, A.E.; Eichinger, K.; Thornton, C.A.; Wang, E.T.; Berglund, J.A. Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 2016, 12, e1006316. [Google Scholar] [CrossRef] [Green Version]
- Charizanis, K.; Lee, K.Y.; Batra, R.; Goodwin, M.; Zhang, C.; Yuan, Y.; Shiue, L.; Cline, M.; Scotti, M.M.; Xia, G.; et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 2012, 75, 437–450. [Google Scholar] [CrossRef] [Green Version]
- Suenaga, K.; Lee, K.Y.; Nakamori, M.; Tatsumi, Y.; Takahashi, M.P.; Fujimura, H.; Jinnai, K.; Yoshikawa, H.; Du, H.; Ares, M., Jr.; et al. Muscleblind-like 1 knockout mice reveal novel splicing defects in the myotonic dystrophy brain. PLoS ONE 2012, 7, e33218. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.S.; Kuyumcu-Martinez, M.N.; Sarma, S.; Mathur, N.; Wehrens, X.H.; Cooper, T.A. PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. J. Clin. Invest. 2009, 119, 3797–3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flower, M.; Lomeikaite, V.; Ciosi, M.; Cumming, S.; Morales, F.; Lo, K.; Hensman Moss, D.; Jones, L.; Holmans, P.; Investigators, T.-H.; et al. MSH3 modifies somatic instability and disease severity in Huntington’s and myotonic dystrophy type 1. Brain 2019, 142, 1876–1886. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.M.; Dragileva, E.; Kirby, A.; Lloret, A.; Lopez, E.; St Claire, J.; Panigrahi, G.B.; Hou, C.; Holloway, K.; Gillis, T.; et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: Genome-wide and candidate approaches. PLoS Genet. 2013, 9, e1003930. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.L.; Gable, D.L.; Samadzadeh-Tarighat, S.; Cheng, L.; Yu, L.; Gillespie, J.L.; Toland, A.E. Differential expression of miR-1, a putative tumor suppressing microRNA, in cancer resistant and cancer susceptible mice. PeerJ 2013, 1, e68. [Google Scholar] [CrossRef] [Green Version]
- Koscianska, E.; Witkos, T.M.; Kozlowska, E.; Wojciechowska, M.; Krzyzosiak, W.J. Cooperation meets competition in microRNA-mediated DMPK transcript regulation. Nucleic Acids Res. 2015, 43, 9500–9518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez Castel, A.; Overby, S.J.; Artero, R. MicroRNA-based therapeutic perspectives in myotonic dystrophy. Int. J. Mol. Sci. 2019, 20, 5600. [Google Scholar] [CrossRef]
- Wang, E.T.; Treacy, D.; Eichinger, K.; Struck, A.; Estabrook, J.; Olafson, H.; Wang, T.T.; Bhatt, K.; Westbrook, T.; Sedehizadeh, S.; et al. Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Hum. Mol. Genet. 2019, 28, 1312–1321. [Google Scholar] [CrossRef]
- Jang, Y.N.; Baik, E.J. JAK-STAT pathway and myogenic differentiation. Jak-stat 2013, 2, e23282. [Google Scholar] [CrossRef] [Green Version]
- Vihola, A.; Bachinski, L.L.; Sirito, M.; Olufemi, S.E.; Hajibashi, S.; Baggerly, K.A.; Raheem, O.; Haapasalo, H.; Suominen, T.; Holmlund-Hampf, J.; et al. Differences in aberrant expression and splicing of sarcomeric proteins in the myotonic dystrophies DM1 and DM2. Acta Neuropathol. 2010, 119, 465–479. [Google Scholar] [CrossRef] [Green Version]
- Champy, M.F.; Selloum, M.; Zeitler, V.; Caradec, C.; Jung, B.; Rousseau, S.; Pouilly, L.; Sorg, T.; Auwerx, J. Genetic background determines metabolic phenotypes in the mouse. Mamm. Genome 2008, 19, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Schneck, K.; Washington, M.; Holder, D.; Lodge, K.; Motzel, S. Hematologic and serum biochemical reference values in nontransgenic FVB mice. Comp Med 2000, 50, 32–35. [Google Scholar] [PubMed]
- Wang, P.Y.; Chang, K.T.; Lin, Y.M.; Kuo, T.Y.; Wang, G.S. Ubiquitination of MBNL1 is required for its cytoplasmic localization and function in promoting neurite outgrowth. Cell Rep. 2018, 22, 2294–2306. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.; Gourrier, N.; Lemercier-Neuillet, C.; Dhaenens, C.-M.; Vautrin, A.; Fernandez-Gomez, F.J.; Arandel, L.; Carpentier, C.; Obriot, H.; Eddarkaoui, S.; et al. Analysis of exonic regions involved in nuclear localization, splicing activity, and dimerization of Muscleblind-like-1 isoforms. J. Biol. Chem. 2011, 286, 16435–16446. [Google Scholar] [CrossRef] [Green Version]
- Konieczny, P.; Stepniak-Konieczna, E.; Sobczak, K. MBNL expression in autoregulatory feedback loops. RNA Biol. 2017, 15, 826–828. [Google Scholar] [CrossRef] [Green Version]
- Gomes-Pereira, M.; Foiry, L.; Nicole, A.; Huguet, A.; Junien, C.; Munnich, A.; Gourdon, G. CTG trinucleotide repeat “big jumps”: Large expansions, small mice. PLoS Genet. 2007, 3, e52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seznec, H.; Agbulut, O.; Sergeant, N.; Savouret, C.; Ghestem, A.; Tabti, N.; Willer, J.C.; Ourth, L.; Duros, C.; Brisson, E.; et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet. 2001, 10, 2717–2726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabaniss, C.D. Creatine kinase. In Clinical Methods: The History, Physical, and Laboratory Examinations; Walker, H.K., Hall, W.D., Hurst, J.W., Eds.; Butterworths: Boston, MA, USA, 1990. [Google Scholar]
- Gowda, S.; Desai, P.B.; Hull, V.V.; Math, A.A.; Vernekar, S.N.; Kulkarni, S.S. A review on laboratory liver function tests. Pan Afr. Med. J. 2009, 3, 17. [Google Scholar]
- Senior, J.R. Alanine aminotransferase: A clinical and regulatory tool for detecting liver injury-past, present, and future. Clin. Pharmacol. Ther. 2012, 92, 332–339. [Google Scholar] [CrossRef]
- Tabas, I. Cholesterol in health and disease. J. Clin. Investig. 2002, 110, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Botros, M.; Sikaris, K.A. The de ritis ratio: The test of time. Clin. Biochem. Rev. 2013, 34, 117–130. [Google Scholar] [PubMed]
- Heatwole, C.; Johnson, N.; Goldberg, B.; Martens, W.; Moxley, R., 3rd. Laboratory abnormalities in patients with myotonic dystrophy type 2. Arch. Neurol. 2011, 68, 1180–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burdick, A.D.; Sciabola, S.; Mantena, S.R.; Hollingshead, B.D.; Stanton, R.; Warneke, J.A.; Zeng, M.; Martsen, E.; Medvedev, A.; Makarov, S.S.; et al. Sequence motifs associated with hepatotoxicity of locked nucleic acid—Modified antisense oligonucleotides. Nucleic Acids Res. 2014, 42, 4882–4891. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Target | Modified Sequence | Concentration |
|---|---|---|
| M1-Block-23b-1 | CbsCbsAbsTbsTbsAbsuscsascsasusususTbsGb | 50 nM |
| M2-Block-23b-1 | AbsuscsascsasusgsasTbsTbsCbsAbsAbsCbsGb | 50 nM |
| M1-Block-218-1 | GbsAbsusgsusgscsusususAbsAbsAbsTbsAbsTb | 200 nM |
| M1-Block-218-2 | GbsususgsusgscsusgsTbsCbsTbsAbsTbsTbsGb | 200 nM |
| M2-Block-218-1 | AbsCbsTbsusgsusgscsususGbsAbsAbsusTbsTb | 200 nM |
| M2-Block-218-2 | GbsTbsTbsGbsusgsusgscsusasasTbsAbsAbsTb | 200 nM |
| M2-Block-218-3 | CbsGbsAbsTbsAbsgsusgscsususAbsAbsAbsAb | 200 nM |
| Scramble | TbsGbscsascscsusususgsTbsTbsAbsTbsTbsTb | 50 nM & 200 nM |
| M1-Block23b-H | CbsCbsAbsTbsTbsAbsuscsascsasusususTbsGb | 12.5 mg/kg |
| M1-Block23b-M | CbsCbsAbsTbsTbsGbsuscsascsasusususTbsGb | 12.5 mg/kg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Overby, S.J.; Cerro-Herreros, E.; Espinosa-Espinosa, J.; González-Martínez, I.; Moreno, N.; Fernández-Costa, J.M.; Balaguer-Trias, J.; Ramón-Azcón, J.; Pérez-Alonso, M.; Møller, T.; et al. BlockmiR AONs as Site-Specific Therapeutic MBNL Modulation in Myotonic Dystrophy 2D and 3D Muscle Cells and HSALR Mice. Pharmaceutics 2023, 15, 1118. https://doi.org/10.3390/pharmaceutics15041118
Overby SJ, Cerro-Herreros E, Espinosa-Espinosa J, González-Martínez I, Moreno N, Fernández-Costa JM, Balaguer-Trias J, Ramón-Azcón J, Pérez-Alonso M, Møller T, et al. BlockmiR AONs as Site-Specific Therapeutic MBNL Modulation in Myotonic Dystrophy 2D and 3D Muscle Cells and HSALR Mice. Pharmaceutics. 2023; 15(4):1118. https://doi.org/10.3390/pharmaceutics15041118
Chicago/Turabian StyleOverby, Sarah J., Estefanía Cerro-Herreros, Jorge Espinosa-Espinosa, Irene González-Martínez, Nerea Moreno, Juan M. Fernández-Costa, Jordina Balaguer-Trias, Javier Ramón-Azcón, Manuel Pérez-Alonso, Thorleif Møller, and et al. 2023. "BlockmiR AONs as Site-Specific Therapeutic MBNL Modulation in Myotonic Dystrophy 2D and 3D Muscle Cells and HSALR Mice" Pharmaceutics 15, no. 4: 1118. https://doi.org/10.3390/pharmaceutics15041118
APA StyleOverby, S. J., Cerro-Herreros, E., Espinosa-Espinosa, J., González-Martínez, I., Moreno, N., Fernández-Costa, J. M., Balaguer-Trias, J., Ramón-Azcón, J., Pérez-Alonso, M., Møller, T., Llamusí, B., & Artero, R. (2023). BlockmiR AONs as Site-Specific Therapeutic MBNL Modulation in Myotonic Dystrophy 2D and 3D Muscle Cells and HSALR Mice. Pharmaceutics, 15(4), 1118. https://doi.org/10.3390/pharmaceutics15041118