The Efficacy and Safety of Polyethylene Glycol Cholesterol- and Tocopherol Polyethylene Glycol 1000 Succinate-Modified Transforming Growth Factor β1 Small Interfering RNA Lipid Nanoparticles in the Treatment of Paclitaxel-Resistant Non-Small-Cell Lung Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of TGFβ1 siRNA LNPs
2.3. Particle Morphology, Size Distribution, and Zeta Potential of TGFβ1 siRNA LNPs
2.4. Detection of the Contents and Encapsulation Efficiencies of siRNA in TGFβ1 siRNA LNPs
2.5. Inhibition of TGFβ1 mRNA Expression in A549/T Cell after siRNA LNP Intervention
2.6. Inhibition of TGFβ1 Protein Expression in A549/T Cell after siRNA LNP Intervention
2.7. In Vitro Proliferation Inhibition of siRNA LNP in A549/T Cells
2.8. Tissue Distributions of siRNA LNPs in Tumor-Bearing Nude Mice Observed Using an In Vivo Mice Imaging System
2.9. Pharmacodynamic Study of siRNA LNPs in Tumor-Bearing Nude Mice
3. Results and Discussion
3.1. The Results of Particle Morphology, Size Distribution, and Zeta Potential of TGFβ1 siRNA LNPs
3.2. The Results of siRNA Contents and Encapsulation Efficiencies in siRNA LNPs
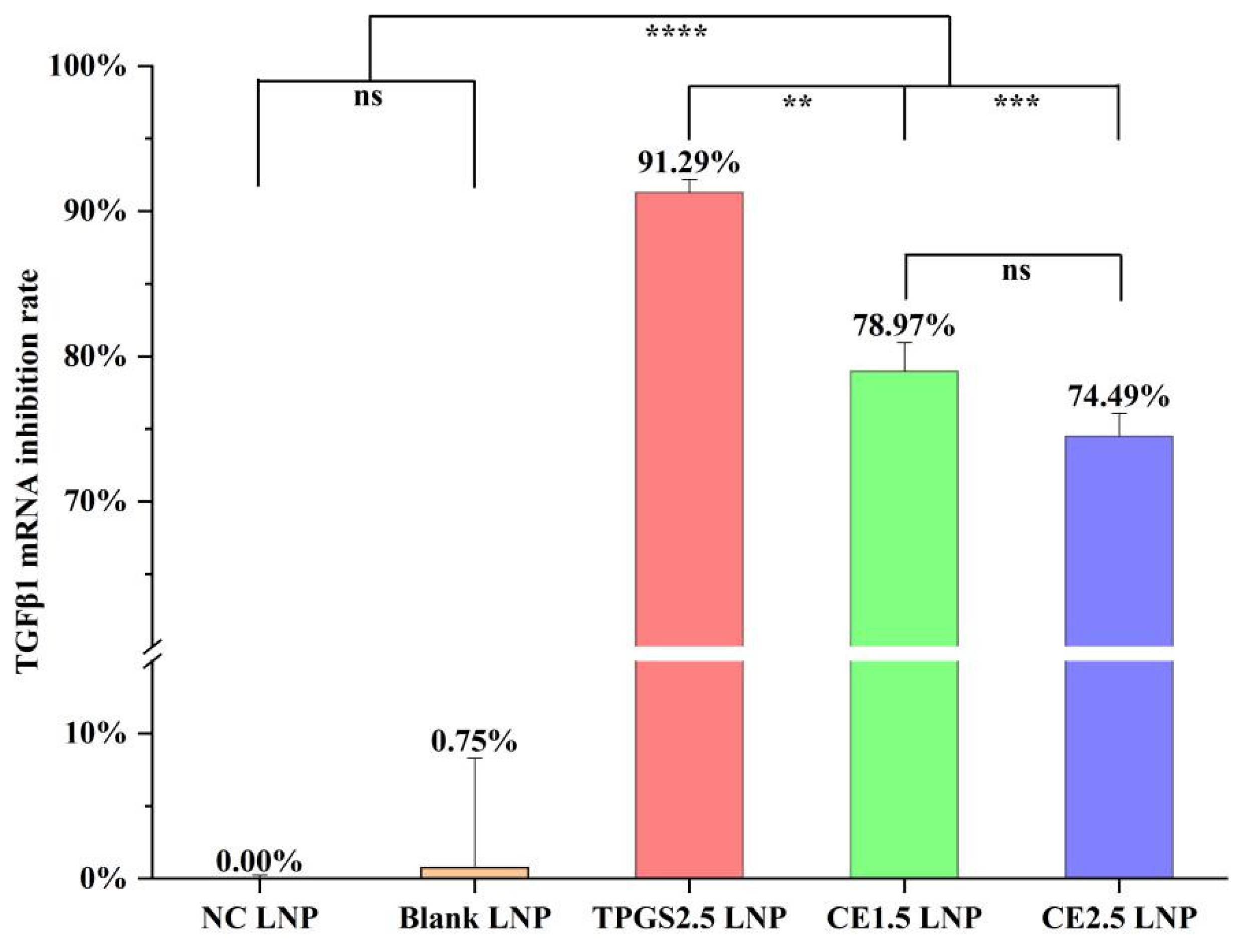
3.3. TGFβ1 mRNA Expression Inhibition by the Three siRNA LNPs
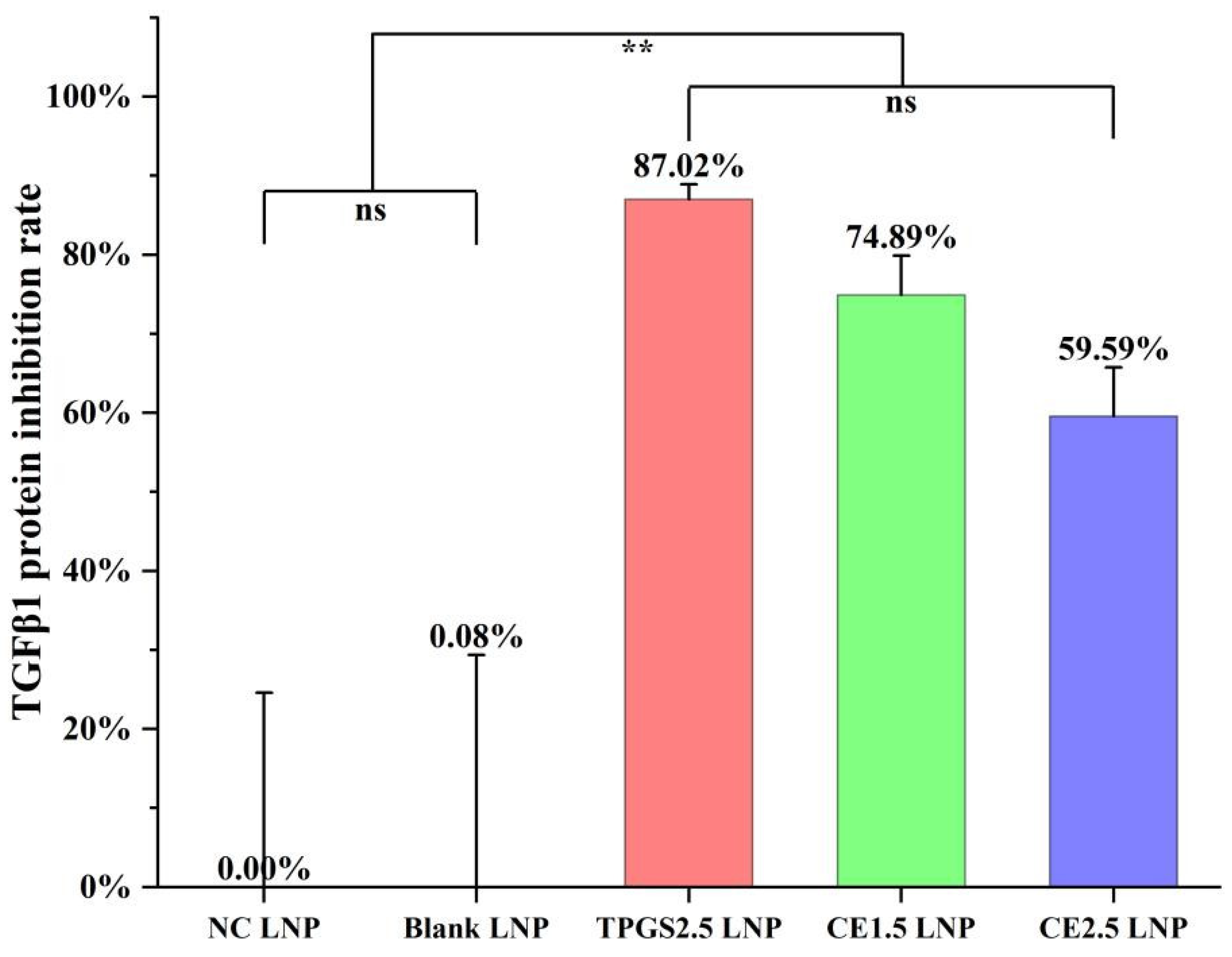
3.4. TGFβ1 Protein Expression Inhibition by the Three siRNA LNPs
3.5. The In Vitro A549/T Cell Proliferation Inhibition for siRNA LNPs
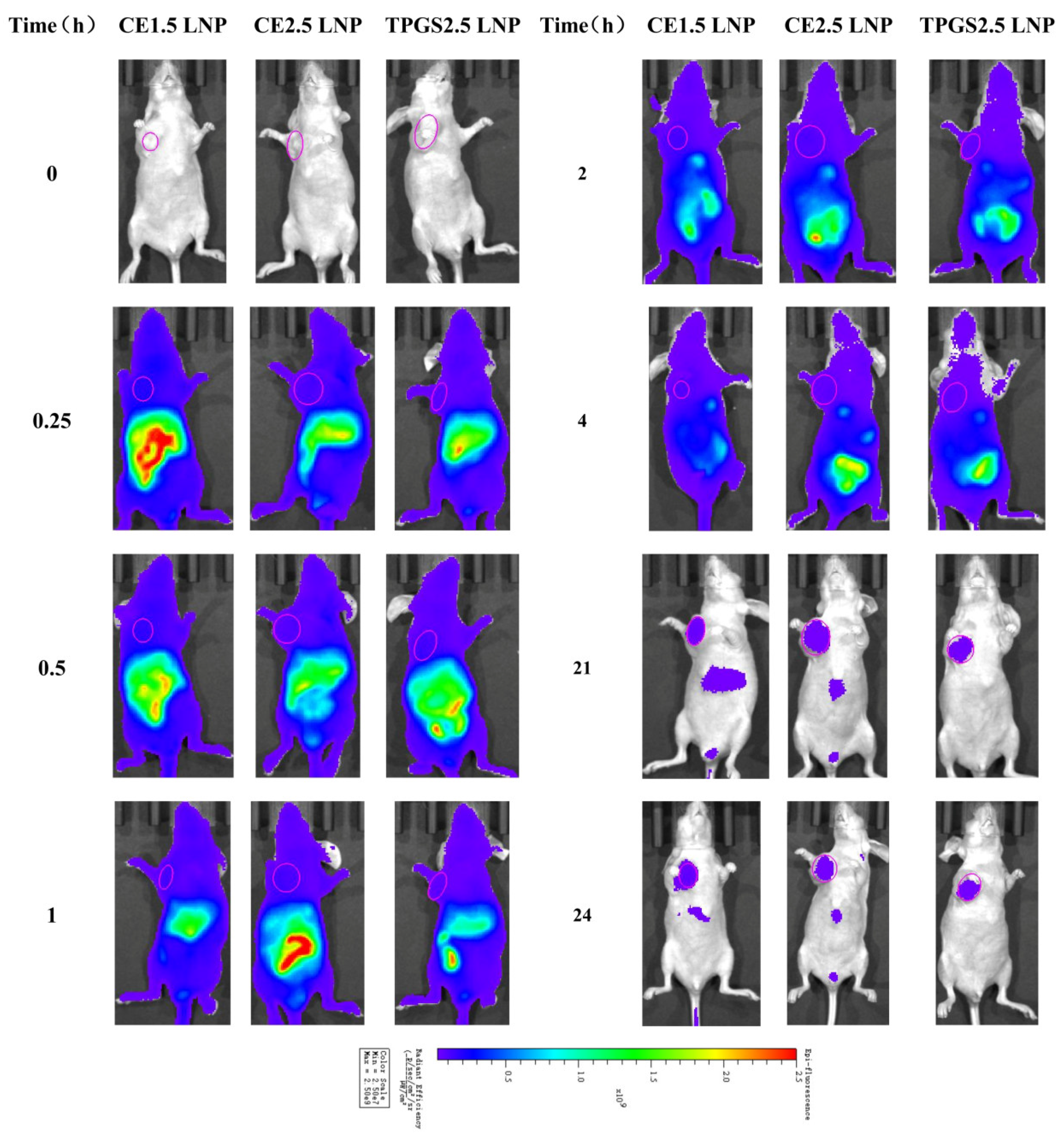
3.6. The Tissue Distribution of siRNA LNP in Tumor-Bearing Nude Mice Determined Using an In Vivo Mice Imaging System
3.6.1. Tissue Distribution Imaging of Cypate-Labeled siRNA LNPs in Tumor Tissue
3.6.2. The Fluorescence Intensity Dynamics of Cypate-Labeled siRNA LNPs in Tumor Tissue
3.7. The Results of Efficacy, Tumor Immunohistochemistry, and Organ Pathology of the Three siRNA LNPs for the Treatment of Paclitaxel-Resistant Lung Adenocarcinoma
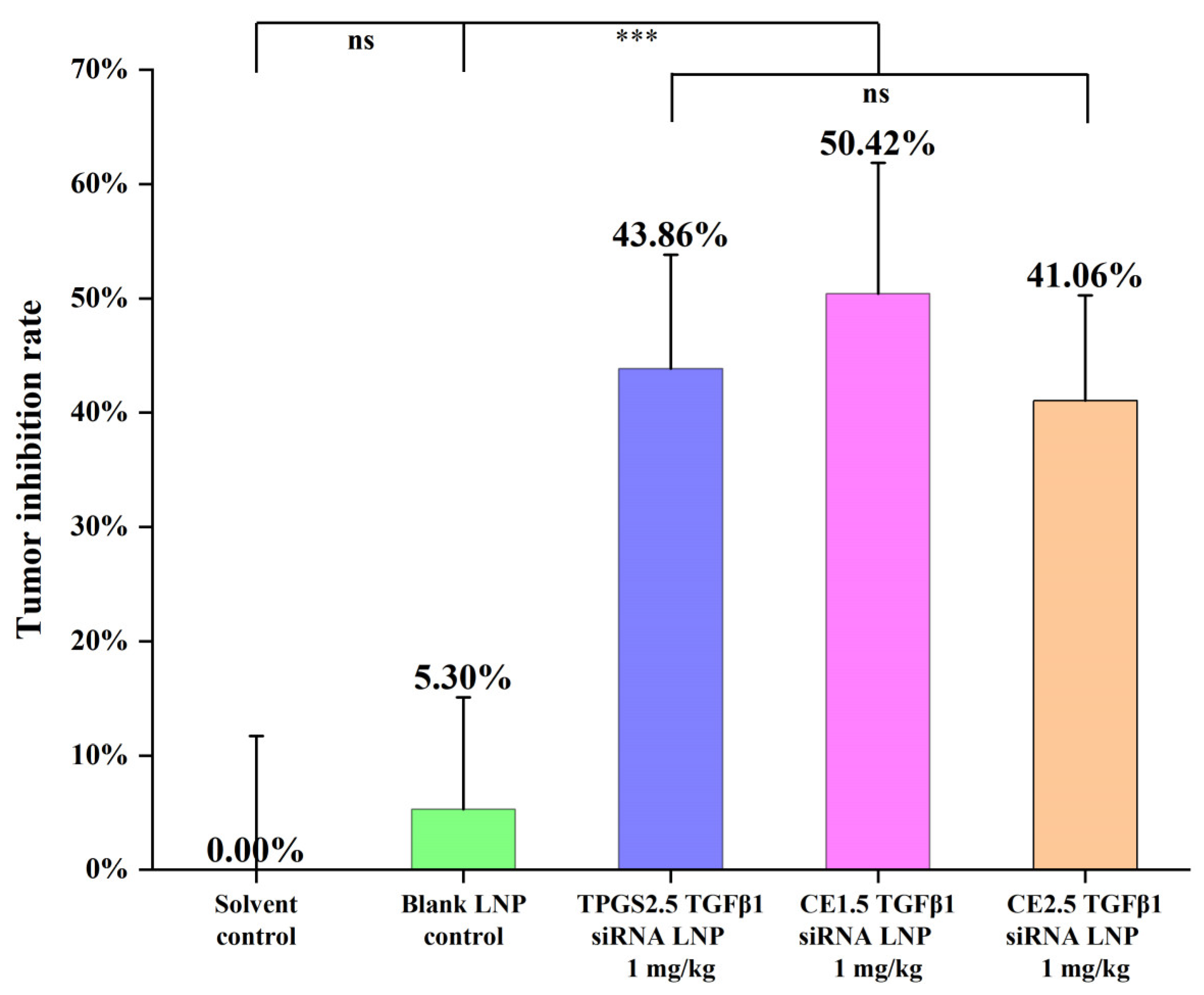
3.7.1. The Efficacies of the Three siRNA LNPs in the Treatment of Paclitaxel-Resistant Lung Adenocarcinoma
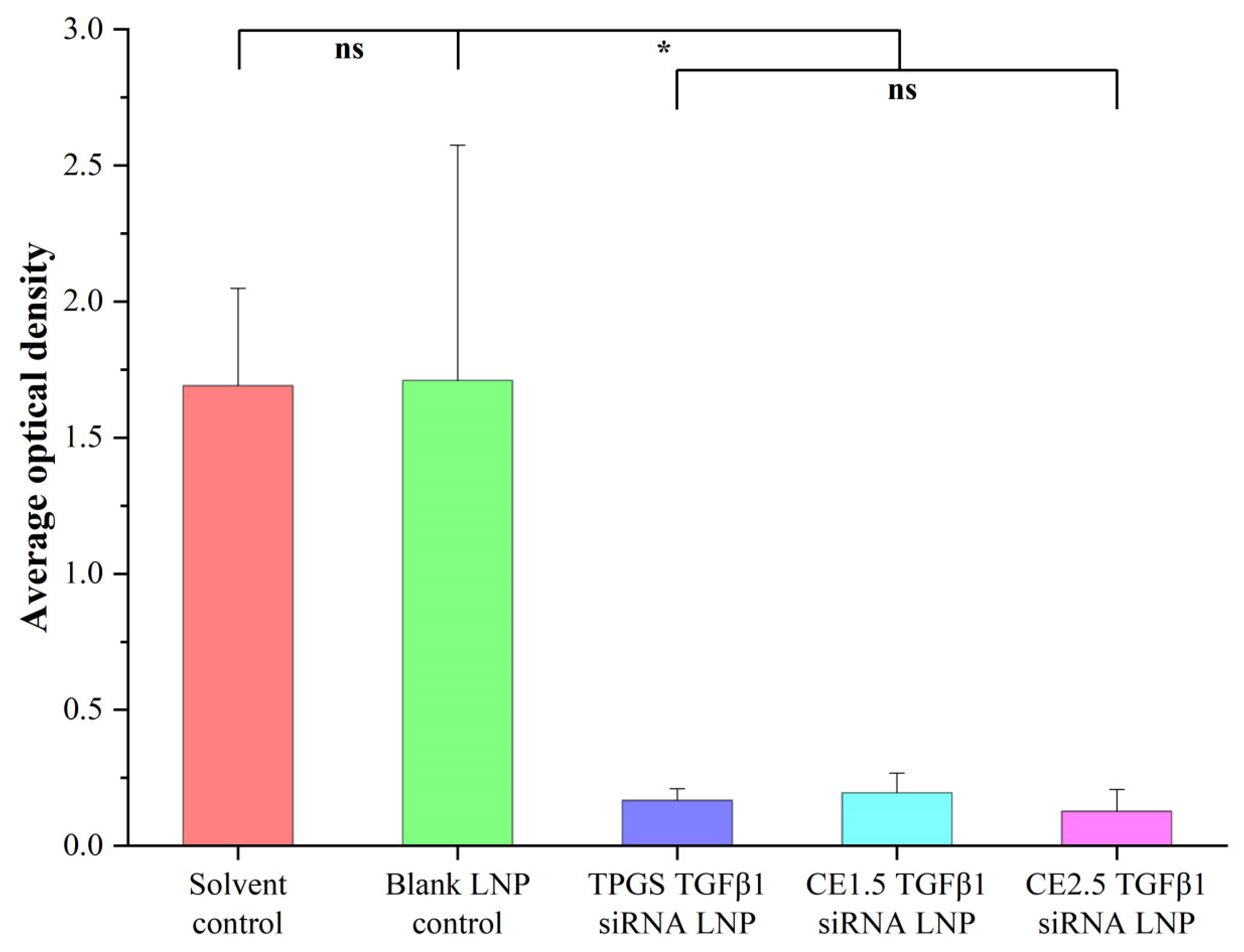
3.7.2. The Tumor Immunohistochemistry Results of the Three siRNA LNPs after the Treatment of Paclitaxel-Resistant Lung Adenocarcinoma
3.7.3. The Organ Pathology Results after the In Vivo Treatment of Paclitaxel-Resistant Lung Adenocarcinoma
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kenicer, J.; Spears, M.; Lyttle, N.; Taylor, K.J.; Liao, L.; Cunningham, C.A.; Lambros, M.; MacKay, A.; Yao, C.; Reis-Filho, J.; et al. Molecular characterisation of isogenic taxane resistant cell lines identify novel drivers of drug resistance. BMC Cancer 2014, 14, 762. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Gifford, C.C.; Samarakoon, R.; Higgins, P.J. Deregulation of negative controls on TGF-β1 signaling in tumor progression. Cancers 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Gentry, L.E.; Nash, B.W. The pro domain of pre-pro-transforming growth factor beta 1 when independently expressed is a functional binding protein for the mature growth factor. Biochemistry 1990, 29, 6851–6857. [Google Scholar] [CrossRef] [PubMed]
- Martelossi Cebinelli, G.C.; Paiva Trugilo, K.; Badaró Garcia, S.; Brajão de Oliveira, K. TGF-β1 functional polymorphisms: A review. Eur. Cytokine Netw. 2016, 27, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Lin, C.; Li, X.; Liang, Y.; Saw, P.E. TME-Responsive multistage nanoplatform for siRNA delivery and effective cancer therapy. Int. J. Nanomed. 2021, 16, 5909–5921. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Li, X.; Jiang, Y.; Ge, Z. Blocking TGF-β expression attenuates tumor growth in lung cancers, potentially mediated by skewing development of neutrophils. J. Oncol. 2022, 2022, 3447185. [Google Scholar] [CrossRef]
- Friedrich, M.; Aigner, A. Therapeutic siRNA: State-of-the-Art and future perspectives. BioDrugs 2022, 36, 549–571. [Google Scholar] [CrossRef]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Wood, H. FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat. Rev. Neurol. 2018, 14, 570. [Google Scholar] [CrossRef]
- Jyotsana, N.; Sharma, A.; Chaturvedi, A.; Budida, R.; Scherr, M.; Kuchenbauer, F.; Lindner, R.; Noyan, F.; Sühs, K.W.; Stangel, M.; et al. Lipid nanoparticle-mediated siRNA delivery for safe targeting of human CML in vivo. Ann. Hematol. 2019, 98, 1905–1918. [Google Scholar] [CrossRef]
- Van Hoeck, J.; Braeckmans, K.; De Smedt, S.C.; Raemdonck, K. Non-viral siRNA delivery to T cells: Challenges and opportunities in cancer immunotherapy. Biomaterials 2022, 286, 121510. [Google Scholar] [CrossRef] [PubMed]
- Chernikov, I.V.; Ponomareva, U.A.; Chernolovskaya, E.L. Structural modifications of siRNA improve its performance in vivo. Int. J. Mol. Sci. 2023, 24, 956. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huang, L.; Yang, M. Lipid-based vehicles for siRNA delivery in biomedical field. Curr. Pharm. Biotechnol. 2020, 21, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.T.; Wang, X.; Liu, Y.; Yang, G.Z.; Falconer, R.J.; Zhao, C.X. Lipid nanoparticles for drug delivery. Adv. Nanobiomed. Res. 2022, 2, 2100109. [Google Scholar] [CrossRef]
- Van der Meel, R.; Chen, S.; Zaifman, J.; Kulkarni, J.A.; Zhang, X.R.S.; Tam, Y.K.; Bally, M.B.; Schiffelers, R.M.; Ciufolini, M.A.; Cullis, P.R.; et al. Modular lipid nanoparticle platform technology for siRNA and lipophilic prodrug delivery. Small 2021, 17, e2103025. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Witzigmann, D.; Leung, J.; Tam, Y.Y.C.; Cullis, P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale 2019, 11, 21733–21739. [Google Scholar] [CrossRef] [PubMed]
- Ferraresso, F.; Strilchuk, A.W.; Juang, L.J.; Poole, L.G.; Luyendyk, J.P.; Kastrup, C.J. Comparison of DLin-MC3-DMA and ALC-0315 for siRNA delivery to hepatocytes and hepatic stellate cells. Mol. Pharm. 2022, 19, 2175–2182. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef]
- Eygeris, Y.; Patel, S.; Jozic, A.; Sahay, G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020, 20, 4543–4549. [Google Scholar] [CrossRef]
- Maeki, M.; Okada, Y.; Uno, S.; Niwa, A.; Ishida, A.; Tani, H.; Tokeshi, M. Production of siRNA-loaded lipid nanoparticles using a microfluidic device. J. Vis. Exp. 2022, 181, e62999. [Google Scholar] [CrossRef]
- Eygeris, Y.; Gupta, M.; Kim, J.; Sahay, G. Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 2022, 55, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Subhan, M.A.; Torchilin, V.P. siRNA based drug design, quality, delivery and clinical translation. Nanomedicine 2020, 29, 102239. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Lin, J.; Huang, Y.; Li, L.; Delcassian, D.; Ge, Y.; Shi, Y.; Anderson, D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020, 11, 2424. [Google Scholar] [CrossRef]
- Coelho, T.; Adams, D.; Silva, A.; Lozeron, P.; Hawkins, P.N.; Mant, T.; Perez, J.; Chiesa, J.; Warrington, S.; Tranter, E.; et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013, 369, 819–829. [Google Scholar] [CrossRef]
- Segalla, G. Apparent cytotoxicity and intrinsic cytotoxicity of lipid nanomaterials contained in a COVID-19 mRNA vaccine. Int. J. Vaccine Theory Pract. Res. 2023, 3, 957–972. [Google Scholar] [CrossRef]
- Wilhelmy, C.; Keil, I.S.; Uebbing, L.; Schroer, M.A.; Franke, D.; Nawroth, T.; Barz, M.; Sahin, U.; Haas, H.; Diken, M.; et al. Polysarcosine-functionalized mRNA lipid nanoparticles tailored for immunotherapy. Pharmaceutics 2023, 15, 2068. [Google Scholar] [CrossRef]
- Tan, T.; Feng, Y.; Wang, W.; Wang, R.; Yin, L.; Zeng, Y.; Zeng, Z.; Xie, T. Cabazitaxel-loaded human serum albumin nanoparticles combined with TGFβ-1 siRNA lipid nanoparticles for the treatment of paclitaxel-resistant non-small cell lung cancer. Cancer Nano 2023, 14, 70. [Google Scholar] [CrossRef]
- Kalita, T.; Dezfouli, S.A.; Pandey, L.M.; Uludag, H. siRNA functionalized lipid nanoparticles (LNPs) in management of diseases. Pharmaceutics 2022, 14, 2520. [Google Scholar] [CrossRef] [PubMed]
- Niemietz, C.; Nadzemova, O.; Zibert, A.; Schmidt, H.H. APOE polymorphism in ATTR amyloidosis patients treated with lipid nanoparticle siRNA. Amyloid 2020, 27, 45–51. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Darjuan, M.M.; Mercer, J.E.; Chen, S.; van der Meel, R.; Thewalt, J.L.; Tam, Y.Y.C.; Cullis, P.R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano 2018, 12, 4787–4795. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Leung, J.; van der Meel, R.; Zaifman, J.; Darjuan, M.M.; Grisch-Chan, H.M.; Thöny, B.; Tam, Y.Y.C.; Cullis, P.R. Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale 2019, 11, 9023–9031. [Google Scholar] [CrossRef] [PubMed]
- Charbe, N.B.; Amnerkar, N.D.; Ramesh, B.; Tambuwala, M.M.; Bakshi, H.A.; Aljabali, A.A.A.; Khadse, S.C.; Satheeshkumar, R.; Satija, S.; Metha, M.; et al. Small interfering RNA for cancer treatment: Overcoming hurdles in delivery. Acta Pharm. Sin. B 2020, 10, 2075–2109. [Google Scholar] [CrossRef] [PubMed]
- Sasayama, Y.; Hasegawa, M.; Taguchi, E.; Kubota, K.; Kuboyama, T.; Naoi, T.; Yabuuchi, H.; Shimai, N.; Asano, M.; Tokunaga, A.; et al. In vivo activation of PEGylated long circulating lipid nanoparticle to achieve efficient siRNA delivery and target gene knock down in solid tumors. J. Control. Release 2019, 311–312, 245–256. [Google Scholar] [CrossRef]
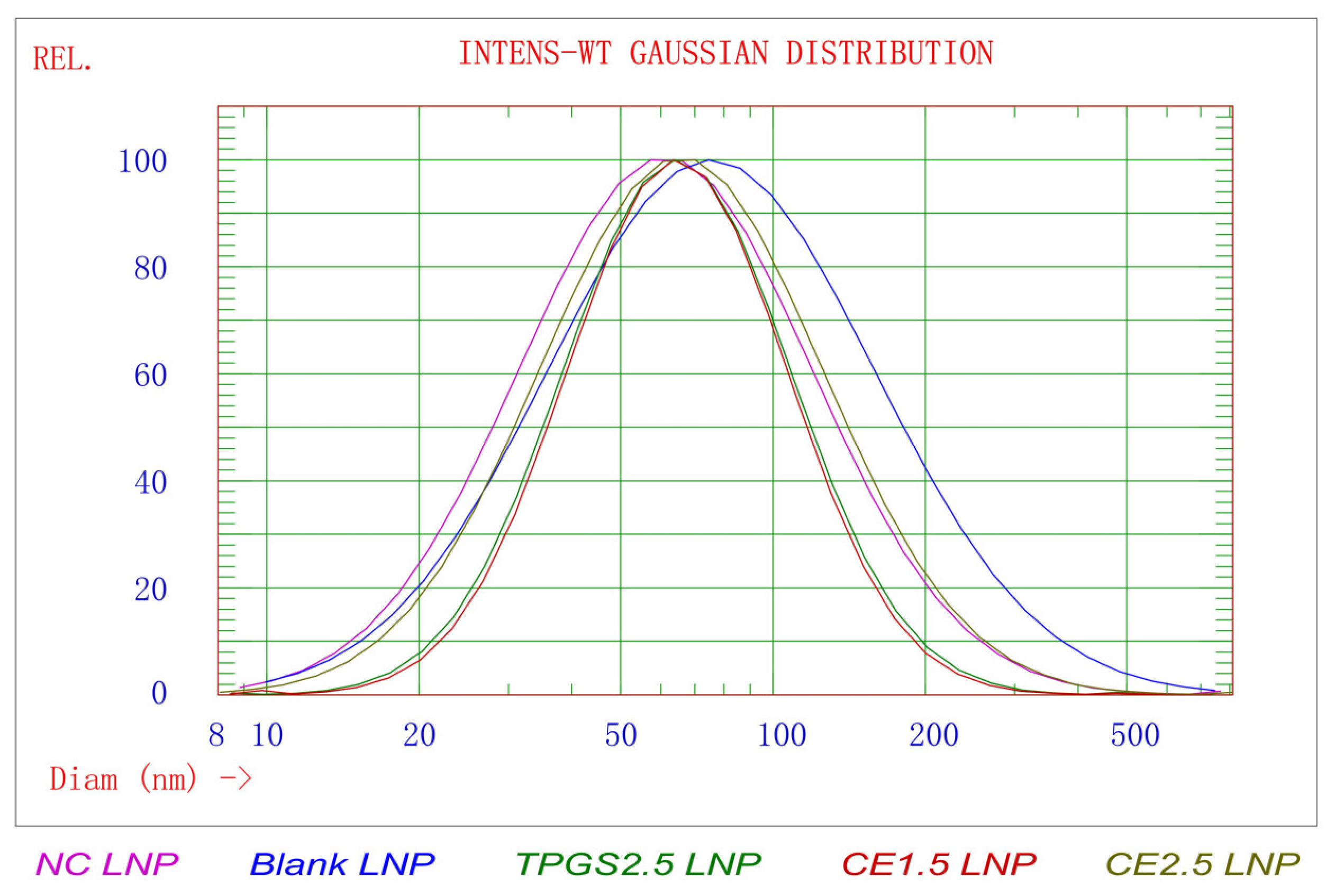


| LNPs | Size (nm) | STD * (nm) | PDI ** | 90% of Distribution < (nm) | Zeta Potential in pH 4 Buffer (mV) | Zeta Potential in pH 7.4 Buffer (mV) |
|---|---|---|---|---|---|---|
| CE1.5 LNP | 74.0 ± 1.6 | 37.6 ± 2.1 | 0.259 ± 0.018 | 132.9 ± 5.3 | 11.71 ± 0.58 | −6.27 ± 0.54 |
| CE2.5 LNP | 81.9 ± 1.9 | 53.2 ± 3.2 | 0.422 ± 0.032 | 169.1 ± 7.9 | 7.73 ± 0.91 | −8.76 ± 0.39 |
| TPGS2.5 LNP | 73.9 ± 0.5 | 38.4 ± 0.6 | 0.270 ± 0.005 | 134.3 ± 1.5 | 45.06 ± 2.05 | −5.11 ± 0.63 |
| NC LNP | 77.1 ± 1.1 | 50.3 ± 0.6 | 0.426 ± 0.020 | 159.9 ± 0.9 | 5.71 ± 0.23 | −9.91 ± 0.73 |
| Blank LNP | 98.5 ± 7.2 | 72.7 ± 6.4 | 0.544 ± 0.018 | 221.1 ± 18.3 | 8.09 ± 0.89 | −6.32 ± 0.31 |
| Parameter | Units | CE1.5 LNP | CE2.5 LNP | TPGS2.5 LNP | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| AUC(0–t) | AFE × h | 1.42 × 109 | 3.04 × 108 | 1.39 × 109 | 1.20 × 108 | 1.22 × 109 | 1.44 × 108 |
| AUC(0–∞) | AFE × h | 2.33 × 109 | 1.33 × 109 | 1.69 × 109 | 1.76 × 108 | 1.48 × 109 | 1.74 × 108 |
| R_AUC(t/∞) | % | 69.68 | 19.671 | 82.7 | 7.276 | 82.82 | 6.687 |
| AUMC(0–t) | h × h × AFE | 1.11 × 1010 | 3.93 × 109 | 8.86 × 109 | 5.78 × 108 | 8.04 × 109 | 7.99 × 108 |
| AUMC(0–∞) | h × h × AFE | 7.16 × 1010 | 1.01 × 1011 | 2.07 × 1010 | 6.60 × 109 | 1.84 × 1010 | 6.44 × 109 |
| MRT(0–t) | h | 7.66 | 1.28 | 6.395 | 0.657 | 6.604 | 0.724 |
| MRT(0–∞) | h | 22.703 | 18.142 | 12.093 | 3.282 | 12.308 | 3.553 |
| VRT(0–t) | h × h | 62.863 | 11.637 | 49.612 | 6.999 | 52.792 | 8.885 |
| VRT(0–∞) | h × h | 896.19 | 1316.742 | 233.055 | 107.471 | 241.478 | 129.526 |
| λz | 1/h | 0.055 | 0.026 | 0.073 | 0.017 | 0.071 | 0.015 |
| C_last | AFE | 2.90 × 107 | 1.43 × 107 | 2.00 × 107 | 5.75 × 106 | 1.70 × 107 | 3.36 × 106 |
| t1/2z | h | 17.001 | 12.691 | 9.917 | 2.327 | 10.218 | 2.709 |
| Tmax | h | 0.9 | 0.224 | 0.8 | 0.274 | 0.6 | 0.224 |
| Vz | AFE | 20.19 | 4.016 | 16.906 | 3.674 | 19.88 | 4.436 |
| CLz | AFE | 1.049 | 0.456 | 1.193 | 0.119 | 1.364 | 0.166 |
| Cmax | AFE × h | 1.41 × 108 | 3.21 × 107 | 1.51 × 108 | 2.45 × 107 | 1.32 × 108 | 1.38 × 107 |
| C0 | AFE × h | 1.35 × 108 | 6.63 × 107 | 1.14 × 108 | 5.13 × 107 | 1.73 × 108 | 8.36 × 107 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Z.; Zeng, X.; Li, X.; Feng, Y.; Kan, Y.; Liu, X.; Zeng, Y. The Efficacy and Safety of Polyethylene Glycol Cholesterol- and Tocopherol Polyethylene Glycol 1000 Succinate-Modified Transforming Growth Factor β1 Small Interfering RNA Lipid Nanoparticles in the Treatment of Paclitaxel-Resistant Non-Small-Cell Lung Cancer. Pharmaceutics 2024, 16, 75. https://doi.org/10.3390/pharmaceutics16010075
Zeng Z, Zeng X, Li X, Feng Y, Kan Y, Liu X, Zeng Y. The Efficacy and Safety of Polyethylene Glycol Cholesterol- and Tocopherol Polyethylene Glycol 1000 Succinate-Modified Transforming Growth Factor β1 Small Interfering RNA Lipid Nanoparticles in the Treatment of Paclitaxel-Resistant Non-Small-Cell Lung Cancer. Pharmaceutics. 2024; 16(1):75. https://doi.org/10.3390/pharmaceutics16010075
Chicago/Turabian StyleZeng, Zhaowu, Xianglong Zeng, Xinyi Li, Yuxin Feng, Yue Kan, Xingyan Liu, and Yiying Zeng. 2024. "The Efficacy and Safety of Polyethylene Glycol Cholesterol- and Tocopherol Polyethylene Glycol 1000 Succinate-Modified Transforming Growth Factor β1 Small Interfering RNA Lipid Nanoparticles in the Treatment of Paclitaxel-Resistant Non-Small-Cell Lung Cancer" Pharmaceutics 16, no. 1: 75. https://doi.org/10.3390/pharmaceutics16010075
APA StyleZeng, Z., Zeng, X., Li, X., Feng, Y., Kan, Y., Liu, X., & Zeng, Y. (2024). The Efficacy and Safety of Polyethylene Glycol Cholesterol- and Tocopherol Polyethylene Glycol 1000 Succinate-Modified Transforming Growth Factor β1 Small Interfering RNA Lipid Nanoparticles in the Treatment of Paclitaxel-Resistant Non-Small-Cell Lung Cancer. Pharmaceutics, 16(1), 75. https://doi.org/10.3390/pharmaceutics16010075






