Sucrose Treatment Enhances the Electrotransfer of DNA by Activating Phospholipase A2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Preparation
2.2. Plasmid Preparation
2.3. Electrotransfer of Plasmid
2.4. Inhibitor Treatment
2.5. RNA Sequencing and Pathway Analysis
2.6. Flow Cytometry Analysis
2.7. Statistical Analyses
3. Results
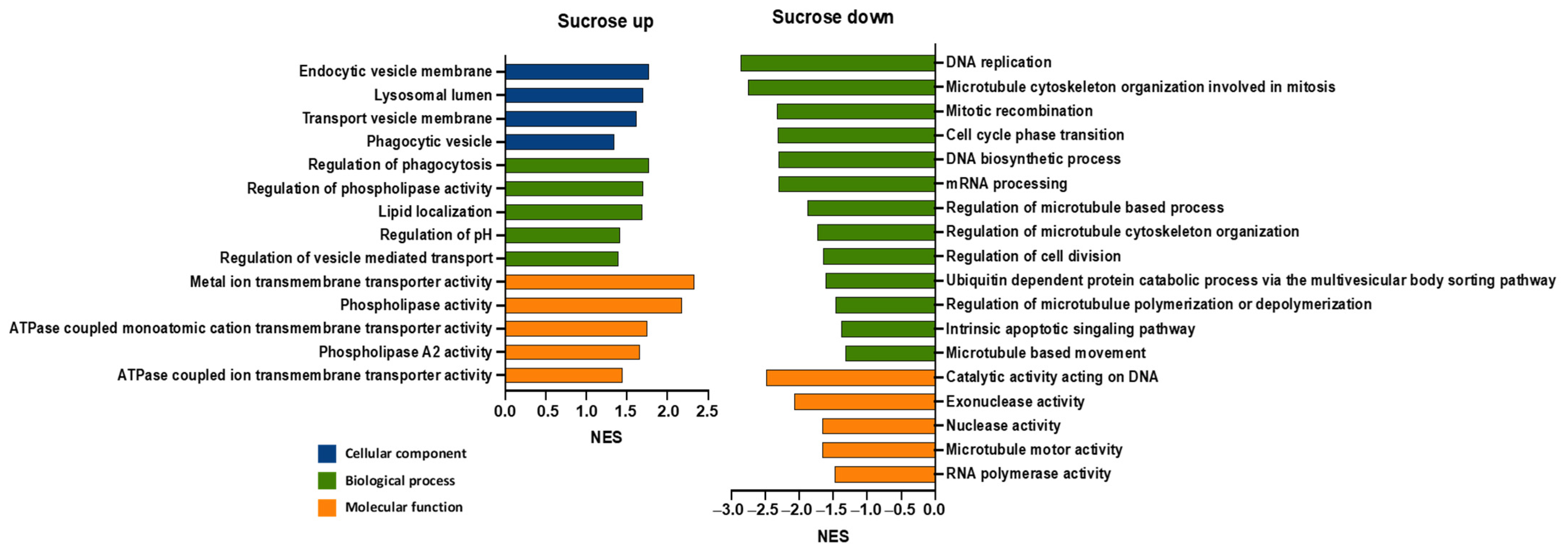
3.1. Sucrose Induces Differential Expression of Genes in Mammalian Cells
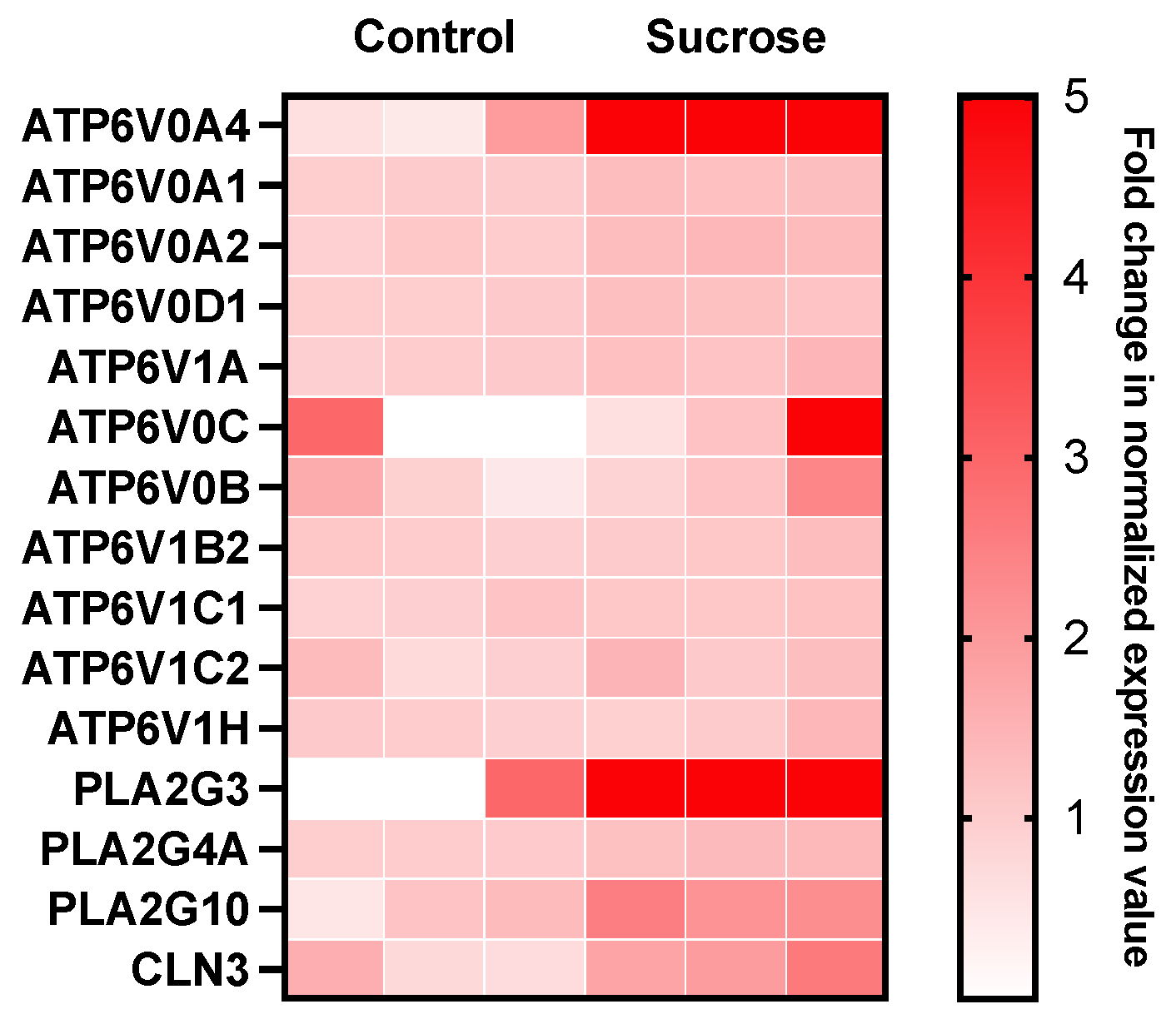
3.2. Sucrose Activates Phospholipase A2 and V-ATPase in Mammalian Cells
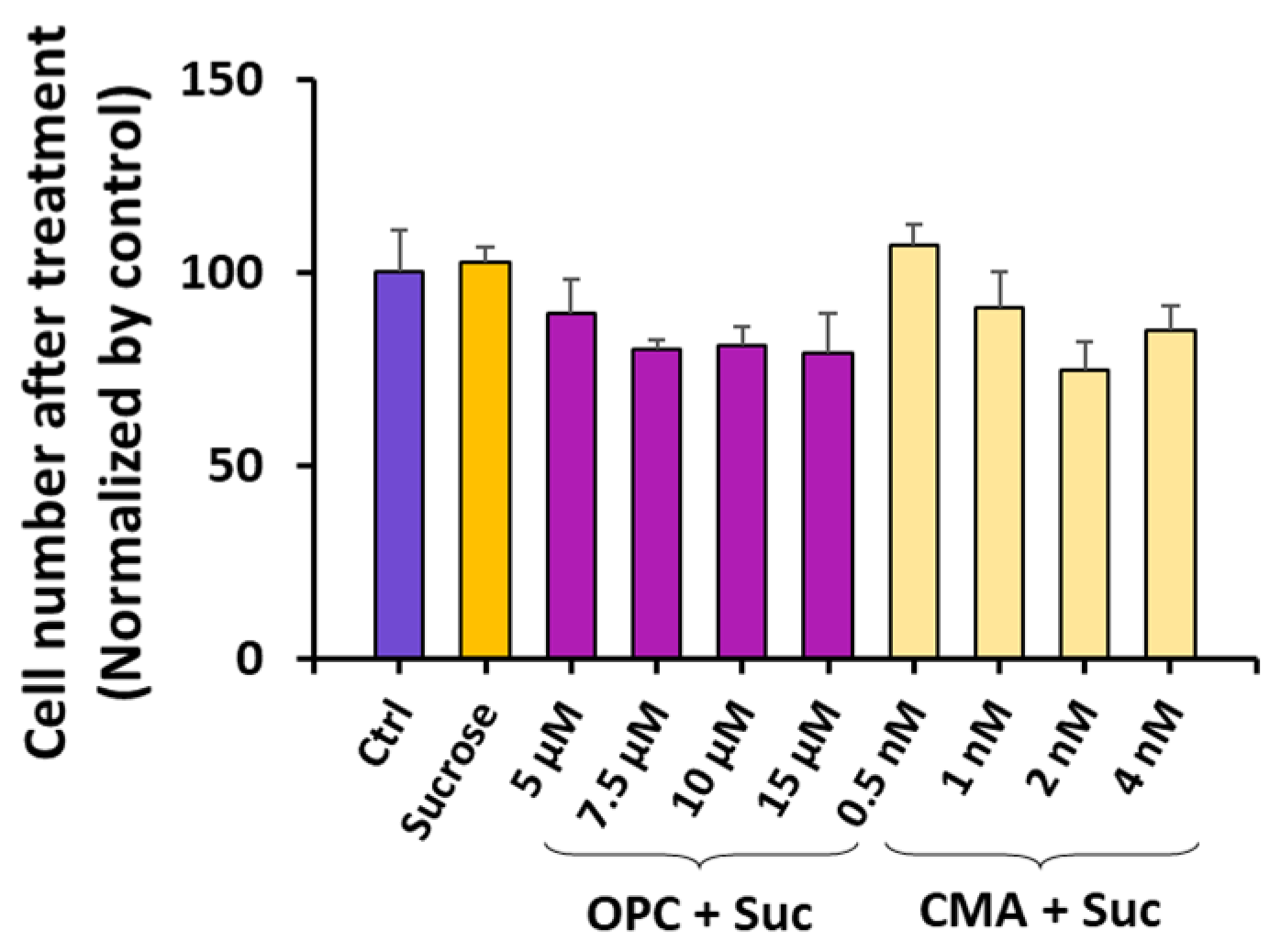
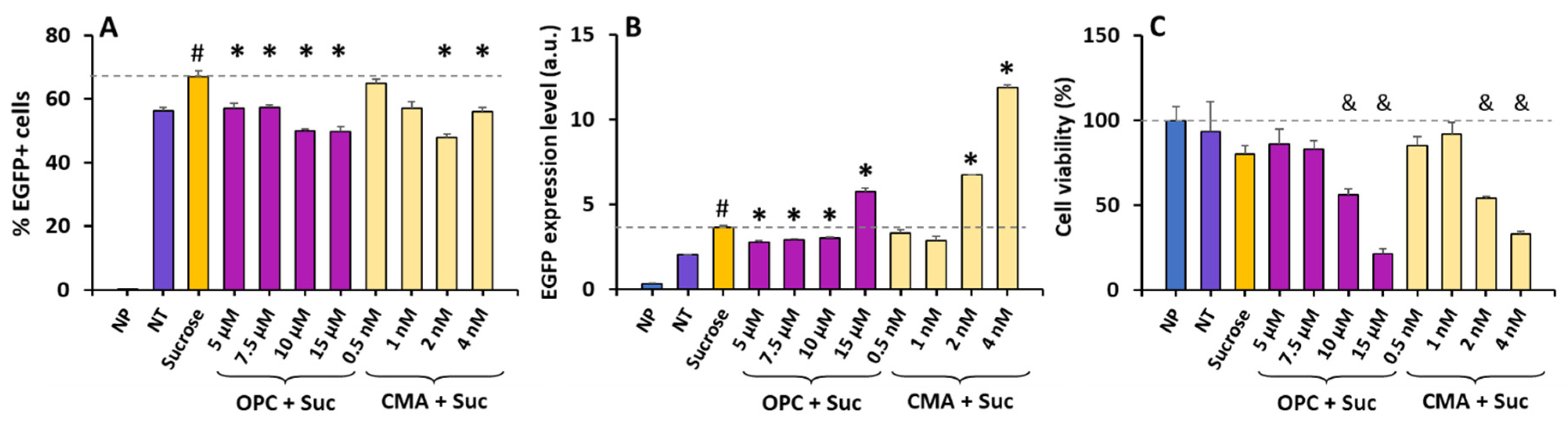
3.3. The Effects of Sucrose on ET Can Be Reversed via Inhibition of Phospholipase A2
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rumsey, S.C.; Galeano, N.F.; Arad, Y.; Deckelbaum, R.J. Cryopreservation with sucrose maintains normal physical and biological properties of human plasma low density lipoproteins. J. Lipid Res. 1992, 33, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, R.; Hubel, A. Interfacial interactions of sucrose during cryopreservation detected by Raman spectroscopy. Langmuir 2018, 35, 7388–7395. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.A.; Davidovich, I.; Yang, Y.; Lai, A.; Goncalves, J.P.; Deliwala, V.; Busatto, S.; Shapiro, S.; Koifman, N.A.; Salomon, C. Sucrose-based cryoprotective storage of extracellular vesicles. Extracell. Vesicle 2022, 1, 100016. [Google Scholar] [CrossRef]
- Fabbri, R.; Pasquinelli, G.; Keane, D.; Magnani, V.; Paradisi, R.; Venturoli, S. Optimization of protocols for human ovarian tissue cryopreservation with sucrose, 1, 2-propanediol and human serum. Reprod. Biomed. Online 2010, 21, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Marsella, T.; Sena, P.; Xella, S.; La Marca, A.; Giulini, S.; De Pol, A.; Volpe, A.; Marzona, L. Human ovarian tissue cryopreservation: Effect of sucrose concentration on morphological features after thawing. Reprod. Biomed. Online 2008, 16, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Hosn, R.R.; Remba, T.; Yun, D.; Li, N.; Abraham, W.; Melo, M.B.; Cortes, M.; Li, B.; Zhang, Y. Optimization of storage conditions for lipid nanoparticle-formulated self-replicating RNA vaccines. J. Control. Release 2023, 353, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Pelliccia, M.; Andreozzi, P.; Paulose, J.; D’Alicarnasso, M.; Cagno, V.; Donalisio, M.; Civra, A.; Broeckel, R.M.; Haese, N.; Jacob Silva, P. Additives for vaccine storage to improve thermal stability of adenoviruses from hours to months. Nat. Commun. 2016, 7, 13520. [Google Scholar] [CrossRef]
- Valkama, A.J.; Oruetxebarria, I.; Lipponen, E.M.; Leinonen, H.M.; Käyhty, P.; Hynynen, H.; Turkki, V.; Malinen, J.; Miinalainen, T.; Heikura, T. Development of large-scale downstream processing for lentiviral vectors. Mol. Ther. Methods Clin. Dev. 2020, 17, 717–730. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Urru, S.A.; Maines, E.; Campomori, A.; Soffiati, M. Safety of SARS-CoV-2 vaccines administration for adult patients with hereditary fructose intolerance. Hum. Vaccines Immunother. 2021, 17, 4112–4114. [Google Scholar] [CrossRef]
- Graham, J.M. Isolation of Golgi membranes from tissues and cells by differential and density gradient centrifugation. Curr. Protoc. Cell Biol. 2001, 10. Chapter 3: Unit 3.9. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Proteomic Profiling Methods Protoc. 2015, 1295, 179–209. [Google Scholar]
- Williams, H.D.; Ward, R.; Hardy, I.J.; Melia, C.D. The effect of sucrose and salts in combination on the drug release behaviour of an HPMC matrix. Eur. J. Pharm. Biopharm. 2010, 76, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Taketo, A. Properties of Electroporation-Mediated DNA Transfer in Escherichia coil. J. Biochem. 1989, 105, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Rubio, T.; Terefe, J. High Efficiency Electroporation Buffer. U.S. Patent US20080268542A1, 30 October 2008. [Google Scholar]
- Sherba, J.J.; Hogquist, S.; Lin, H.; Shan, J.W.; Shreiber, D.I.; Zahn, J.D. The effects of electroporation buffer composition on cell viability and electro-transfection efficiency. Sci. Rep. 2020, 10, 3053. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Nihonyanagi, H.; Numano, R.; Shibata, T.; Takashima, K.; Kurita, H. Influence of electroporation medium on delivery of cell-impermeable small molecules by electrical short-circuiting via an aqueous droplet in dielectric oil: A comparison of different fluorescent tracers. Sensors 2022, 22, 2494. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.C. Electroporation of cells and tissues. IEEE Trans. Plasma Sci. 2000, 28, 24–33. [Google Scholar] [CrossRef]
- Kotnik, T.; Frey, W.; Sack, M.; Meglič, S.H.; Peterka, M.; Miklavčič, D. Electroporation-based applications in biotechnology. Trends Biotechnol. 2015, 33, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.C. Electroporation: A general phenomenon for manipulating cells and tissues. J. Cell. Biochem. 1993, 51, 426–435. [Google Scholar] [CrossRef]
- Heller, L.; Thinard, R.; Chevalier, M.; Arpag, S.; Jing, Y.; Greferath, R.; Heller, R.; Nicolau, C. Secretion of proteins and antibody fragments from transiently transfected endothelial progenitor cells. J. Cell. Mol. Med. 2020, 24, 8772–8778. [Google Scholar] [CrossRef]
- Bolhassani, A.; Khavari, A.; Orafa, Z. Electroporation-advantages and drawbacks for delivery of drug, gene and vaccine. In Application of Nanotechnology in Drug Delivery; IntechOpen: London, UK, 2014; pp. 369–397. [Google Scholar]
- Jordan, E.T.; Collins, M.; Terefe, J.; Ugozzoli, L.; Rubio, T. Optimizing electroporation conditions in primary and other difficult-to-transfect cells. J. Biomol. Tech. JBT 2008, 19, 328. [Google Scholar]
- Mao, M.; Chang, C.C.; Pickar-Oliver, A.; Cervia, L.D.; Wang, L.; Ji, J.; Liton, P.B.; Gersbach, C.A.; Yuan, F. Redirecting vesicular transport to improve nonviral delivery of molecular cargo. Adv. Biosyst. 2020, 4, 2000059. [Google Scholar] [CrossRef]
- Cervia, L.D. Enhancement of Electrotransfection Efficiency through Understanding of Underlying Biological Mechanisms. Ph.D. Thesis, Duke University, Durham, NC, USA, 2017. [Google Scholar]
- Wang, C.; Karlsson, A.; Oguin III, T.H.; Macintyre, A.N.; Sempowski, G.D.; McCarthy, K.R.; Wang, Y.; Moody, M.A.; Yuan, F. Transient inhibition of lysosomal functions potentiates nucleic acid vaccines. Proc. Natl. Acad. Sci. USA 2023, 120, e2306465120. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Cervia, L.D.; Chang, C.C.; Wang, L.; Yuan, F. Distinct effects of endosomal escape and inhibition of endosomal trafficking on gene delivery via electrotransfection. PLoS ONE 2017, 12, e0171699. [Google Scholar] [CrossRef]
- Chang, C.C.; Mao, M.; Liu, Y.; Wu, M.; Vo-Dinh, T.; Yuan, F. Improvement in Electrotransfection of Cells Using Carbon-Based Electrodes. Cell Mol. Bioeng. 2016, 9, 538–545. [Google Scholar] [CrossRef]
- Hafner, M.; Niepel, M.; Chung, M.; Sorger, P.K. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods 2016, 13, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Cervia, L.D.; Yuan, F. Current Progress in Electrotransfection as a Nonviral Method for Gene Delivery. Mol. Pharm. 2018, 15, 3617–3624. [Google Scholar] [CrossRef] [PubMed]
- Rosazza, C.; Deschout, H.; Buntz, A.; Braeckmans, K.; Rols, M.P.; Zumbusch, A. Endocytosis and Endosomal Trafficking of DNA after Gene Electrotransfer In Vitro. Mol. Ther. Nucleic Acids 2016, 5, e286. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Wang, L.; Chang, C.C.; Rothenberg, K.E.; Huang, J.; Wang, Y.; Hoffman, B.D.; Liton, P.B.; Yuan, F. Involvement of a Rac1-Dependent Macropinocytosis Pathway in Plasmid DNA Delivery by Electrotransfection. Mol. Ther. 2017, 25, 803–815. [Google Scholar] [CrossRef]
- Huss, M.; Ingenhorst, G.; Konig, S.; Gassel, M.; Drose, S.; Zeeck, A.; Altendorf, K.; Wieczorek, H. Concanamycin A, the specific inhibitor of V-ATPases, binds to the V(o) subunit c. J. Biol. Chem. 2002, 277, 40544–40548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Fang, R.H.; Gao, W.; Zhang, L. A Biomimetic Nanoparticle to “Lure and Kill” Phospholipase A2. Angew. Chem. Int. Ed. Engl. 2020, 59, 10461–10465. [Google Scholar] [CrossRef]
- Nishihara, T.; Akifusa, S.; Koseki, T.; Kato, S.; Muro, M.; Hanada, N. Specific inhibitors of vacuolar type H+-ATPases induce apoptotic cell death. Biochem. Biophys. Res. Commun. 1995, 212, 255–262. [Google Scholar] [CrossRef]
- Sales Conniff, A.; Tur, J.; Kohena, K.; Zhang, M.; Gibbons, J.; Heller, L.C. Transcriptomic Analysis of the Acute Skeletal Muscle Effects after Intramuscular DNA Electroporation Reveals Inflammatory Signaling. Vaccines 2022, 10, 2037. [Google Scholar] [CrossRef] [PubMed]
- Dabral, D.; Coorssen, J.R. Phospholipase A2: Potential roles in native membrane fusion. Int. J. Biochem. Cell Biol. 2017, 85, 1–5. [Google Scholar] [CrossRef]
- Merz, A.J. What are the roles of V-ATPases in membrane fusion? Proc. Natl. Acad. Sci. USA 2015, 112, 8–9. [Google Scholar] [CrossRef]
- Mauvezin, C.; Nagy, P.; Juhász, G.; Neufeld, T.P. Autophagosome–lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 2015, 6, 7007. [Google Scholar] [CrossRef] [PubMed]
- Gleize, V.; Boisselier, B.; Marie, Y.; Poea-Guyon, S.; Sanson, M.; Morel, N. The renal v-ATPase a4 subunit is expressed in specific subtypes of human gliomas. Glia 2012, 60, 1004–1012. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Chang, C.-C.; Chi, J.-T.; Yuan, F. Sucrose Treatment Enhances the Electrotransfer of DNA by Activating Phospholipase A2. Pharmaceutics 2024, 16, 475. https://doi.org/10.3390/pharmaceutics16040475
Wang C, Chang C-C, Chi J-T, Yuan F. Sucrose Treatment Enhances the Electrotransfer of DNA by Activating Phospholipase A2. Pharmaceutics. 2024; 16(4):475. https://doi.org/10.3390/pharmaceutics16040475
Chicago/Turabian StyleWang, Chunxi, Chun-Chi Chang, Jen-Tsan Chi, and Fan Yuan. 2024. "Sucrose Treatment Enhances the Electrotransfer of DNA by Activating Phospholipase A2" Pharmaceutics 16, no. 4: 475. https://doi.org/10.3390/pharmaceutics16040475
APA StyleWang, C., Chang, C.-C., Chi, J.-T., & Yuan, F. (2024). Sucrose Treatment Enhances the Electrotransfer of DNA by Activating Phospholipase A2. Pharmaceutics, 16(4), 475. https://doi.org/10.3390/pharmaceutics16040475





