Ensemble-Learning and Feature Selection Techniques for Enhanced Antisense Oligonucleotide Efficacy Prediction in Exon Skipping
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Description
2.2. Feature Description
2.3. Problem Formulation and Model Input
2.4. Machine-Learning Libraries and Regressors
2.5. Model Assessment and Selection
2.6. Feature Importance Analysis
2.7. Model Comparison and Generalizability
3. Results
4. Discussions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crooke, S.T.; Liang, X.-H.; Baker, B.F.; Crooke, R.M. Antisense technology: A review. J. Biol. Chem. 2021, 296, 100416. [Google Scholar] [CrossRef]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 285–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.H.; Lim, S.; Wong, W.F. Antisense oligonucleotides: From design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 533–540. [Google Scholar] [CrossRef]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip. Rev. RNA 2020, 11, e1594. [Google Scholar] [CrossRef]
- Rinaldi, C.; Wood, M.J. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018, 14, 9–21. [Google Scholar] [CrossRef]
- Inoue, H.; Hayase, Y.; Iwai, S.; Ohtsuka, E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987, 215, 327–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundin, K.E.; Gissberg, O.; Smith, C.E. Oligonucleotide therapies: The past and the present. Hum. Gene Ther. 2015, 26, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Walder, J.A.; Walder, R.Y. Nucleic Acid Hybridization and Amplification Method for Detection of Specific Sequences in Which a Complementary Labeled Nucleic Acid Probe is Cleaved. U.S. Patent 5,403,711, 4 April 1995. [Google Scholar]
- Lim, S.R.; Hertel, K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 2001, 276, 45476–45483. [Google Scholar] [CrossRef] [Green Version]
- Gilden, D. The changing treatment options for CMV retinitis. GMHC Treat Issues 1995, 9, 1–8. [Google Scholar]
- Aartsma-Rus, A. FDA Approval of Nusinersen for Spinal Muscular Atrophy Makes 2016 the Year of Splice Modulating Oligonucleotides. Nucleic Acid Ther. 2017, 27, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.A. Eteplirsen Approved for Duchenne Muscular Dystrophy: The FDA Faces a Difficult Choice. Mol. Ther. 2016, 24, 1884–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirley, M. Casimersen: First Approval. Drugs 2021, 81, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.F.; Miceli, M.C. FDA Approval of Eteplirsen for Muscular Dystrophy. JAMA 2017, 317, 1480. [Google Scholar] [CrossRef] [PubMed]
- Roshmi, R.R.; Yokota, T. Viltolarsen: From Preclinical Studies to FDA Approval. Methods Mol. Biol. 2023, 2587, 31–41. [Google Scholar]
- Aartsma-Rus, A.; Corey, D.R. The 10th Oligonucleotide Therapy Approved: Golodirsen for Duchenne Muscular Dystrophy. Nucleic Acid Ther. 2020, 30, 67–70. [Google Scholar] [CrossRef] [Green Version]
- Brudvig, J.J.; Weimer, J.M. On the cusp of cures: Breakthroughs in Batten disease research. Curr. Opin. Neurobiol. 2022, 72, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hu, C.; Moufawad El Achkar, C.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [Google Scholar] [CrossRef]
- Huizing, M.; Gahl, W.A. Inherited disorders of lysosomal membrane transporters. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183336. [Google Scholar] [CrossRef]
- Kim, Y.J.; Sivetz, N.; Layne, J.; Voss, D.M.; Yang, L.; Zhang, Q.; Krainer, A.R. Exon-skipping antisense oligonucleotides for cystic fibrosis therapy. Proc. Natl. Acad. Sci. USA 2022, 119, e2114858118. [Google Scholar] [CrossRef]
- Covello, G.; Ibrahim, G.H.; Bacchi, N.; Casarosa, S.; Denti, M.A. Exon skipping through chimeric antisense U1 snRNAs to correct retinitis pigmentosa GTPase-regulator (RPGR) splice defect. Nucleic Acid Ther. 2022, 32, 333–349. [Google Scholar] [CrossRef]
- Wyatt, E.J.; Demonbreun, A.R.; Kim, E.Y.; Puckelwartz, M.J.; Vo, A.H.; Dellefave-Castillo, L.M.; Gao, Q.Q.; Vainzof, M.; Pavanello, R.C.M.; Zatz, M.; et al. Efficient exon skipping of SGCG mutations mediated by phosphorodiamidate morpholino oligomers. JCI Insight 2018, 3, e99357. [Google Scholar] [CrossRef] [PubMed]
- Demonbreun, A.R.; Wyatt, E.J.; Fallon, K.S.; Oosterbaan, C.C.; Page, P.G.; Hadhazy, M.; Quattrocelli, M.; Barefield, D.Y.; McNally, E.M. A gene-edited mouse model of limb-girdle muscular dystrophy 2C for testing exon skipping. Dis. Model Mech. 2019, 13, dmm040832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barthelemy, F.; Blouin, C.; Wein, N.; Mouly, V.; Courrier, S.; Dionnet, E.; Kergourlay, V.; Mathieu, Y.; Garcia, L.; Butler-Browne, G.; et al. Exon 32 Skipping of Dysferlin Rescues Membrane Repair in Patients’ Cells. J. Neuromuscul. Dis. 2015, 2, 281–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwar, S.; Yokota, T. Morpholino-Mediated Exons 28-29 Skipping of Dysferlin and Characterization of Multiexon-skipped Dysferlin using RT-PCR, Immunoblotting, and Membrane Wounding Assay. Methods Mol. Biol. 2023, 2587, 183–196. [Google Scholar] [PubMed]
- Lee, J.J.A.; Maruyama, R.; Duddy, W.; Sakurai, H.; Yokota, T. Identification of Novel Antisense-Mediated Exon Skipping Targets in DYSF for Therapeutic Treatment of Dysferlinopathy. Mol. Ther. Nucleic Acids 2018, 13, 596–604. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, R.; Yokota, T. Morpholino-Mediated Exon Skipping Targeting Human ACVR1/ALK2 for Fibrodysplasia Ossificans Progressiva. Methods Mol. Biol. 2018, 1828, 497–502. [Google Scholar]
- Shi, S.; Cai, J.; de Gorter, D.J.; Sanchez-Duffhues, G.; Kemaladewi, D.U.; Hoogaars, W.M.; Aartsma-Rus, A.; ‘t Hoen, P.A.; ten Dijke, P. Antisense-oligonucleotide mediated exon skipping in activin-receptor-like kinase 2: Inhibiting the receptor that is overactive in fibrodysplasia ossificans progressiva. PLoS ONE 2013, 8, e69096. [Google Scholar] [CrossRef] [Green Version]
- Vermeer, F.C.; Bremer, J.; Sietsma, R.J.; Sandilands, A.; Hickerson, R.P.; Bolling, M.C.; Pasmooij, A.M.G.; Lemmink, H.H.; Swertz, M.A.; Knoers, N.; et al. Therapeutic Prospects of Exon Skipping for Epidermolysis Bullosa. Int. J. Mol. Sci. 2021, 22, 12222. [Google Scholar] [CrossRef]
- Bornert, O.; Kuhl, T.; Bremer, J.; van den Akker, P.C.; Pasmooij, A.M.; Nystrom, A. Analysis of the functional consequences of targeted exon deletion in COL7A1 reveals prospects for dystrophic epidermolysis bullosa therapy. Mol. Ther. 2016, 24, 1302–1311. [Google Scholar] [CrossRef] [Green Version]
- Siva, K.; Covello, G.; Denti, M.A. Exon-skipping antisense oligonucleotides to correct missplicing in neurogenetic diseases. Nucleic Acid Ther. 2014, 24, 69–86. [Google Scholar] [CrossRef] [Green Version]
- Kalbfuss, B.; Mabon, S.A.; Misteli, T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J. Biol. Chem. 2001, 276, 42986–42993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J. Antisense-mediated exon skipping to shift alternative splicing to treat cancer. Methods Mol. Biol. 2012, 867, 201–208. [Google Scholar] [PubMed]
- Maruyama, R.; Yokota, T. Tips to Design Effective Splice-Switching Antisense Oligonucleotides for Exon Skipping and Exon Inclusion. Methods Mol. Biol. 2018, 1828, 79–90. [Google Scholar]
- Sciabola, S.; Xi, H.; Cruz, D.; Cao, Q.; Lawrence, C.; Zhang, T.; Rotstein, S.; Hughes, J.D.; Caffrey, D.R.; Stanton, R.V. PFRED: A computational platform for siRNA and antisense oligonucleotides design. PLoS ONE 2021, 16, e0238753. [Google Scholar] [CrossRef]
- Shimo, T.; Maruyama, R.; Yokota, T. Designing effective antisense oligonucleotides for exon skipping. In Duchenne Muscular Dystrophy: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2018; pp. 143–155. [Google Scholar]
- Chiba, S.; Lim, K.R.Q.; Sheri, N.; Anwar, S.; Erkut, E.; Shah, M.N.A.; Aslesh, T.; Woo, S.; Sheikh, O.; Maruyama, R.; et al. eSkip-Finder: A machine learning-based web application and database to identify the optimal sequences of antisense oligonucleotides for exon skipping. Nucleic Acids Res. 2021, 49, W193–W198. [Google Scholar] [CrossRef]
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4. [Google Scholar]
- Chandra, A.; Yao, X. Ensemble learning using multi-objective evolutionary algorithms. J. Math. Model. Algorithms 2006, 5, 417–445. [Google Scholar] [CrossRef]
- Sahin, E.K. Assessing the predictive capability of ensemble tree methods for landslide susceptibility mapping using XGBoost, gradient boosting machine, and random forest. SN Appl. Sci. 2020, 2, 1308. [Google Scholar] [CrossRef]
- Malueka, R.G.; Takaoka, Y.; Yagi, M.; Awano, H.; Lee, T.; Dwianingsih, E.K.; Nishida, A.; Takeshima, Y.; Matsuo, M. Categorization of 77 dystrophin exons into 5 groups by a decision tree using indexes of splicing regulatory factors as decision markers. BMC Genet. 2012, 13, 23. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn Res. 2011, 12, 2825–2830. [Google Scholar]
- Ramraj, S.; Uzir, N.; Sunil, R.; Banerjee, S. Experimenting XGBoost algorithm for prediction and classification of different datasets. Int. J. Control. Theory Appl. 2016, 9, 651–662. [Google Scholar]
- Crunkhorn, S. Exon skipping combats Batten disease. Nat. Rev. Drug Discov. 2020, 19, 588. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Clemens, P.R.; Hoffman, E.P. Exon-Skipping in Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2021, 8, S343–S358. [Google Scholar] [CrossRef] [PubMed]
- Dulla, K.; Slijkerman, R.; van Diepen, H.C.; Albert, S.; Dona, M.; Beumer, W.; Turunen, J.J.; Chan, H.L.; Schulkens, I.A.; Vorthoren, L.; et al. Antisense oligonucleotide-based treatment of retinitis pigmentosa caused by USH2A exon 13 mutations. Mol. Ther. 2021, 29, 2441–2455. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, H.; Matsutani, T.; Yamada, K.; Iwano, N.; Sumi, S.; Hosoda, S.; Zhao, S.; Fukunaga, T.; Hamada, M. Representation learning applications in biological sequence analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3198–3208. [Google Scholar] [CrossRef]
- Chen, W.; Liu, X.; Zhang, S.; Chen, S. Artificial intelligence for drug discovery: Resources, methods, and applications. Mol. Ther. Nucleic Acids 2023, 31, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Echigoya, Y.; Mouly, V.; Garcia, L.; Yokota, T.; Duddy, W. In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS ONE 2015, 10, e0120058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
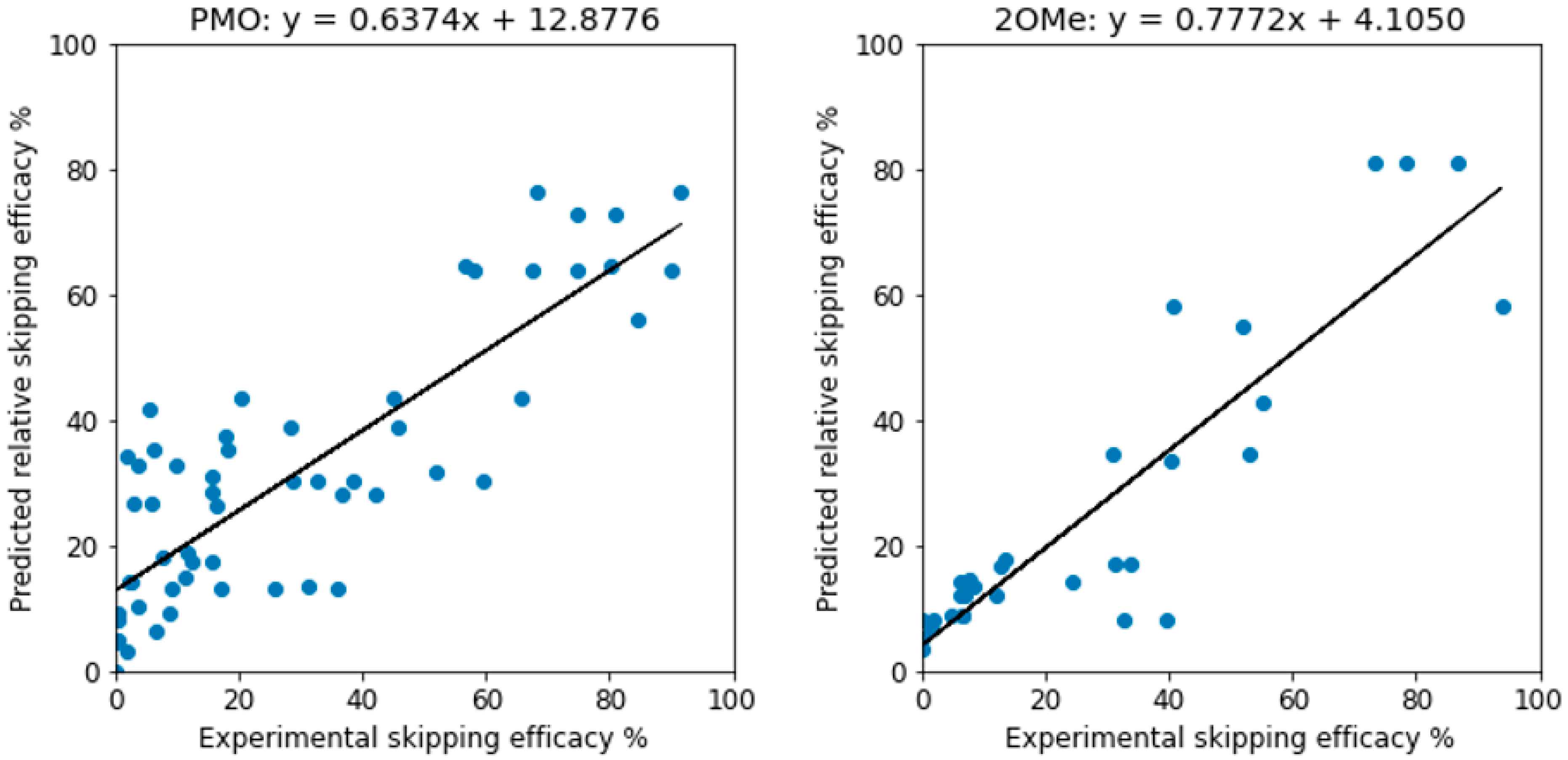

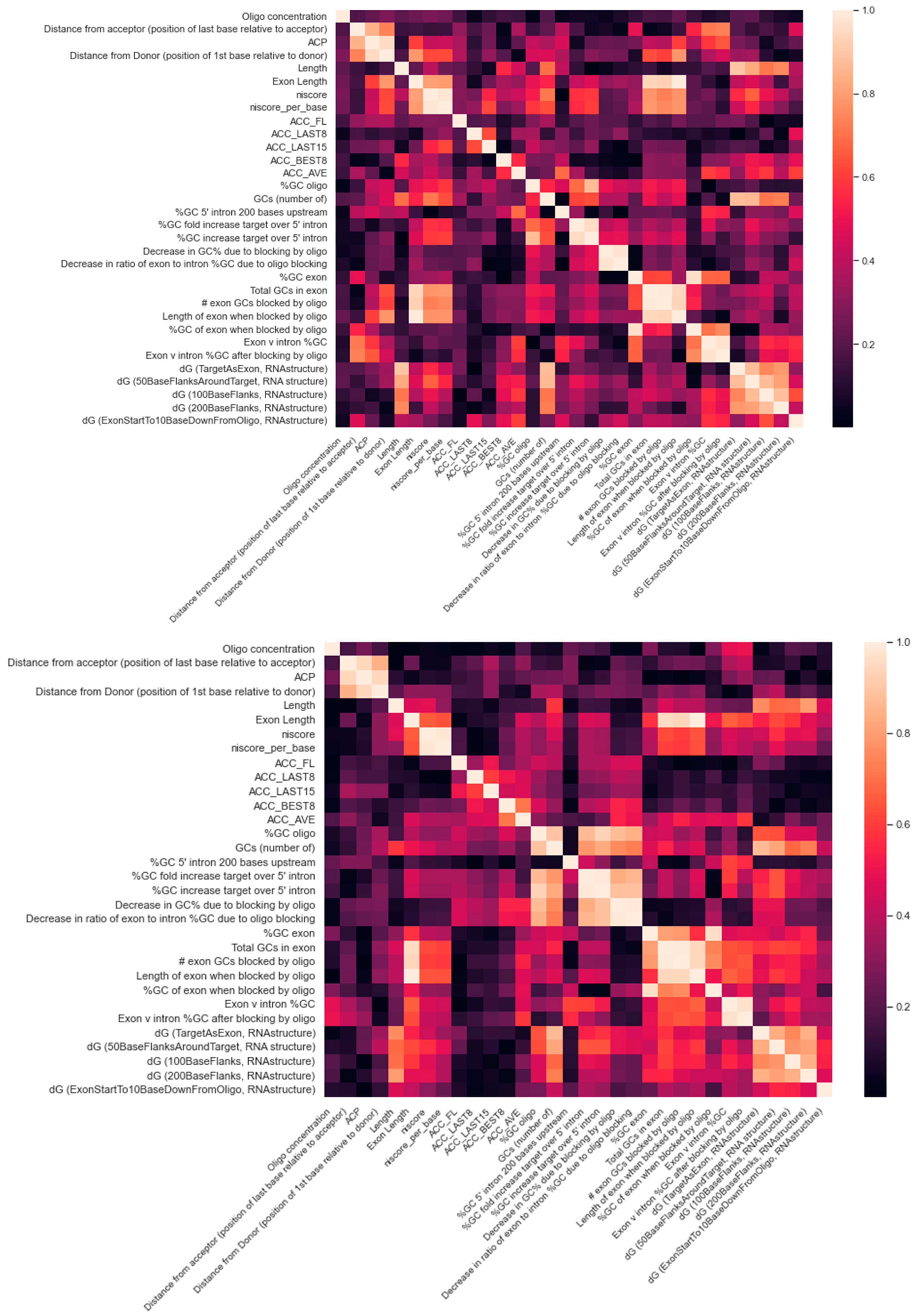
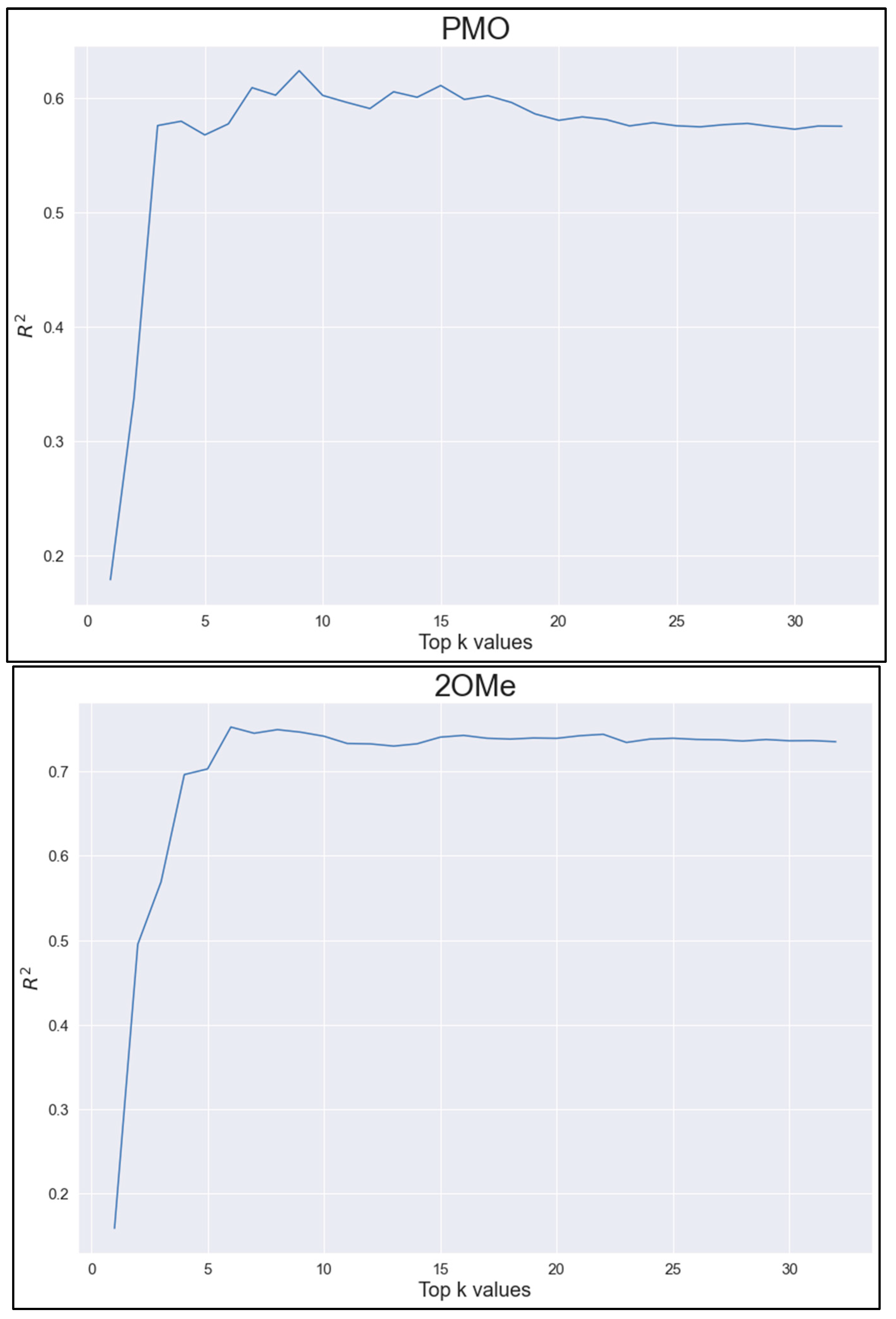
| Methods | PMO | 2OMe | ||
|---|---|---|---|---|
| R2 | MAE | R2 | MAE | |
| Support Vector | 0.138 ± 0.076 | 22.06 ± 4.02 | 0.558 ± 0.093 | 17.70 ± 5.32 |
| Random Forest | 0.555 ± 0.247 | 15.39 ± 4.84 | 0.729 ± 0.169 | 10.59 ± 3.31 |
| Gradient Boosting | 0.564 ± 0.234 | 14.97 ± 4.58 | 0.721 ± 0.152 | 10.13 ± 2.77 |
| XGBoost | 0.530 ± 0.214 | 15.58 ± 3.87 | 0.717 ± 0.164 | 10.56 ± 3.49 |
| Three-way Voting | 0.576 ± 0.244 | 14.87 ± 4.63 | 0.740 ± 0.157 | 10.07 ± 3.29 |
| ASO Name | Voting Predicted | eSkip Predicted | Experimental [14] |
|---|---|---|---|
| H73A (+16 + 40) | 63% (ranked #1) | 60% (ranked #1) | 100% (ranked #1) |
| H73A (+16 + 35) | 37% (ranked #3) | 23% (ranked #3) | 40% (ranked #3) |
| H73A (+21 + 40) | 42% (ranked #2) | 48% (ranked #2) | 85% (ranked #2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, A.; Chiba, S.; Shimizu, Y.; Kunitake, K.; Okuno, Y.; Aoki, Y.; Yokota, T. Ensemble-Learning and Feature Selection Techniques for Enhanced Antisense Oligonucleotide Efficacy Prediction in Exon Skipping. Pharmaceutics 2023, 15, 1808. https://doi.org/10.3390/pharmaceutics15071808
Zhu A, Chiba S, Shimizu Y, Kunitake K, Okuno Y, Aoki Y, Yokota T. Ensemble-Learning and Feature Selection Techniques for Enhanced Antisense Oligonucleotide Efficacy Prediction in Exon Skipping. Pharmaceutics. 2023; 15(7):1808. https://doi.org/10.3390/pharmaceutics15071808
Chicago/Turabian StyleZhu, Alex, Shuntaro Chiba, Yuki Shimizu, Katsuhiko Kunitake, Yasushi Okuno, Yoshitsugu Aoki, and Toshifumi Yokota. 2023. "Ensemble-Learning and Feature Selection Techniques for Enhanced Antisense Oligonucleotide Efficacy Prediction in Exon Skipping" Pharmaceutics 15, no. 7: 1808. https://doi.org/10.3390/pharmaceutics15071808
APA StyleZhu, A., Chiba, S., Shimizu, Y., Kunitake, K., Okuno, Y., Aoki, Y., & Yokota, T. (2023). Ensemble-Learning and Feature Selection Techniques for Enhanced Antisense Oligonucleotide Efficacy Prediction in Exon Skipping. Pharmaceutics, 15(7), 1808. https://doi.org/10.3390/pharmaceutics15071808







