Lipid Nanoparticles as a Shuttle for Anti-Adipogenic miRNAs to Human Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of miRNA-Complexed Pegylated Lipid Nanoparticles (PEG-LNPs)
2.2. Particle Size and Zeta Potential Measurements
2.3. Complexation Capacity
2.4. Cell Culture Experiments
2.5. RNA Isolation and Reverse Transcription
2.6. Real-Time Quantitative PCR (RT-qPCR)
2.7. Oil Red O (ORO) Staining
2.8. Immunofluorescence Staining
2.9. Image Acquisition and Quantitative Image Analysis
2.10. Confocal Laser Scanning Microscopy (cLSM)
2.11. Cell Viability
2.12. Statistical Analysis
3. Results
3.1. Preparation and Characterisation of miRNA-Complexed PEG-LNPs
3.2. Cellular Uptake of Anti-Adipogenic miRNAs Complexed with PEG-LNPs
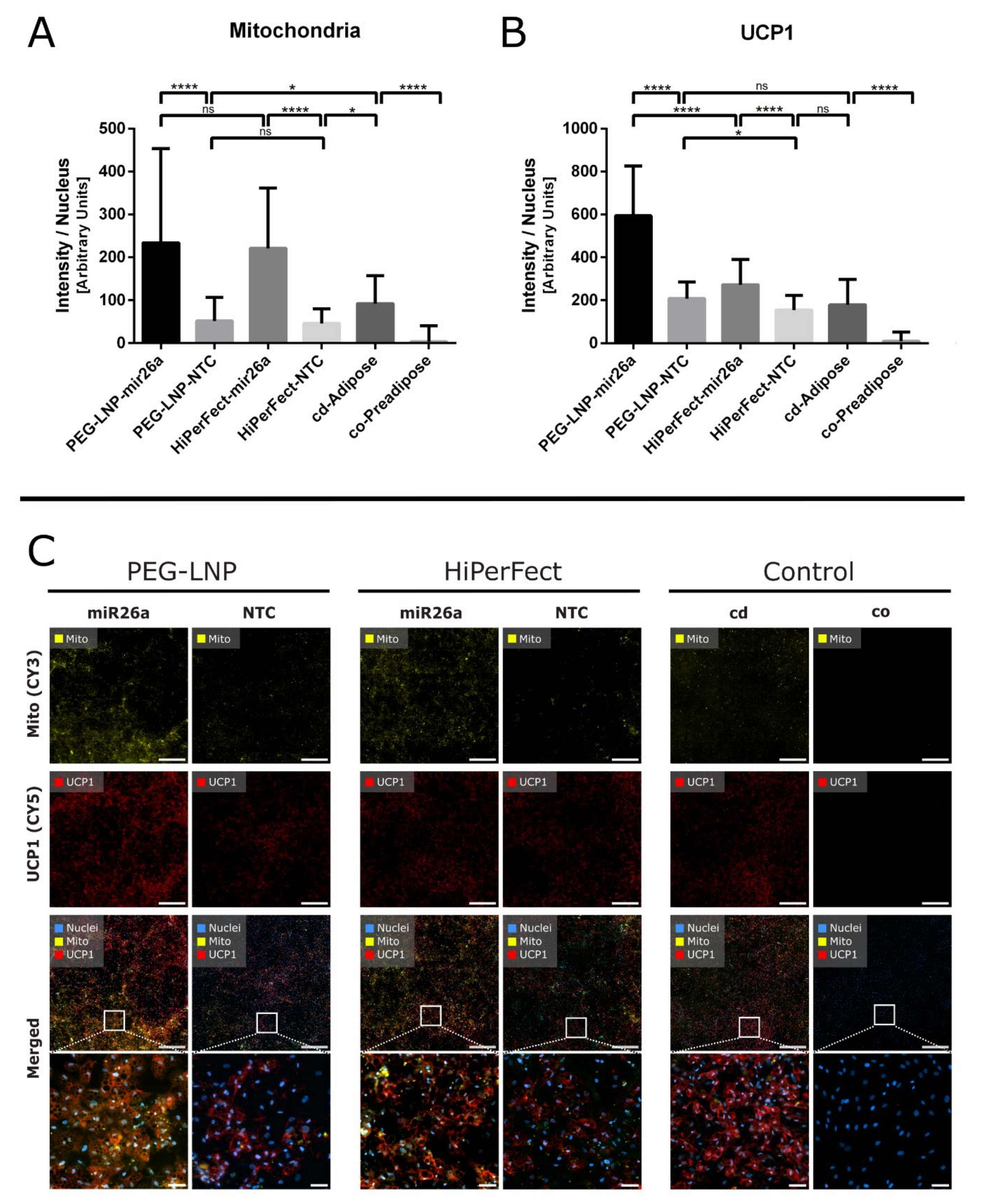
3.3. Biological Effects of Anti-Adipogenic miRNAs Complexed with PEG-LNPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef] [Green Version]
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Hruby, A.; Hu, F.B. The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics 2015, 33, 673–689. [Google Scholar] [CrossRef]
- Cignarella, A.; Busetto, L.; Vettor, R. Pharmacotherapy of obesity: An update. Pharmacol. Res. 2021, 169, 105649. [Google Scholar] [CrossRef]
- Hiradate, R.; Khalil, I.A.; Matsuda, A.; Sasaki, M.; Hida, K.; Harashima, H. A novel dual-targeted rosiglitazone-loaded nanoparticle for the prevention of diet-induced obesity via the browning of white adipose tissue. J. Control. Release Off. J. Control. Release Soc. 2021, 329, 665–675. [Google Scholar] [CrossRef]
- Giordano, A.; Frontini, A.; Cinti, S. Convertible visceral fat as a therapeutic target to curb obesity. Nat. Rev. Drug Discov. 2016, 15, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Kusminski, C.M.; Bickel, P.E.; Scherer, P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov. 2016, 15, 639–660. [Google Scholar] [CrossRef]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Boström, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroon, T.; Harms, M.; Maurer, S.; Bonnet, L.; Alexandersson, I.; Lindblom, A.; Ahnmark, A.; Nilsson, D.; Gennemark, P.; O’Mahony, G.; et al. PPARγ and PPARα synergize to induce robust browning of white fat in vivo. Mol. Metab. 2020, 36, 100964. [Google Scholar] [CrossRef]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Himms-Hagen, J.; Melnyk, A.; Zingaretti, M.C.; Ceresi, E.; Barbatelli, G.; Cinti, S. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol. 2000, 279, C670–C681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbatelli, G.; Murano, I.; Madsen, L.; Hao, Q.; Jimenez, M.; Kristiansen, K.; Giacobino, J.P.; De Matteis, R.; Cinti, S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E1244–E1253. [Google Scholar] [CrossRef] [Green Version]
- Rosenwald, M.; Perdikari, A.; Rülicke, T.; Wolfrum, C. Bi-directional interconversion of brite and white adipocytes. Nat. Cell Biol. 2013, 15, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Scheideler, M. Regulatory Small and Long Noncoding RNAs in Brite/Brown Adipose Tissue. In Brown Adipose Tissue; Pfeifer, A.K.M., Herzig, S., Eds.; Handbook of Experimental Pharmacology; Springer: Cham, Germany, 2018; Volume 251, pp. 215–236. [Google Scholar]
- Thyagarajan, B.; Foster, M.T. Beiging of white adipose tissue as a therapeutic strategy for weight loss in humans. Horm. Mol. Biol. Clin. Investig. 2017, 31, 20170016. [Google Scholar] [CrossRef]
- Herz, C.T.; Kiefer, F.W. Adipose tissue browning in mice and humans. J. Endocrinol. 2019, 241, R97–R109. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, J.; Dai, H.; Duan, Y.; An, Y.; Shi, L.; Lv, Y.; Li, H.; Wang, C.; Ma, Q.; et al. Brown and beige adipose tissue: A novel therapeutic strategy for obesity and type 2 diabetes mellitus. Adipocyte 2021, 10, 48–65. [Google Scholar] [CrossRef]
- Trayhurn, P. Brown Adipose Tissue—A Therapeutic Target in Obesity? Front. Physiol. 2018, 9, 1672. [Google Scholar] [CrossRef]
- Lizcano, F. The Beige Adipocyte as a Therapy for Metabolic Diseases. Int. J. Mol. Sci. 2019, 20, 5058. [Google Scholar] [CrossRef] [Green Version]
- Kornmueller, K.; Amri, E.Z.; Scheideler, M.; Prassl, R. Delivery of miRNAs to the adipose organ for metabolic health. Adv. Drug Deliv. Rev. 2022, 181, 114110. [Google Scholar] [CrossRef]
- Gharanei, S.; Shabir, K.; Brown, J.E.; Weickert, M.O.; Barber, T.M.; Kyrou, I.; Randeva, H.S. Regulatory microRNAs in Brown, Brite and White Adipose Tissue. Cells 2020, 9, 2489. [Google Scholar] [CrossRef] [PubMed]
- Heyn, G.S.; Corrêa, L.H.; Magalhães, K.G. The Impact of Adipose Tissue-Derived miRNAs in Metabolic Syndrome, Obesity, and Cancer. Front. Endocrinol. 2020, 11, 563816. [Google Scholar] [CrossRef]
- Lorente-Cebrián, S.; González-Muniesa, P.; Milagro, F.I.; Martínez, J.A. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: Emerging roles as biomarkers and therapeutic targets. Clin. Sci. 2019, 133, 23–40. [Google Scholar] [CrossRef]
- Goody, D.; Pfeifer, A. MicroRNAs in brown and beige fat. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Gilani, A.; Yi, Q.; Tang, L. MicroRNAs as Mediators of Adipose Thermogenesis and Potential Therapeutic Targets for Obesity. Biology 2022, 11, 1657. [Google Scholar] [CrossRef]
- Karbiener, M.; Pisani, D.F.; Frontini, A.; Oberreiter, L.M.; Lang, E.; Vegiopoulos, A.; Mössenböck, K.; Bernhardt, G.A.; Mayr, T.; Hildner, F.; et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 2014, 32, 1578–1590. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, Q.G. The role of miR-26 in tumors and normal tissues (Review). Oncol. Lett. 2011, 2, 1019–1023. [Google Scholar] [CrossRef] [Green Version]
- Carpentier, A.C.; Blondin, D.P.; Virtanen, K.A.; Richard, D.; Haman, F.; Turcotte, É.E. Brown Adipose Tissue Energy Metabolism in Humans. Front. Endocrinol. 2018, 9, 447. [Google Scholar] [CrossRef] [Green Version]
- Serino, M.; Menghini, R.; Fiorentino, L.; Amoruso, R.; Mauriello, A.; Lauro, D.; Sbraccia, P.; Hribal, M.L.; Lauro, R.; Federici, M. Mice Heterozygous for Tumor Necrosis Factor-α Converting Enzyme Are Protected from Obesity-Induced Insulin Resistance and Diabetes. Diabetes 2007, 56, 2541–2546. [Google Scholar] [CrossRef] [Green Version]
- Gelling, R.W.; Yan, W.; Al-Noori, S.; Pardini, A.; Morton, G.J.; Ogimoto, K.; Schwartz, M.W.; Dempsey, P.J. Deficiency of TNFα Converting Enzyme (TACE/ADAM17) Causes a Lean, Hypermetabolic Phenotype in Mice. Endocrinology 2008, 149, 6053–6064. [Google Scholar] [CrossRef] [Green Version]
- Karbiener, M.; Fischer, C.; Nowitsch, S.; Opriessnig, P.; Papak, C.; Ailhaud, G.; Dani, C.; Amri, E.Z.; Scheideler, M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 2009, 390, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef]
- Urbich, C.; Kaluza, D.; Frömel, T.; Knau, A.; Bennewitz, K.; Boon, R.A.; Bonauer, A.; Doebele, C.; Boeckel, J.N.; Hergenreider, E.; et al. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood 2012, 119, 1607–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, I.; Boudreau, A.; Stephens, J.M. Adipose tissue in health and disease. Open Biol. 2020, 10, 200291. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPARγ Is Required for the Differentiation of Adipose Tissue in Vivo and in Vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Kajimoto, K.; Naraba, H.; Iwai, N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA 2006, 12, 1626–1632. [Google Scholar] [CrossRef] [Green Version]
- Schachner-Nedherer, A.L.; Werzer, O.; Kornmueller, K.; Prassl, R.; Zimmer, A. Biological Activity Of miRNA-27a Using Peptide-based Drug Delivery Systems. Int. J. Nanomed. 2019, 14, 7795–7808. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Gao, Z.; Alarcon, R.M.; Ye, J.; Yun, Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009, 276, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Kleefeldt, J.M.; Pozarska, A.; Nardiello, C.; Pfeffer, T.; Vadász, I.; Herold, S.; Seeger, W.; Morty, R.E. Commercially available transfection reagents and negative control siRNA are not inert. Anal. Biochem. 2020, 606, 113828. [Google Scholar] [CrossRef]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
- Scheideler, M.; Vidakovic, I.; Prassl, R. Lipid nanocarriers for microRNA delivery. Chem. Phys. Lipids 2020, 226, 104837. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, P.; Addison, M.L.; Dear, J.W.; Webb, D.J. Small interfering RNA: Discovery, pharmacology and clinical development—An introductory review. Br. J. Pharmacol. 2022. [Google Scholar] [CrossRef]
- Kim, J.; Eygeris, Y.; Gupta, M.; Sahay, G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021, 170, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef]
- Tenorio, J.; Arias, P.; Martínez-Glez, V.; Santos, F.; García-Miñaur, S.; Nevado, J.; Lapunzina, P. Simpson-Golabi-Behmel syndrome types I and II. Orphanet J. Rare Dis. 2014, 9, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer-Posovszky, P.; Newell, F.S.; Wabitsch, M.; Tornqvist, H.E. Human SGBS cells—A unique tool for studies of human fat cell biology. Obes. Facts 2008, 1, 184–189. [Google Scholar] [CrossRef]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Dhawan, V.; Holm, R.; Nagarsenker, M.S.; Perrie, Y. Liposomes: Advancements and innovation in the manufacturing process. Adv. Drug Deliv. Rev. 2020, 154–155, 102–122. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, S.J.; Warzecha, C.C.; Yadavali, S.; El-Mayta, R.; Alameh, M.-G.; Wang, L.; Weissman, D.; Wilson, J.M.; Issadore, D.; Mitchell, M.J. Scalable mRNA and siRNA Lipid Nanoparticle Production Using a Parallelized Microfluidic Device. Nano Lett. 2021, 21, 5671–5680. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release Off. J. Control. Release Soc. 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. 2012, 51, 8529–8533. [Google Scholar] [CrossRef]
- Carrasco, M.J.; Alishetty, S.; Alameh, M.-G.; Said, H.; Wright, L.; Paige, M.; Soliman, O.; Weissman, D.; Cleveland, T.E.; Grishaev, A.; et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021, 4, 956. [Google Scholar] [CrossRef] [PubMed]
- McGregor, R.A.; Choi, M.S. microRNAs in the regulation of adipogenesis and obesity. Curr. Mol. Med. 2011, 11, 304–316. [Google Scholar] [CrossRef]
- Gojanovich, A.D.; Gimenez, M.C.; Masone, D.; Rodriguez, T.M.; Dewey, R.A.; Delgui, L.R.; Bustos, D.M.; Uhart, M. Human Adipose-Derived Mesenchymal Stem/Stromal Cells Handling Protocols. Lipid Droplets and Proteins Double-Staining. Front. Cell Dev. Biol. 2018, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Koopman, R.; Schaart, G.; Hesselink, M.K. Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem. Cell Biol. 2001, 116, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Nonn, O.; Daxboeck, C.; Groiss, S.; Moser, G.; Gauster, M.; Lang-Olip, I.; Brislinger, D. Automated Quantitative Image Evaluation of Antigen Retrieval Methods for 17 Antibodies in Placentation and Implantation Diagnostic and Research. Microsc. Microanal. 2021, 27, 1506–1517. [Google Scholar] [CrossRef]
- Nezami, E.; Butcher, J.N. Chapter 16—Objective Personality Assessment. In Handbook of Psychological Assessment, 3rd ed.; Goldstein, G., Hersen, M., Eds.; Pergamon: Amsterdam, The Netherlands, 2000; pp. 413–435. [Google Scholar] [CrossRef]
- Gertych, A.; Ing, N.; Ma, Z.; Fuchs, T.J.; Salman, S.; Mohanty, S.; Bhele, S.; Velásquez-Vacca, A.; Amin, M.B.; Knudsen, B.S. Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput. Med. Imaging Graph. 2015, 46, 197–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilstra, G.; Couture-Senécal, J.; Lau, Y.M.A.; Manning, A.M.; Wong, D.S.M.; Janaeska, W.W.; Wuraola, T.A.; Pang, J.; Khan, O.F. Iterative Design of Ionizable Lipids for Intramuscular mRNA Delivery. J. Am. Chem. Soc. 2023, 145, 2294–2304. [Google Scholar] [CrossRef]
- Wood, H. FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat. Rev. Neurol. 2018, 14, 570. [Google Scholar] [CrossRef] [PubMed]
- Tews, D.; Brenner, R.E.; Siebert, R.; Debatin, K.-M.; Fischer-Posovszky, P.; Wabitsch, M. 20 Years with SGBS cells—A versatile in vitro model of human adipocyte biology. Int. J. Obes. 2022, 46, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
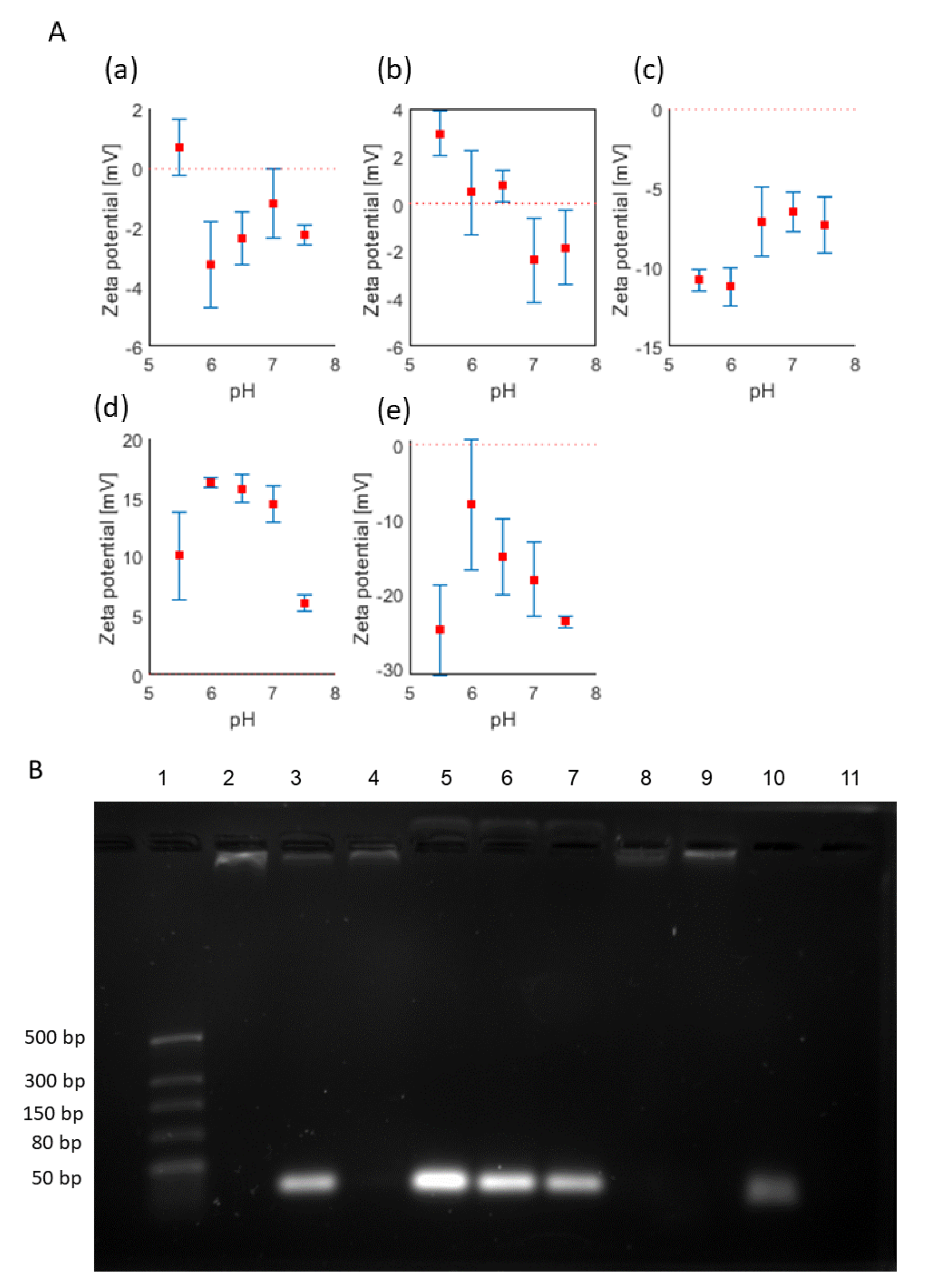




| Sample | Size [nm] | PDI | |
|---|---|---|---|
| PEG-LNP miRNA | before dialysis | 105.6 ± 2.6 | 0.233 |
| after dialysis pH 5.5 | 113.7 ± 1.9 | 0.246 | |
| before dialysis | 87.9 ± 2.3 | 0.102 | |
| after dialysis pH 7.5 | 81.5 ± 1.8 | 0.094 | |
| PEG-LNP empty | before dialysis | 109.6 ± 2.0 | 0.356 |
| after dialysis pH 5.5 | 111.4 ± 1.6 | 0.375 | |
| before dialysis | 87.9 ± 2.3 | 0.564 | |
| after dialysis pH 7.5 | 93.4 ± 1.8 | 0.441 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schachner-Nedherer, A.-L.; Fuchs, J.; Vidakovic, I.; Höller, O.; Schratter, G.; Almer, G.; Fröhlich, E.; Zimmer, A.; Wabitsch, M.; Kornmueller, K.; et al. Lipid Nanoparticles as a Shuttle for Anti-Adipogenic miRNAs to Human Adipocytes. Pharmaceutics 2023, 15, 1983. https://doi.org/10.3390/pharmaceutics15071983
Schachner-Nedherer A-L, Fuchs J, Vidakovic I, Höller O, Schratter G, Almer G, Fröhlich E, Zimmer A, Wabitsch M, Kornmueller K, et al. Lipid Nanoparticles as a Shuttle for Anti-Adipogenic miRNAs to Human Adipocytes. Pharmaceutics. 2023; 15(7):1983. https://doi.org/10.3390/pharmaceutics15071983
Chicago/Turabian StyleSchachner-Nedherer, Anna-Laurence, Julia Fuchs, Ivan Vidakovic, Oliver Höller, Gebhard Schratter, Gunter Almer, Eleonore Fröhlich, Andreas Zimmer, Martin Wabitsch, Karin Kornmueller, and et al. 2023. "Lipid Nanoparticles as a Shuttle for Anti-Adipogenic miRNAs to Human Adipocytes" Pharmaceutics 15, no. 7: 1983. https://doi.org/10.3390/pharmaceutics15071983
APA StyleSchachner-Nedherer, A.-L., Fuchs, J., Vidakovic, I., Höller, O., Schratter, G., Almer, G., Fröhlich, E., Zimmer, A., Wabitsch, M., Kornmueller, K., & Prassl, R. (2023). Lipid Nanoparticles as a Shuttle for Anti-Adipogenic miRNAs to Human Adipocytes. Pharmaceutics, 15(7), 1983. https://doi.org/10.3390/pharmaceutics15071983









