Effect of Micromixer Design on Lipid Nanocarriers Manufacturing for the Delivery of Proteins and Nucleic Acids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Microfluidic Production Station for Lipid Nanocarriers
2.3. Microfluidic Formulation of Liposomes
2.4. Effect of the Formulation Variables
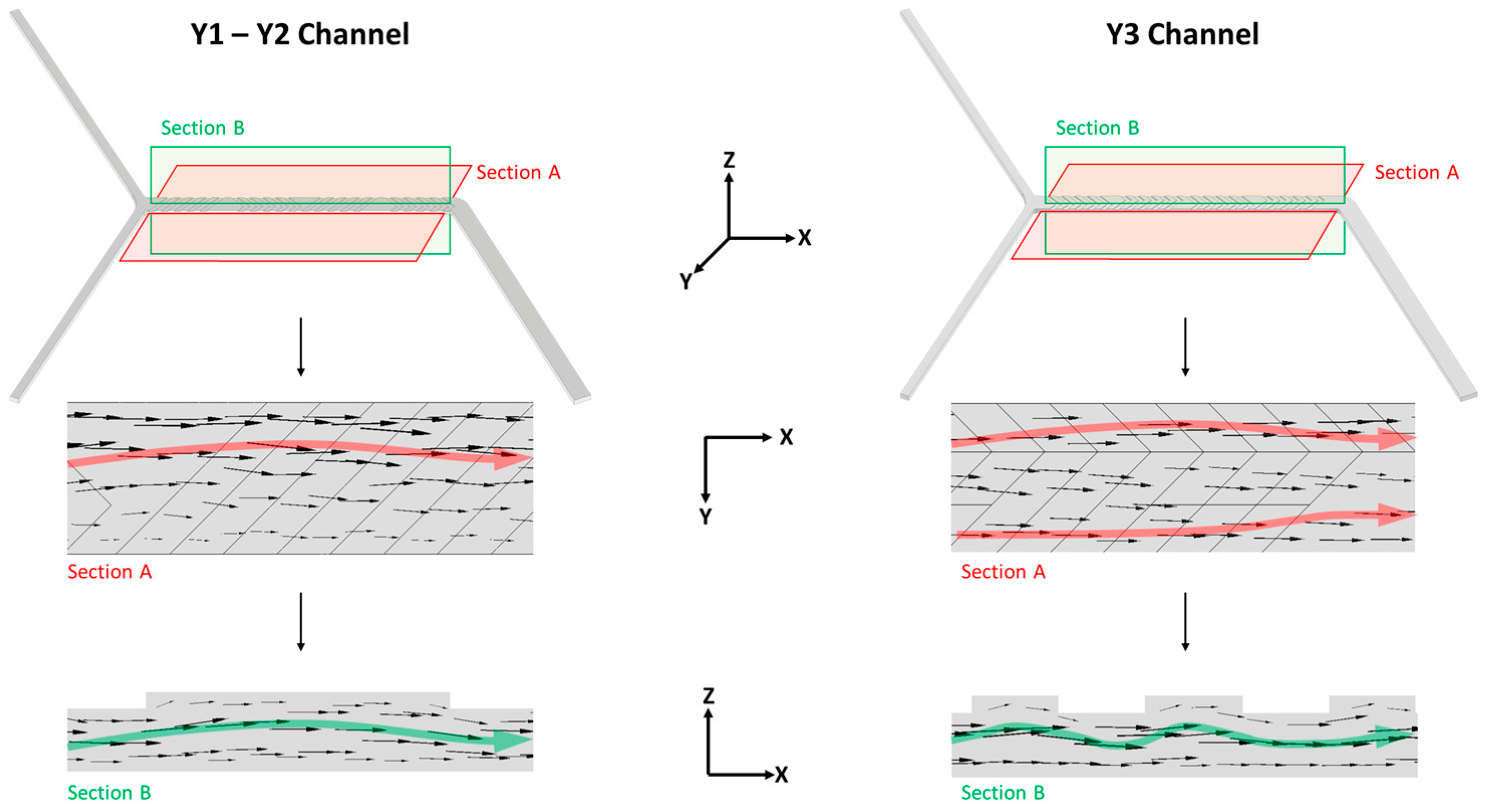
2.4.1. Effect of Channels Geometry
2.4.2. Purification and Buffer Exchange
2.5. Nanocarriers Characterization: Size, Surface Charge and Morphology
2.6. Encapsulation Efficiency of Biomolecules
2.7. Statistical Analysis
3. Results
3.1. Effect of the Formulation Parameters
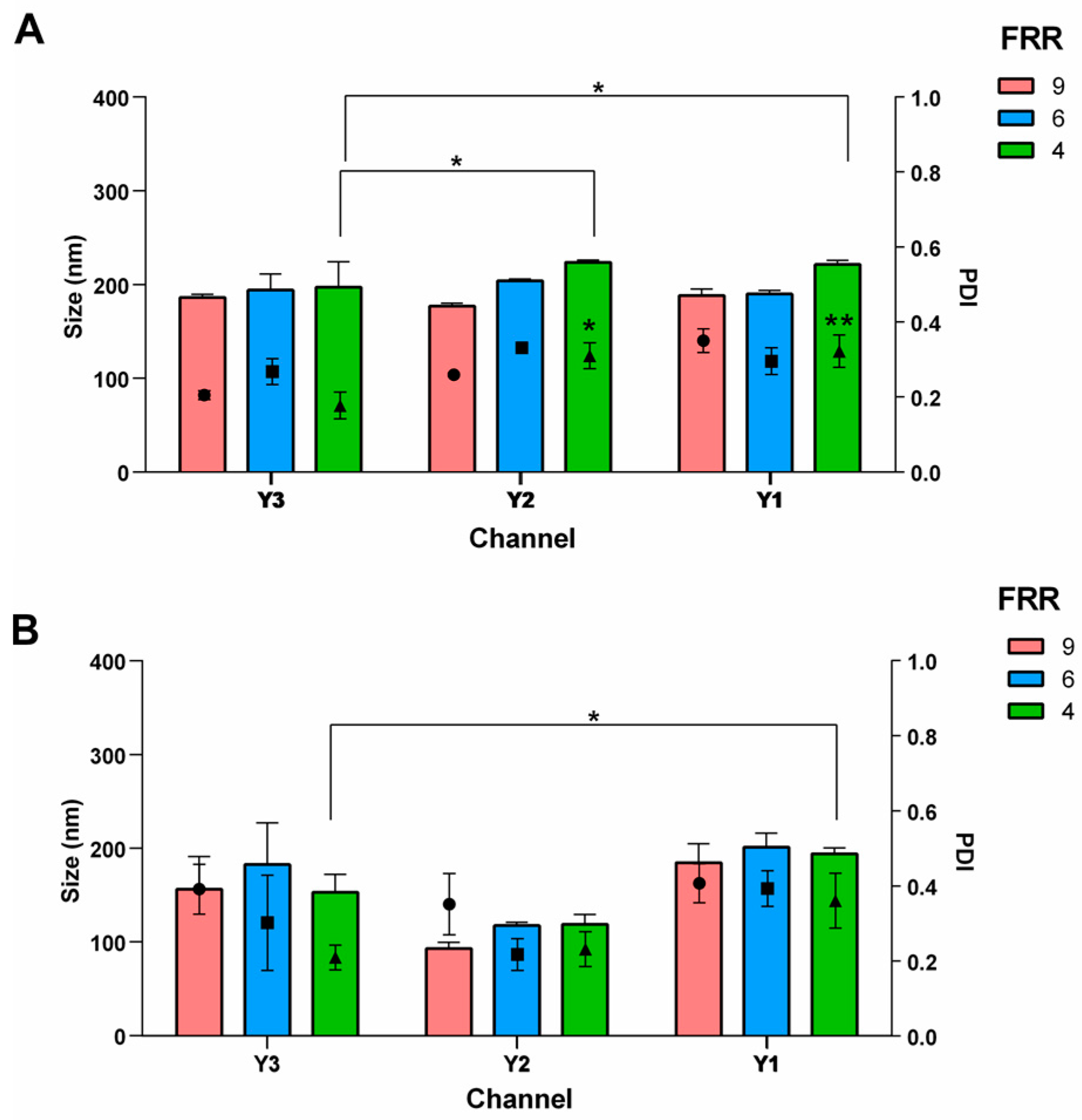
3.2. Effect of the Channel Geometry
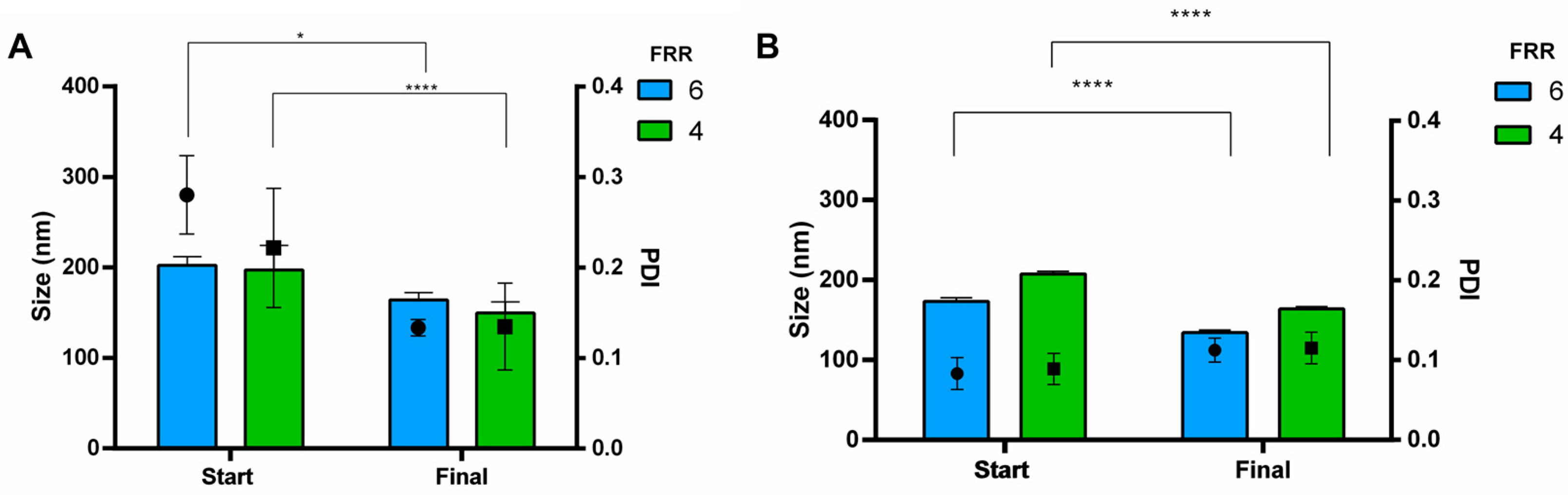
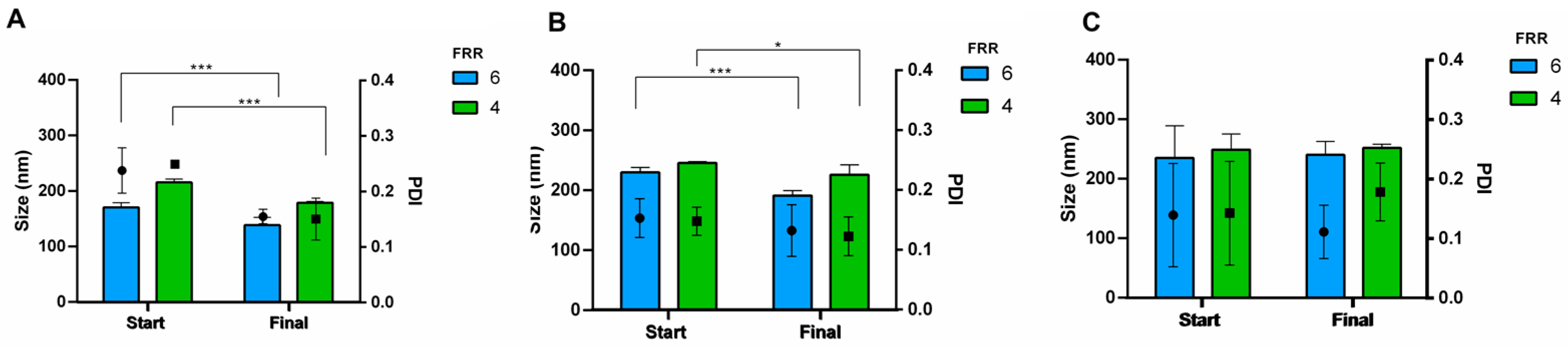
3.3. Purification and Buffer Exchange of Liposomes
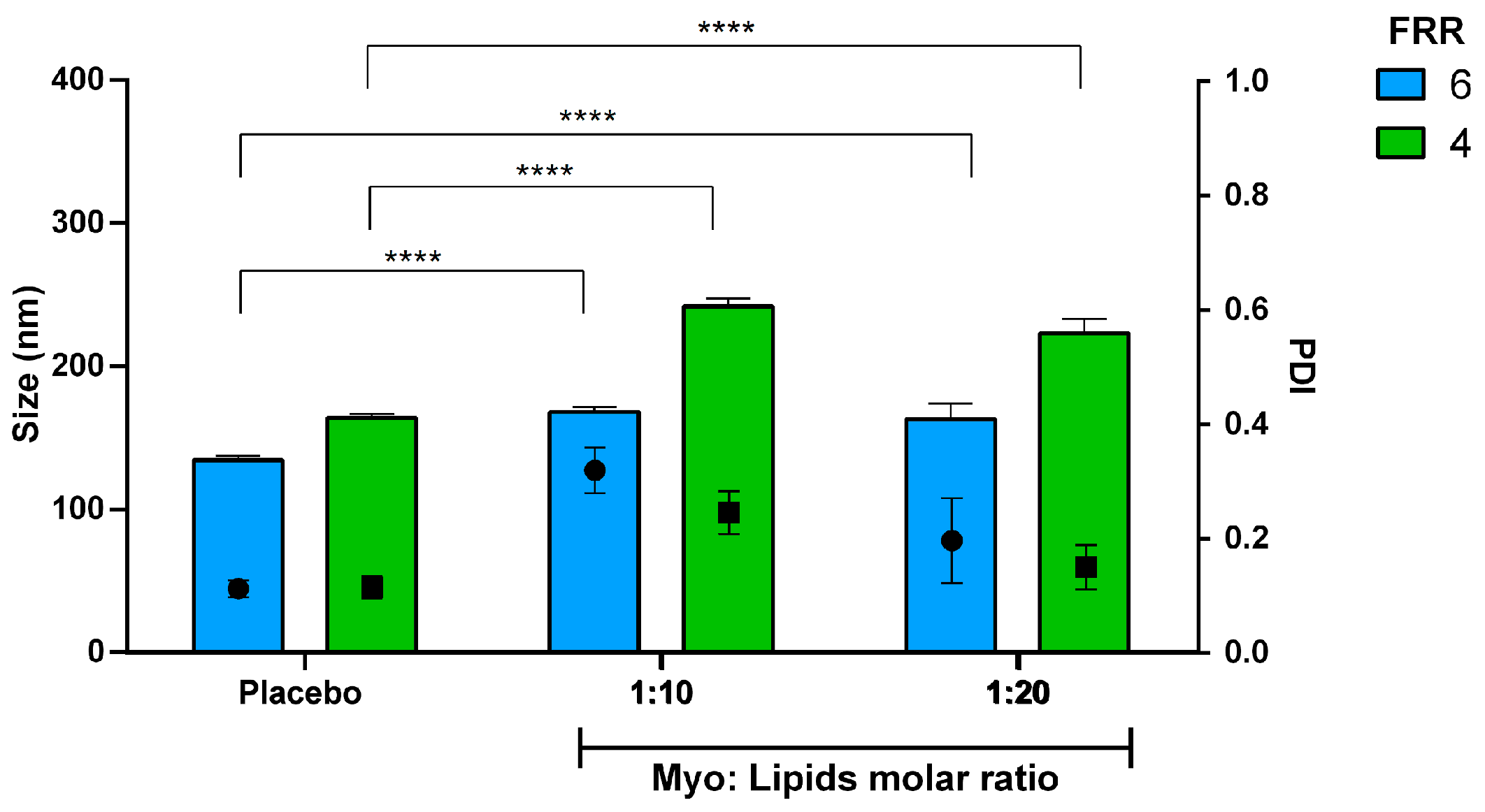
3.4. Encapsulation Efficiency of Biomolecules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vargason, A.; Anselmo, A.; Mitragotri, S. The evolution of commercial drug delivery technologies. Nat. Biomed. Eng. 2021, 5, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Durán-Lobato, M.; López-Estévez, A.M.; Cordeiro, A.S.; Dacoba, T.G.; Crecente-Campo, J.; Torres, D.; Alonso, M.J. Nanotechnologies for the delivery of biologicals: Historical perspective and current landscape. Adv. Drug Deliv. Rev. 2021, 176, 113899. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Langer, R. Impact of nanotechnology on drug delivery. ACS Nano 2009, 3, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Colonna, C.; Dorati, R.; Conti, B.; Caliceti, P.; Genta, I. Sub-unit vaccine against S. aureus-mediated infections: Set-up of nanosized polymeric adjuvant. Int. J. Pharm. 2013, 452, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hoorn, D.; Aitha, A.; Breier, D.; Peer, D. The immunostimulatory nature of mRNA lipid nanoparticles. Adv. Drug Deliv. Rev. 2024, 205, 115175. [Google Scholar] [CrossRef] [PubMed]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles—From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef] [PubMed]
- Maeki, M.; Uno, S.; Niwa, A.; Okada, Y.; Tokeshi, M. Microfluidic technologies and devices for lipid nanoparticle-based RNA delivery. J. Control. Release 2022, 344, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.; Cristovao, J.; Silverio, V.; Lino, P.; Fonte, P. Microfluidic production of mRNA-loaded lipid nanoparticles for vaccine applications. Expert Opin. Drug Deliv. 2022, 19, 1381–1395. [Google Scholar] [CrossRef]
- Pattni, B.; Chupin, V.; Torchilin, V. New Developments in Liposomal Drug Delivery. Chem. Rev. 2015, 115, 10938–10966. [Google Scholar] [CrossRef]
- Martins, J.; Torrieri, G.; Santos, H. The importance of microfluidics for the preparation of nanoparticles as advanced drug delivery systems. Expert Opin. Drug Deliv. 2018, 15, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Ballacchino, G.; Weaver, E.; Mathew, E.; Dorati, R.; Genta, I.; Conti, B.; Lamprou, D. Manufacturing of 3D-Printed Microfluidic Devices for the Synthesis of Drug-Loaded Liposomal Formulations. Int. J. Mol. Sci. 2021, 22, 8064. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, Y.; Mendelsohn, B.; Matsuda, Y. Antibody-Drug Conjugate Synthesis Using Continuous Flow Microreactor Technology. Org. Process Res. Dev. 2022, 26, 2766–2770. [Google Scholar] [CrossRef]
- Wong, S.; Bryant, P.; Ward, M.; Wharton, C. Investigation of mixing in a cross-shaped micromixer with static mixing elements for reaction kinetics studies. Sens. Actuators B-Chem. 2003, 95, 414–424. [Google Scholar] [CrossRef]
- Tomeh, M.; Zhao, X. Recent Advances in Microfluidics for the Preparation of Drug and Gene Delivery Systems. Mol. Pharm. 2020, 17, 4421–4434. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, E.; Bellotti, M.; Caimi, A.; Conti, B.; Dorati, R.; Conti, M.; Genta, I.; Auricchio, F. Development and optimization of microfluidic assisted manufacturing process to produce PLGA nanoparticles. Int. J. Pharm. 2022, 629, 122368. [Google Scholar] [CrossRef] [PubMed]
- Dimov, N.; Kastner, E.; Hussain, M.; Perrie, Y.; Szita, N. Formation and purification of tailored liposomes for drug delivery using a module-based micro continuous-flow system. Sci. Rep. 2017, 7, 12045. [Google Scholar] [CrossRef] [PubMed]
- Roces, C.; Lou, G.; Jain, N.; Abraham, S.; Thomas, A.; Halbert, G.; Perrie, Y. Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics. Pharmaceutics 2020, 12, 1095. [Google Scholar] [CrossRef] [PubMed]
- Pisani, S.; Chiesa, E.; Genta, I.; Dorati, R.; Gregorini, M.; Grignano Maria, A.; Ramus, M.; Ceccarelli, G.; Croce, S.; Valsecchi, C.; et al. Liposome Formulation and In Vitro Testing in Non-Physiological Conditions Addressed to Ex Vivo Kidney Perfusion. Int. J. Mol. Sci. 2022, 23, 7999. [Google Scholar] [CrossRef]
- Balasubramaniam, L.; Arayanarakool, R.; Marshall, S.; Li, B.; Lee, P.; Chen, P. Impact of cross-sectional geometry on mixing performance of spiral microfluidic channels characterized by swirling strength of Dean-vortices. J. Micromech. Microeng. 2017, 27, 095016. [Google Scholar] [CrossRef]
- Saravanakumar, S.M.; Cicek, P.V. Microfluidic Mixing: A Physics-Oriented Review. Micromachines 2023, 14, 1827. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, E.; Weaver, E.; Meziane, A.; Lamprou, D. Synthesis and Characterization of Paclitaxel-Loaded PEGylated Liposomes by the Microfluidics Method. Mol. Pharm. 2023, 20, 6184–6196. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.; Forbes, N.; Roces, C.; Anderluzzi, G.; Lou, G.; Abraham, S.; Ingalls, L.; Marshall, K.; Leaver, T.; Watts, J.; et al. Using microfluidics for scalable manufacturing of nanomedicines from bench to GMP: A case study using protein-loaded liposomes. Int. J. Pharm. 2020, 582, 119266. [Google Scholar] [CrossRef] [PubMed]
- Roces, C.B.; Port, E.C.; Daskalakis, N.N.; Watts, J.A.; Aylott, J.W.; Halbert, G.W.; Perrie, Y. Rapid scale-up and production of active-loaded PEGylated liposomes. Int. J. Pharm. 2020, 586, 119566. [Google Scholar] [CrossRef]
- Nag, K.; Sarker, M.; Kumar, S.; Khan, H.; Chakraborty, S.; Islam, M.; Baray, J.; Khan, M.; Mahmud, A.; Barman, U.; et al. DoE-derived continuous and robust process for manufacturing of pharmaceutical-grade wide-range LNPs for RNA-vaccine/drug delivery. Sci. Rep. 2022, 12, 9394. [Google Scholar] [CrossRef] [PubMed]
- Terada, T.; Kulkarni, J.; Huynh, A.; Chen, S.; van der Meel, R.; Tam, Y.; Cullis, P. Characterization of Lipid Nanoparticles Containing Ionizable Cationic Lipids Using Design-of-Experiments Approach. Langmuir 2021, 37, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Hashiba, A.; Toyooka, M.; Sato, Y.; Maeki, M.; Tokeshi, M.; Harashima, H. The use of design of experiments with multiple responses to determine optimal formulations for in vivo hepatic mRNA delivery. J. Control. Release 2020, 327, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Briuglia, M.; Rotella, C.; McFarlane, A.; Lamprou, D. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015, 5, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Forssen, E. The design and development of DaunoXome(R) for solid tumor targeting in vivo. Adv. Drug Deliv. Rev. 1997, 24, 133–150. [Google Scholar] [CrossRef]
- Webb, C.; Khadke, S.; Schmidt, S.T.; Roces, C.B.; Forbes, N.; Berrie, G.; Perrie, Y. The Impact of Solvent Selection: Strategies to Guide the Manufacturing of Liposomes Using Microfluidics. Pharmaceutics 2019, 11, 653. [Google Scholar] [CrossRef] [PubMed]
- Forbes, N.; Hussain, M.T.; Briuglia, M.L.; Edwards, D.P.; Horst, J.H.T.; Szita, N.; Perrie, Y. Rapid and scale-independent microfluidic manufacture of liposomes entrapping protein incorporating in-line purification and at-line size monitoring. Int. J. Pharm. 2019, 556, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Zook, J.; Vreeland, W. Effects of temperature, acyl chain length, and flow-rate ratio on liposome formation and size in a microfluidic hydrodynamic focusing device. Soft Matter 2010, 6, 1352–1360. [Google Scholar] [CrossRef]
- Moghaddam, B.; Ali, M.; Wilkhu, J.; Kirby, D.; Mohammed, A.; Zheng, Q.; Perrie, Y. The application of monolayer studies in the understanding of liposomal formulations. Int. J. Pharm. 2011, 417, 235–244. [Google Scholar] [CrossRef]
- Anderluzzi, G.; Lou, G.; Gallorini, S.; Brazzoli, M.; Johnson, R.; O’Hagan, D.; Baudner, B.; Perrie, Y. Investigating the Impact of Delivery System Design on the Efficacy of Self-Amplifying RNA Vaccines. Vaccines 2020, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- EMA. Information for the Package Leaflet Regarding Ethanol. Used as an Excipient in Medicinal Products for Human Use; EMA: Amsterdam, The Netherlands, 2018; p. 34. [Google Scholar]
- Das, S.; Tilekar, S.; Wangikar, S.; Patowari, P. Numerical and experimental study of passive fluids mixing in micro-channels of different configurations. Microsyst. Technol.-Micro-Nanosyst.-Inf. Storage Process. Syst. 2017, 23, 5977–5988. [Google Scholar] [CrossRef]
- Yang, J.; Huang, K.; Lin, Y. Geometric effects on fluid mixing in passive grooved micromixers. Lab Chip 2005, 5, 1140–1147. [Google Scholar] [CrossRef] [PubMed]

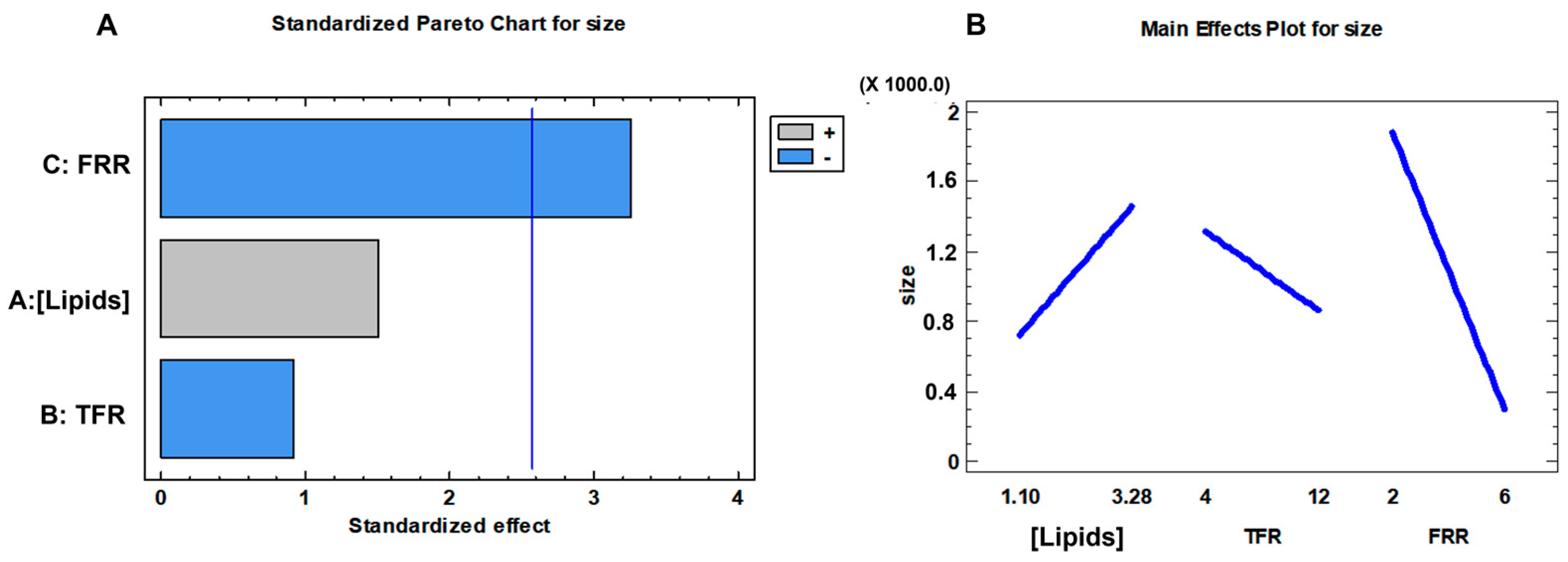

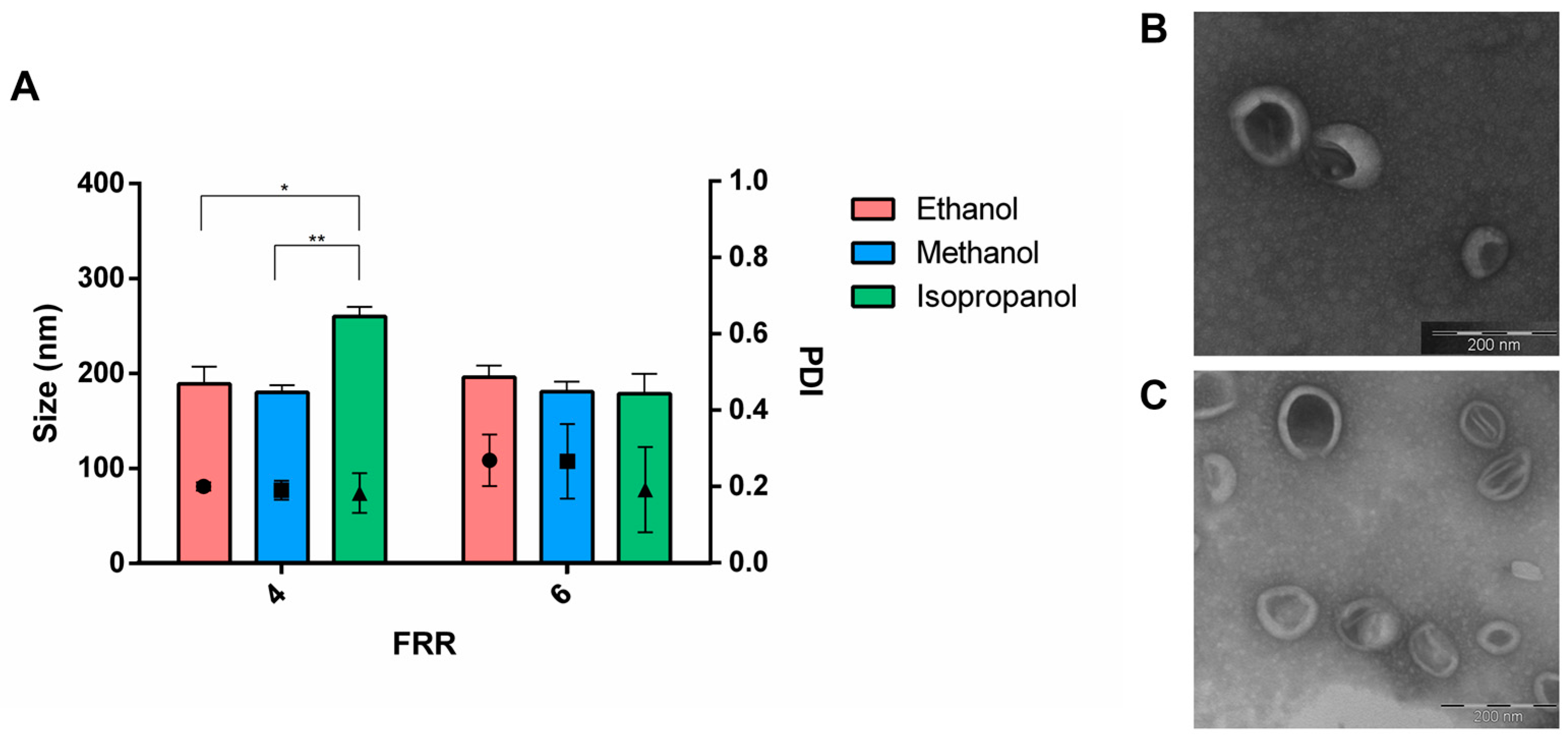
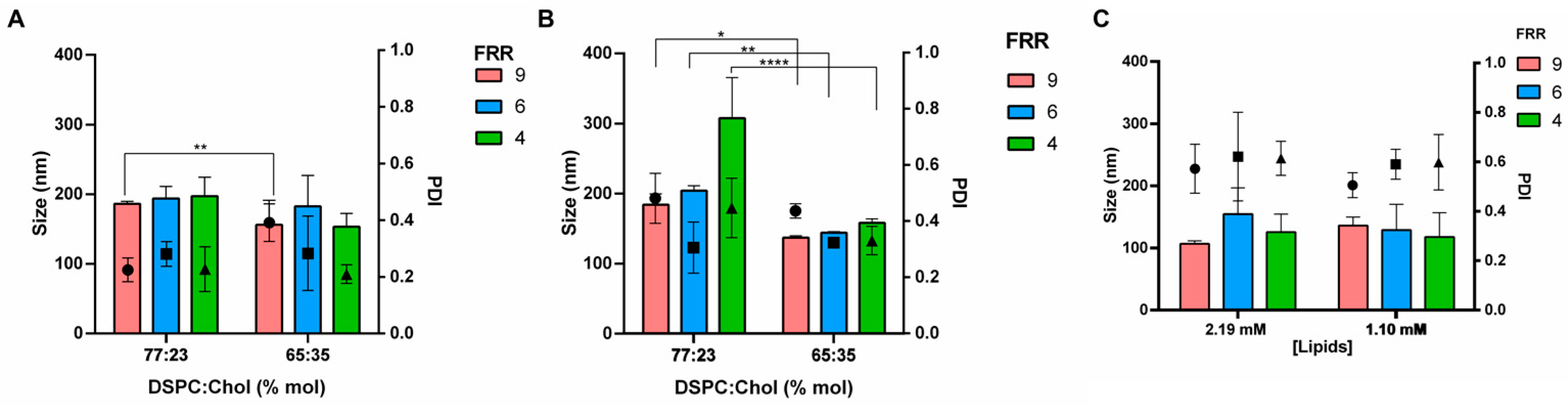
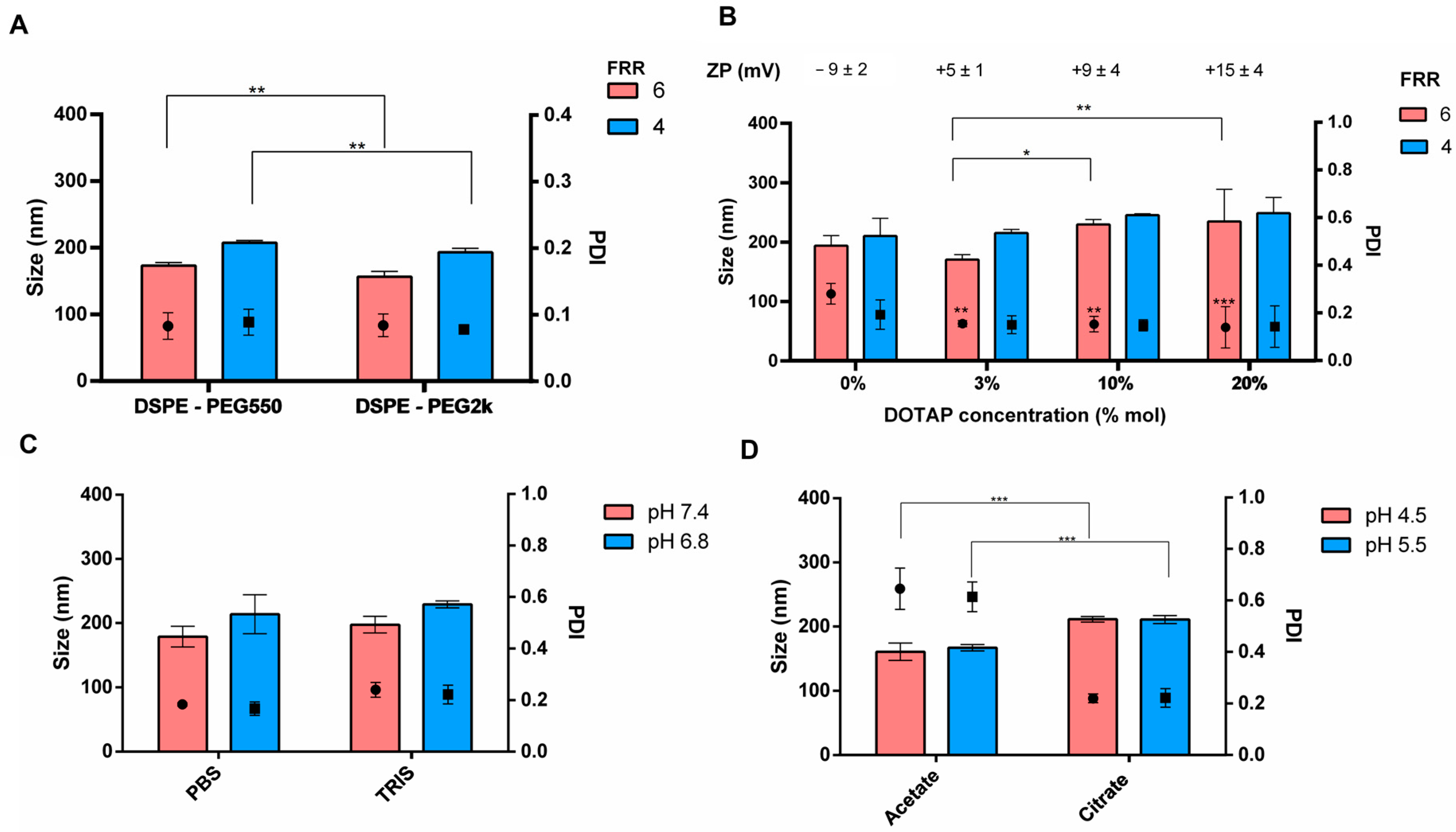
| Factors | Level | |||
|---|---|---|---|---|
| Category | −1 | 0 | +1 | |
| Lipid concentration | Continuous | 1.10 mM | 2.19 mM | 3.28 mM |
| Total flow rate, TFR | Continuous | 4 mL/min | 8 mL/min | 12 mL/min |
| Aqueous-to-ethanol flow rate ratio, FRR | Continuous | 2 | 4 | 6 |
| Batch Code | Lipid Concentration (mM) | TFR (mL/min) | FRR (v:v) |
|---|---|---|---|
| DoE 1 | 3.28 (+1) | 4 (−1) | 6 (+1) |
| DoE 2 | 3.28 (+1) | 4 (−1) | 2 (−1) |
| DoE 3 | 1.10 (−1) | 4 (−1) | 6 (+1) |
| DoE 4 | 1.10 (−1) | 12 (+1) | 6 (+1) |
| DoE 5 | 3.28 (+1) | 12 (+1) | 2 (−1) |
| DoE 6 | 1.10 (−1) | 4 (−1) | 2 (−1) |
| DoE 7 | 2.19 (0) | 8 (0) | 4 (0) |
| DoE 8 | 3.28 (+1) | 12 (+1) | 6 (+1) |
| DoE 9 | 1.10 (−1) | 12 (+1) | 2 (−1) |
| Batch Code | Lipid Concentration (mM) | TFR (mL/min) | FRR (v:v) |
|---|---|---|---|
| DoE 10 | 3.28 (+1) | 8 (0) | 2 (−1) |
| DoE 11 | 2.19 (0) | 12 (+1) | 6 (+1) |
| DoE 12 | 3.28 (+1) | 4 (−1) | 4 (0) |
| DoE 13 | 3.28 (+1) | 12 (+1) | 4 (0) |
| DoE 14 | 2.19 (0) | 8 (0) | 4 (0) |
| DoE 15 | 2.19 (0) | 4 (−1) | 6 (+1) |
| DoE 16 | 1.10 (−1) | 8 (0) | 2 (−1) |
| DoE 17 | 1.10 (−1) | 8 (0) | 6 (+1) |
| DoE 18 | 2.19 (0) | 4 (−1) | 2 (−1) |
| DoE 19 | 3.28 (+1) | 8 (0) | 6 (+1) |
| DoE 20 | 1.10 (−1) | 4 (−1) | 4 (0) |
| DoE 21 | 2.19 (0) | 8 (0) | 4 (0) |
| DoE 22 | 2.19 (0) | 12 (+1) | 2 (−1) |
| DoE 23 | 1.10 (−1) | 12 (+1) | 4 (0) |
| DoE 24 | 2.19 (0) | 8 (0) | 4 (0) |
| Molar Ratio Myo:Lipids | FRR 4 | FRR 6 |
|---|---|---|
| 1:10 | 0.323 mg/mL | 0.484 mg/mL |
| 1:20 | 0.162 mg/mL | 0.242 mg/mL |
| Batch Code | [Lipids] (mM) | TFR (mL/min) | FRR (v:v) | Mean Size ± SD (nm) | PDI ± SD |
|---|---|---|---|---|---|
| DoE 1 | 3.28 | 4 | 6 | 877 ± 20 | 0.71 ± 0.01 |
| DoE 2 | 3.28 | 4 | 2 | 3383 ± 141 | 0.21 ± 0.-11 |
| DoE 3 | 1.10 | 4 | 6 | 224 ± 10 | 0.48 ± 0.01 |
| DoE 4 | 1.10 | 12 | 6 | 222 ± 3 | 0.29 ± 0.04 |
| DoE 5 | 3.28 | 12 | 2 | 1686 ± 55 | 0.93 ± 0.10 |
| DoE 6 | 1.10 | 4 | 2 | 1157 ± 117 | 0.69 ± 0.01 |
| DoE 7 | 2.19 | 8 | 4 | 308 ± 58 | 0.45 ± 0.11 |
| DoE 8 | 3.28 | 12 | 6 | 266 ± 2 | 0.35 ± 0.01 |
| DoE 9 | 1.10 | 12 | 2 | 1688 ± 102 | 0.76 ± 0.05 |
| p = 0.19 | p = 0.40 | p = 0.02 |
| Batch Code | [Lipids] (mM) | TFR (mL/min) | FRR (v:v) | Mean Size ± SD (nm) | PDI ± SD |
|---|---|---|---|---|---|
| DoE 10 | 3.28 | 8 | 2 | 814 ± 50 | 0.29 ± 0.01 |
| DoE 11 | 2.19 | 12 | 6 | 216 ± 10 | 0.30 ± 0.06 |
| DoE 12 | 3.28 | 4 | 4 | 1230 ± 30 | 0.45 ± 0.02 |
| DoE 13 | 3.28 | 12 | 4 | 306 ± 8 | 0.43 ± 0.02 |
| DoE 14 | 2.19 | 8 | 4 | 308 ± 58 | 0.45 ± 0.11 |
| DoE 15 | 2.19 | 4 | 6 | 367 ± 11 | 0.71 ± 0.04 |
| DoE 16 | 1.10 | 8 | 2 | 964 ± 169 | 0.38 ± 0.01 |
| DoE 17 | 1.10 | 8 | 6 | 175 ± 8 | 0.29 ± 0.05 |
| DoE 18 | 2.19 | 4 | 2 | 1305 ± 152 | 0.49 ± 0.07 |
| DoE 19 | 3.28 | 8 | 6 | 247 ± 58 | 0.60 ± 0.22 |
| DoE 20 | 1.10 | 4 | 4 | 383 ± 1 | 0.48 ± 0.01 |
| DoE 21 | 2.19 | 8 | 4 | 258 ± 6 | 0.45 ± 0.11 |
| DoE 22 | 2.19 | 12 | 2 | 1484 ± 359 | 0.34 ± 0.07 |
| DoE 23 | 1.10 | 12 | 4 | 289 ± 3 | 0.43 ± 0.02 |
| DoE 24 | 2.19 | 8 | 4 | 358 ± 9 | 0.45 ± 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiesa, E.; Caimi, A.; Bellotti, M.; Giglio, A.; Conti, B.; Dorati, R.; Auricchio, F.; Genta, I. Effect of Micromixer Design on Lipid Nanocarriers Manufacturing for the Delivery of Proteins and Nucleic Acids. Pharmaceutics 2024, 16, 507. https://doi.org/10.3390/pharmaceutics16040507
Chiesa E, Caimi A, Bellotti M, Giglio A, Conti B, Dorati R, Auricchio F, Genta I. Effect of Micromixer Design on Lipid Nanocarriers Manufacturing for the Delivery of Proteins and Nucleic Acids. Pharmaceutics. 2024; 16(4):507. https://doi.org/10.3390/pharmaceutics16040507
Chicago/Turabian StyleChiesa, Enrica, Alessandro Caimi, Marco Bellotti, Alessia Giglio, Bice Conti, Rossella Dorati, Ferdinando Auricchio, and Ida Genta. 2024. "Effect of Micromixer Design on Lipid Nanocarriers Manufacturing for the Delivery of Proteins and Nucleic Acids" Pharmaceutics 16, no. 4: 507. https://doi.org/10.3390/pharmaceutics16040507






