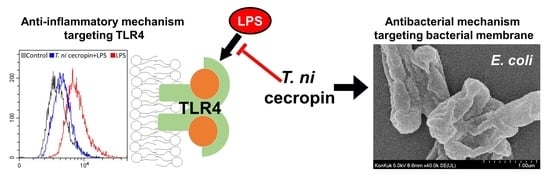
Characterization of the Antimicrobial Activities of Trichoplusia ni Cecropin A as a High-Potency Therapeutic against Colistin-Resistant Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
2.2. Bacteria Strains
2.3. Minimum Inhibitory Concentration (MIC)
2.4. Cytotoxicity
2.5. Hemolysis
2.6. Biofilm Inhibition Assay
2.7. Bacteria Outer Membrane Permeability Test
2.8. Circular Dichroism (CD) Analysis
2.9. Antimicrobial Activity Time-Course Assay
2.10. Bacteria Morphology Imaging
2.11. Limulus Amebocyte Lysate (LAL) Assay
2.12. BODIPY-TR-Cadaverine (BC) Displacement Assay
2.13. Isothermal Titration Calorimetry (ITC)
2.14. Saturation Transfer Difference (STD)-Nuclear Magnetic Resonance (NMR)
2.15. Suppression of LPS-Induced Inflammatory Cytokines
2.16. Suppression of Colistin-Resistant Bacteria-Induced Inflammatory Cytokines
2.17. Inhibition of Nitric Oxide (NO) Production by T. ni Cecropin in Response to Various Toll-like Receptors (TLRs)
2.18. Secreted Embryonic Alkaline Phosphatase (SEAP) Assay
2.19. Surface Plasmon Resonance (SPR)
2.20. Flow Cytometry
2.21. Animal Study Information
2.22. Mouse Model of LPS-Induced Endotoxemia
2.23. Histological Analysis of Lung Tissue
2.24. Data Analysis
3. Results
3.1. Antibacterial Activities of T. ni Cecropin
3.2. Toxicity of T. ni Cecropin to Mammalian Cells
3.3. T. ni Cecropin Inhibits ColREC Biofilm Formation
3.4. Antibacterial Mechanisms of T. ni Cecropin against Gram-Negative Bacteria
3.4.1. LPS-Neutralizing Capacity of T. ni Cecropin
3.4.2. Membrane Depolarization Ability of T. ni Cecropin against E. coli
3.4.3. T. ni Cecropin Induces E. coli Cell Membrane Damage
3.4.4. T. ni Cecropin Directly Interacts with LPS
3.4.5. Secondary Structure of T. ni Cecropin
3.5. Inhibition of Cytokine Production in RAW 264.7 Cells Stimulated by LPS or ColREC
3.6. T. ni Cecropin Selectively Targets the TLR4-Inflammatory Signaling Pathway
3.7. T. ni Cecropin Significantly Attenuates LPS-Induced Endotoxemia in a Mouse Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, C.L.; Anderson, M.T.; Mobley, H.L.T.; Bachman, M.A. Pathogenesis of Gram-Negative Bacteremia. Clin. Microbiol. Rev. 2021, 34, e00234-20. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum beta-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Walas, N.; Slown, S.; Amato, H.K.; Lloyd, T.; Bender, M.; Varghese, V.; Pandori, M.; Graham, J.P. The role of plasmids in carbapenem resistant E. coli in Alameda County, California. BMC Microbiol. 2023, 23, 147. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Peghin, M.; Vena, A.; Giacobbe, D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 2019, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Benedict, R.G.; Langlykke, A.F. Antibiotic activity of Bacillus polymyxa. J. Bacteriol. 1947, 54, 24. [Google Scholar] [PubMed]
- Sisay, M.; Hagos, B.; Edessa, D.; Tadiwos, Y.; Mekuria, A.N. Polymyxin-induced nephrotoxicity and its predictors: A systematic review and meta-analysis of studies conducted using RIFLE criteria of acute kidney injury. Pharmacol. Res. 2021, 163, 105328. [Google Scholar] [CrossRef]
- Mohapatra, S.S.; Dwibedy, S.K.; Padhy, I. Polymyxins, the last-resort antibiotics: Mode of action, resistance emergence, and potential solutions. J. Biosci. 2021, 46, 85. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Ayoub Moubareck, C. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 2020, 10, 181. [Google Scholar] [CrossRef]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old’ class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef]
- Domingues, M.M.; Inacio, R.G.; Raimundo, J.M.; Martins, M.; Castanho, M.A.; Santos, N.C. Biophysical characterization of polymyxin B interaction with LPS aggregates and membrane model systems. Biopolymers 2012, 98, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Godoy, A.; Hansford, K.A.; Muldoon, C.; Becker, B.; Elliott, A.G.; Huang, J.X.; Pelingon, R.; Butler, M.S.; Blaskovich, M.A.T.; Cooper, M.A. Structure-Function Studies of Polymyxin B Lipononapeptides. Molecules 2019, 24, 553. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wan, M.; Zhang, S.; Gao, L.; Fang, W. Polymyxin B Loosens Lipopolysaccharide Bilayer but Stiffens Phospholipid Bilayer. Biophys. J. 2020, 118, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Goode, A.; Yeh, V.; Bonev, B.B. Interactions of polymyxin B with lipopolysaccharide-containing membranes. Faraday Discuss. 2021, 232, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Srinivas, S.; Xu, Y.; Wei, W.; Feng, Y. Genetic and Biochemical Mechanisms for Bacterial Lipid A Modifiers Associated with Polymyxin Resistance. Trends Biochem. Sci. 2019, 44, 973–988. [Google Scholar] [CrossRef]
- Hamel, M.; Rolain, J.M.; Baron, S.A. The History of Colistin Resistance Mechanisms in Bacteria: Progress and Challenges. Microorganisms 2021, 9, 442. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462. [Google Scholar] [CrossRef]
- Son, S.J.; Huang, R.; Squire, C.J.; Leung, I.K.H. MCR-1: A promising target for structure-based design of inhibitors to tackle polymyxin resistance. Drug. Discov. Today 2019, 24, 206–216. [Google Scholar] [CrossRef]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Wang, J.; Thompson, P.E.; Li, J. Teaching ‘old’ polymyxins new tricks: New-generation lipopeptides targeting gram-negative ‘superbugs’. ACS Chem. Biol. 2014, 9, 1172–1177. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Han, M.; Tran, K.; Patil, N.A.; Ma, W.; Roberts, K.D.; Xiao, M.; Sommer, B.; Schreiber, F.; Wang, L.; et al. An Intelligent Strategy with All-Atom Molecular Dynamics Simulations for the Design of Lipopeptides against Multidrug-Resistant Pseudomonas aeruginosa. J. Med. Chem. 2022, 65, 10001–10013. [Google Scholar] [CrossRef] [PubMed]
- Rodvold, K.A.; Bader, J.; Gupta, V.K.; Lister, T.; Srivastava, P.; Bruss, J. 625. SPR206 Pharmacokinetics (PK) in Plasma, Epithelial Lining Fluid (ELF), and Alveolar Macrophages (AM) in Healthy Adult Subjects. Open Forum Infect. Dis. 2022, 9. [Google Scholar] [CrossRef]
- Griffith, D.; Carmeli, Y.; Gehrke, S.; Morgan, E.; Dudley, M.; Loutit, J. 217. A Phase 1 Study of the Safety, Tolerability, and Pharmacokinetics of Multiple Doses of the Lipopeptide QPX9003 in Healthy Adult Subjects. Open Forum Infect. Dis. 2022, 9. [Google Scholar] [CrossRef]
- Lepak, A.J.; Wang, W.; Andes, D.R. Pharmacodynamic Evaluation of MRX-8, a Novel Polymyxin, in the Neutropenic Mouse Thigh and Lung Infection Models against Gram-Negative Pathogens. Antimicrob. Agents Chemother. 2020, 64, e01517-20. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug. Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides With Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdacs, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J.J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engstrom, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef]
- Morishima, I.; Suginaka, S.; Ueno, T.; Hirano, H. Isolation and structure of cecropins, inducible antibacterial peptides, from the silkworm, Bombyx mori. Comp. Biochem. Physiol. B 1990, 95, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Hong, M.Y.; Park, S.W.; Choi, K.H.; Yun, E.Y.; Goo, T.W.; Kang, S.W.; Suh, H.J.; Kim, I.; Hwang, J.S. Characterization and cDNA cloning of a cecropin-like antimicrobial peptide, papiliocin, from the swallowtail butterfly, Papilio xuthus. Mol. Cells 2010, 29, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Lee, J.H.; Kim, I.; Seo, S.J.; Son, S.M.; Lee, K.Y.; Lee, I.H. Purification and cDNA cloning of a cecropin-like peptide from the great wax moth, Galleria mellonella. Mol. Cells 2004, 17, 262–266. [Google Scholar] [PubMed]
- Qu, Z.; Steiner, H.; Engstrom, A.; Bennich, H.; Boman, H.G. Insect immunity: Isolation and structure of cecropins B and D from pupae of the Chinese oak silk moth, Antheraea pernyi. Eur. J. Biochem. 1982, 127, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Martin, F.; Herrera-Leon, C.; D’Amelio, N. Bombyx mori Cecropin D could trigger cancer cell apoptosis by interacting with mitochondrial cardiolipin. Biochim. Biophys. Acta Biomembr. 2022, 1864, 184003. [Google Scholar] [CrossRef]
- Kalsy, M.; Tonk, M.; Hardt, M.; Dobrindt, U.; Zdybicka-Barabas, A.; Cytrynska, M.; Vilcinskas, A.; Mukherjee, K. The insect antimicrobial peptide cecropin A disrupts uropathogenic Escherichia coli biofilms. NPJ Biofilms Microbiomes 2020, 6, 6. [Google Scholar] [CrossRef]
- Krishnan, M.; Choi, J.; Jang, A.; Choi, S.; Yeon, J.; Jang, M.; Lee, Y.; Son, K.; Shin, S.Y.; Jeong, M.S.; et al. Molecular mechanism underlying the TLR4 antagonistic and antiseptic activities of papiliocin, an insect innate immune response molecule. Proc. Natl. Acad. Sci. USA 2022, 119, e2115669119. [Google Scholar] [CrossRef]
- Peng, C.; Liu, Y.; Shui, L.; Zhao, Z.; Mao, X.; Liu, Z. Mechanisms of Action of the Antimicrobial Peptide Cecropin in the Killing of Candida albicans. Life 2022, 12, 1581. [Google Scholar] [CrossRef]
- Mikonranta, L.; Dickel, F.; Mappes, J.; Freitak, D. Lepidopteran species have a variety of defence strategies against bacterial infections. J. Invertebr. Pathol. 2017, 144, 88–96. [Google Scholar] [CrossRef]
- Wei, L.; Yang, Y.; Zhou, Y.; Li, M.; Yang, H.; Mu, L.; Qian, Q.; Wu, J.; Xu, W. Anti-inflammatory activities of Aedes aegypti cecropins and their protection against murine endotoxin shock. Parasit. Vectors 2018, 11, 470. [Google Scholar] [CrossRef]
- Lee, E.; Shin, A.; Kim, Y. Anti-inflammatory activities of cecropin A and its mechanism of action. Arch. Insect Biochem. Physiol. 2015, 88, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, E.; Shin, S.; Jeong, K.W.; Lee, J.Y.; Bae, S.Y.; Kim, S.H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. J. Biol. Chem. 2011, 286, 41296–41311. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Kim, J.K.; Jeon, D.; Jeong, K.W.; Shin, A.; Kim, Y. Functional Roles of Aromatic Residues and Helices of Papiliocin in its Antimicrobial and Anti-inflammatory Activities. Sci. Rep. 2015, 5, 12048. [Google Scholar] [CrossRef] [PubMed]
- Holak, T.A.; Engstrom, A.; Kraulis, P.J.; Lindeberg, G.; Bennich, H.; Jones, T.A.; Gronenborn, A.M.; Clore, G.M. The solution conformation of the antibacterial peptide cecropin A: A nuclear magnetic resonance and dynamical simulated annealing study. Biochemistry 1988, 27, 7620–7629. [Google Scholar] [CrossRef]
- Ramos-Martin, F.; Herrera-Leon, C.; D’Amelio, N. Molecular basis of the anticancer, apoptotic and antibacterial activities of Bombyx mori Cecropin A. Arch. Biochem. Biophys. 2022, 715, 109095. [Google Scholar] [CrossRef]
- Zheng, Z.; Tharmalingam, N.; Liu, Q.; Jayamani, E.; Kim, W.; Fuchs, B.B.; Zhang, R.; Vilcinskas, A.; Mylonakis, E. Synergistic Efficacy of Aedes aegypti Antimicrobial Peptide Cecropin A2 and Tetracycline against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017, 61, e00686-17. [Google Scholar] [CrossRef]
- Brady, D.; Grapputo, A.; Romoli, O.; Sandrelli, F. Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications. Int. J. Mol. Sci. 2019, 20, 5862. [Google Scholar] [CrossRef]
- Kang, D.; Liu, G.; Gunne, H.; Steiner, H. PCR differential display of immune gene expression in Trichoplusia ni. Insect Biochem. Mol. Biol. 1996, 26, 177–184. [Google Scholar] [CrossRef]
- Freitak, D.; Wheat, C.W.; Heckel, D.G.; Vogel, H. Immune system responses and fitness costs associated with consumption of bacteria in larvae of Trichoplusia ni. BMC Biol. 2007, 5, 56. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kawahara, R.; Hamamoto, K.; Hirai, I.; Khong, D.T.; Nguyen, T.N.; Tran, H.T.; Motooka, D.; Nakamura, S.; Yamamoto, Y. High Prevalence of Colistin-Resistant Escherichia coli with Chromosomally Carried mcr-1 in Healthy Residents in Vietnam. mSphere 2020, 5, e00117-20. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.; Choi, J.; Jang, A.; Kim, Y. A Novel Peptide Antibiotic, Pro10-1D, Designed from Insect Defensin Shows Antibacterial and Anti-Inflammatory Activities in Sepsis Models. Int. J. Mol. Sci. 2020, 21, 6216. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Durai, P.; Jeon, D.; Jung, I.D.; Lee, S.J.; Park, Y.M.; Kim, Y. Phloretin as a Potent Natural TLR2/1 Inhibitor Suppresses TLR2-Induced Inflammation. Nutrients 2018, 10, 868. [Google Scholar] [CrossRef] [PubMed]
- Cheon, D.; Kim, J.; Jeon, D.; Shin, H.C.; Kim, Y. Target Proteins of Phloretin for Its Anti-Inflammatory and Antibacterial Activities Against Propionibacterium acnes-Induced Skin Infection. Molecules 2019, 24, 1319. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jacob, B.; Jang, M.; Kwak, C.; Lee, Y.; Son, K.; Lee, S.; Jung, I.D.; Jeong, M.S.; Kwon, S.H.; et al. Development of a novel short 12-meric papiliocin-derived peptide that is effective against Gram-negative sepsis. Sci. Rep. 2019, 9, 3817. [Google Scholar] [CrossRef] [PubMed]
- Manniello, M.D.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect antimicrobial peptides: Potential weapons to counteract the antibiotic resistance. Cell Mol. Life Sci. 2021, 78, 4259–4282. [Google Scholar] [CrossRef]
- Lee, E.; Jeong, K.W.; Lee, J.; Shin, A.; Kim, J.K.; Lee, J.; Lee, D.G.; Kim, Y. Structure-activity relationships of cecropin-like peptides and their interactions with phospholipid membrane. BMB Rep. 2013, 46, 282–287. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS): The Detection and Reporting of Colistin Resistance; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Ferreira, A.R.; Teixeira, C.; Sousa, C.F.; Bessa, L.J.; Gomes, P.; Gameiro, P. How Insertion of a Single Tryptophan in the N-Terminus of a Cecropin A-Melittin Hybrid Peptide Changes Its Antimicrobial and Biophysical Profile. Membranes 2021, 11, 48. [Google Scholar] [CrossRef]
- Vergis, J.; Malik, S.V.S.; Pathak, R.; Kumar, M.; Kurkure, N.V.; Barbuddhe, S.B.; Rawool, D.B. Exploring Galleria mellonella larval model to evaluate antibacterial efficacy of Cecropin A (1-7)-Melittin against multi-drug resistant enteroaggregative Escherichia coli. Pathog. Dis. 2021, 79, ftab010. [Google Scholar] [CrossRef]
- Lee, J.K.; Seo, C.H.; Luchian, T.; Park, Y. Antimicrobial Peptide CMA3 Derived from the CA-MA Hybrid Peptide: Antibacterial and Anti-inflammatory Activities with Low Cytotoxicity and Mechanism of Action in Escherichia coli. Antimicrob. Agents Chemother. 2016, 60, 495–506. [Google Scholar] [CrossRef]
- Namvar Erbani, S.; Madanchi, H.; Ajodani Far, H.; Rostamian, M.; Rahmati, S.; Shabani, A.A. First report of antifungal activity of CecropinA-Magenin2 (CE-MA) hybrid peptide and its truncated derivatives. Biochem. Biophys. Res. Commun. 2021, 549, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jang, A.; Yoon, Y.K.; Kim, Y. Development of Novel Peptides for the Antimicrobial Combination Therapy against Carbapenem-Resistant Acinetobacter baumannii Infection. Pharmaceutics 2021, 13, 1800. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, Y.; Xue, Z.; Jia, Y.; Li, R.; He, C.; Chen, H. The structure-mechanism relationship and mode of actions of antimicrobial peptides: A review. Trends Food Sci. Technol. 2021, 109, 103–115. [Google Scholar] [CrossRef]











| Minimal Inhibitory Concentration (μM) | |||||
|---|---|---|---|---|---|
| Microorganism | Cecropin A | T. ni Cecropin | Polymyxin B | Colistin | Melittin |
| E. coli | 2 | 2 | 0.25 | 0.25 | 8 |
| A. baumannii | 2 | 1 | 0.5 | 0.25 | 4 |
| P. aeruginosa | 8 | 4 | 2 | 1 | 32 |
| K. pneumoniae | 1 | 1 | 0.25 | 0.25 | 32 |
| ColREC 1557 | 1 | 1 | 8 | 8 | 2 |
| ColREC 12 | 1 | 1 | 4 | 4 | 2 |
| ColRAB 1915 | 1 | 1 | 64 | >64 | 2 |
| ColRKP 139 | 2 | 2 | 64 | >64 | 32 |
| GM * | 2.25 | 1.63 | 17.88 | 33.72 | 14.25 |
| HC10 † | 200 | 200 | 200 | 200 | 3.1 |
| Relative selective Index ** | 88.89 | 123.08 | 11.19 | 5.93 | 0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Kim, B.; Kim, M.; Yoo, S.; Lee, J.; Hwang, E.; Kim, Y. Characterization of the Antimicrobial Activities of Trichoplusia ni Cecropin A as a High-Potency Therapeutic against Colistin-Resistant Escherichia coli. Pharmaceutics 2023, 15, 1752. https://doi.org/10.3390/pharmaceutics15061752
Lee H, Kim B, Kim M, Yoo S, Lee J, Hwang E, Kim Y. Characterization of the Antimicrobial Activities of Trichoplusia ni Cecropin A as a High-Potency Therapeutic against Colistin-Resistant Escherichia coli. Pharmaceutics. 2023; 15(6):1752. https://doi.org/10.3390/pharmaceutics15061752
Chicago/Turabian StyleLee, Hyeju, Byeongkwon Kim, Minju Kim, Seoyeong Yoo, Jinkyeong Lee, Eunha Hwang, and Yangmee Kim. 2023. "Characterization of the Antimicrobial Activities of Trichoplusia ni Cecropin A as a High-Potency Therapeutic against Colistin-Resistant Escherichia coli" Pharmaceutics 15, no. 6: 1752. https://doi.org/10.3390/pharmaceutics15061752
APA StyleLee, H., Kim, B., Kim, M., Yoo, S., Lee, J., Hwang, E., & Kim, Y. (2023). Characterization of the Antimicrobial Activities of Trichoplusia ni Cecropin A as a High-Potency Therapeutic against Colistin-Resistant Escherichia coli. Pharmaceutics, 15(6), 1752. https://doi.org/10.3390/pharmaceutics15061752