Molecular Ballet: Investigating the Complex Interaction between Self-Assembling Dendrimers and Human Serum Albumin via Computational and Experimental Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
2.2. Sample Preparation and Analytical Procedures
2.3. Far-UV Circular Dichroism Spectroscopy
2.4. Steady-State and 3D Fluorescence Spectroscopy
2.5. Isothermal Titration Calorimetry Studies
2.6. Dynamic Light Scattering and Zeta Potential Measurements
2.7. Transmission Electron Microscopy
2.8. Molecular Simulations
3. Results and Discussion
3.1. Experimental Binding Analysis of the HSA/AD Complex
3.1.1. Fluorescence Spectra and Binding Constants of the HSA/AD Complexes
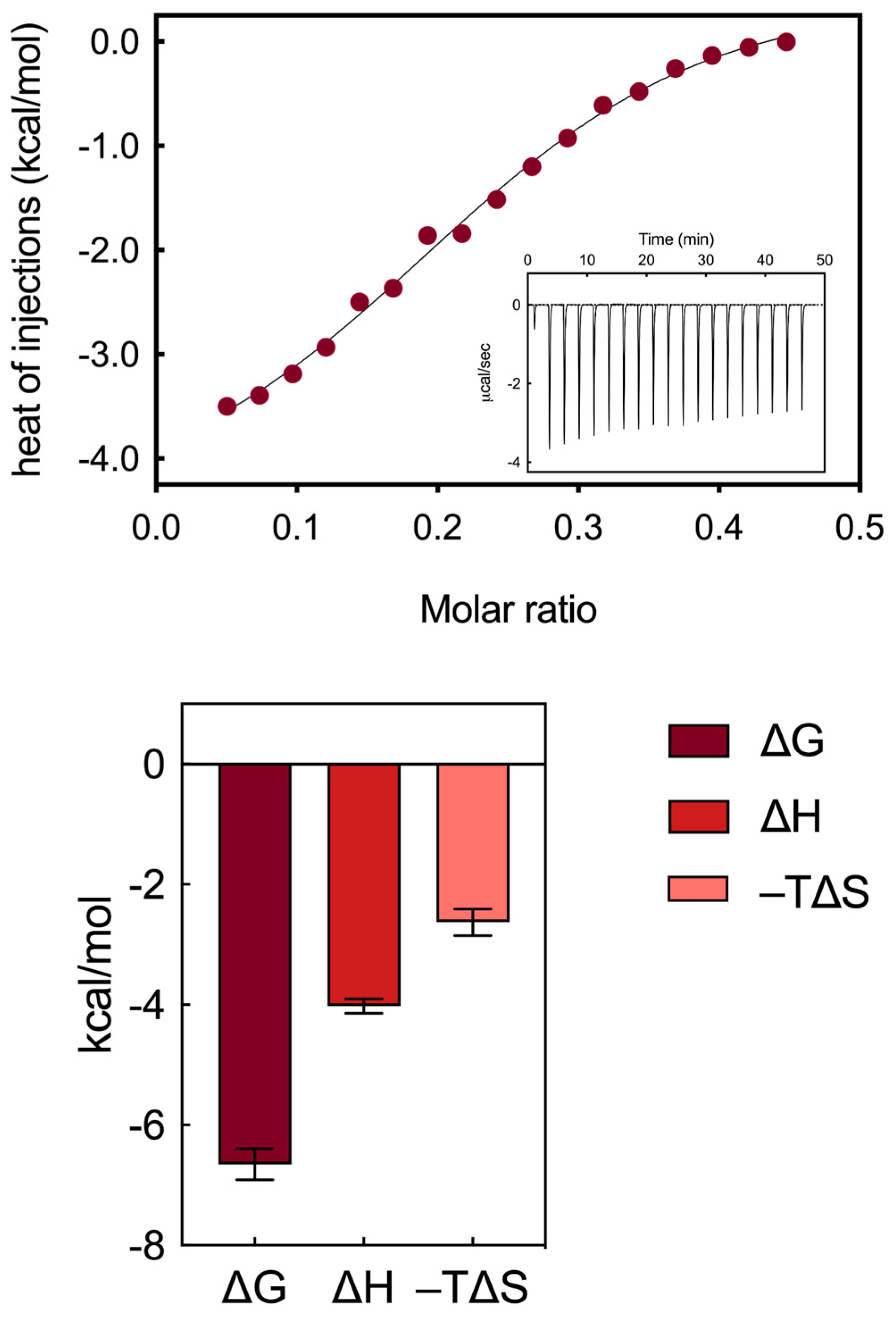
3.1.2. Isothermal Titration Calorimetry Binding Analysis
- robust electrostatic interactions are established between the positively charged terminal groups of the AD NMs and the side chains of some aspartic and glutamic acid residues within HSA;
- a limited network of hydrogen bonds (HBs) forms between the internal groups of the AD molecules and specific amino acid residues of the serum protein, facilitating the exposure of side chains that serve as counterparts for the formation of proton bridges. Notably, within the AD dendron, the oxygens of the internal amide groups serve a hydrogen bond acceptor, with the -NH moieties of the same groups acting as HB donors;
- these two types of interactions are mainly enthalpic in nature, as indicated by the favorable ΔH value obtained from the titrations (−4.02 ± 0.12 kcal/mol, Figure 3).
- weak hydrophobic interactions occur between the hydrocarbon/aliphatic components of the nanocarrier heads and the side chain of hydrophobic amino acids of HSA;
- secondary hydrophobic alterations occur due to bond formation at the respective surfaces, leading to the release of water molecules, ions and counterions. The latter two interactions contribute to favorable entropic changes, enhancing the stability of the interaction between the protein and the NMs, as demonstrated by the value of −TΔS = −2.63 ± 0.22 kcal/mol (Figure 3).
3.2. Structural and Thermal Perturbation of HSA upon AD Binding
3.2.1. Determination of the HSA Secondary Structure and Melting Temperature through Circular Dichroism Spectroscopy
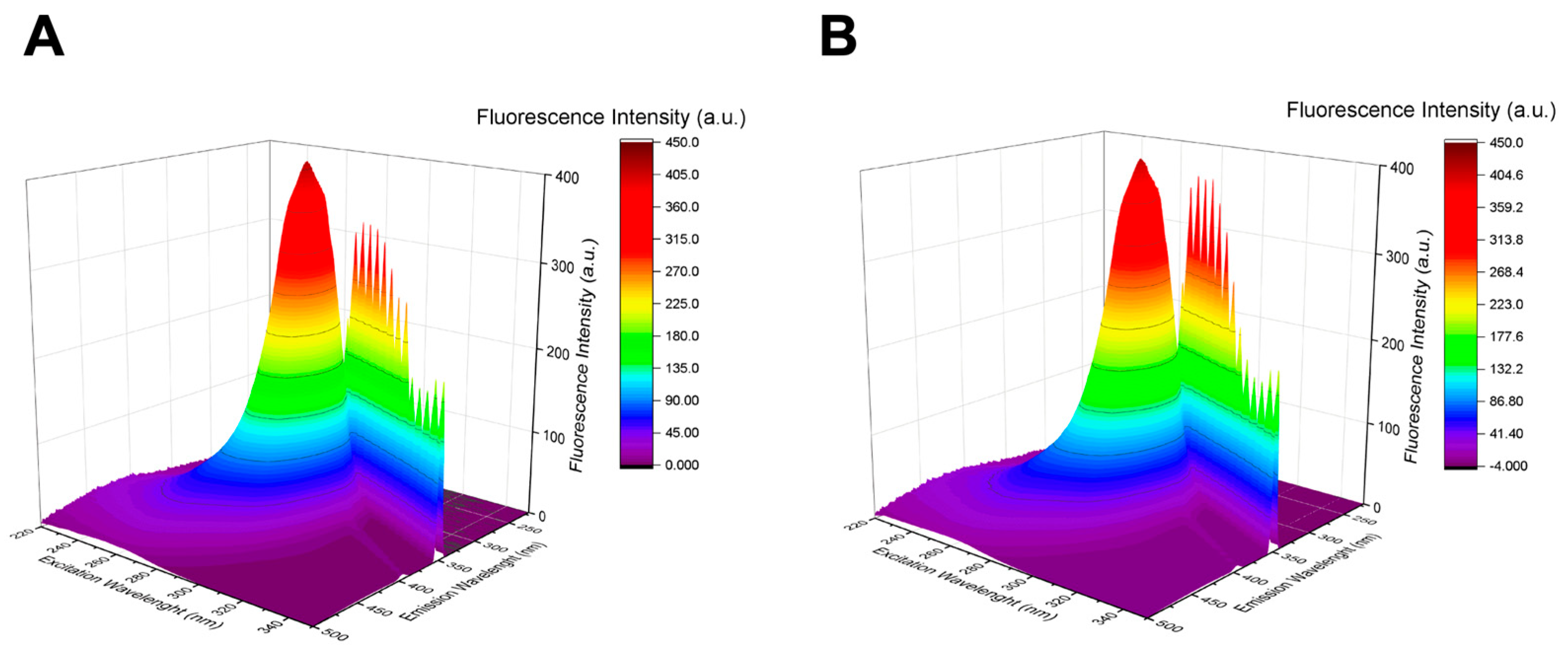
3.2.2. Three-Dimensional Fluorescence Spectroscopy
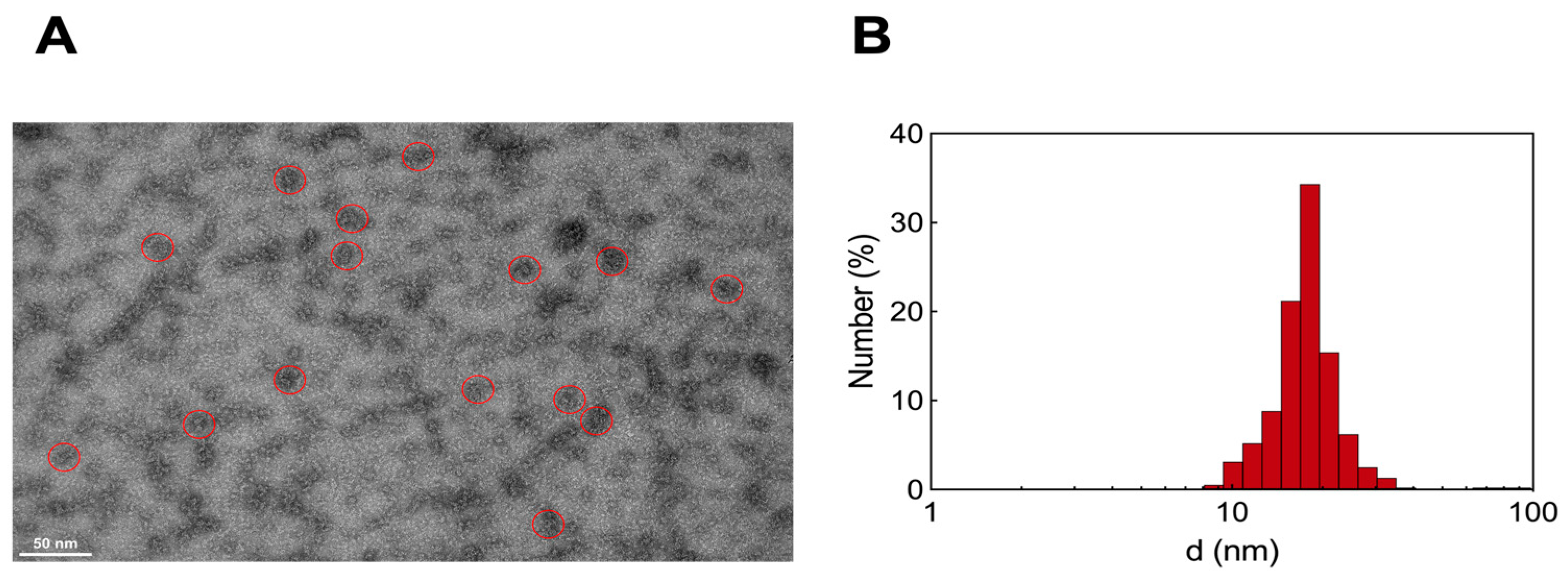
3.2.3. Transmission Electron Microscopy and Dynamic Light Scattering Analysis
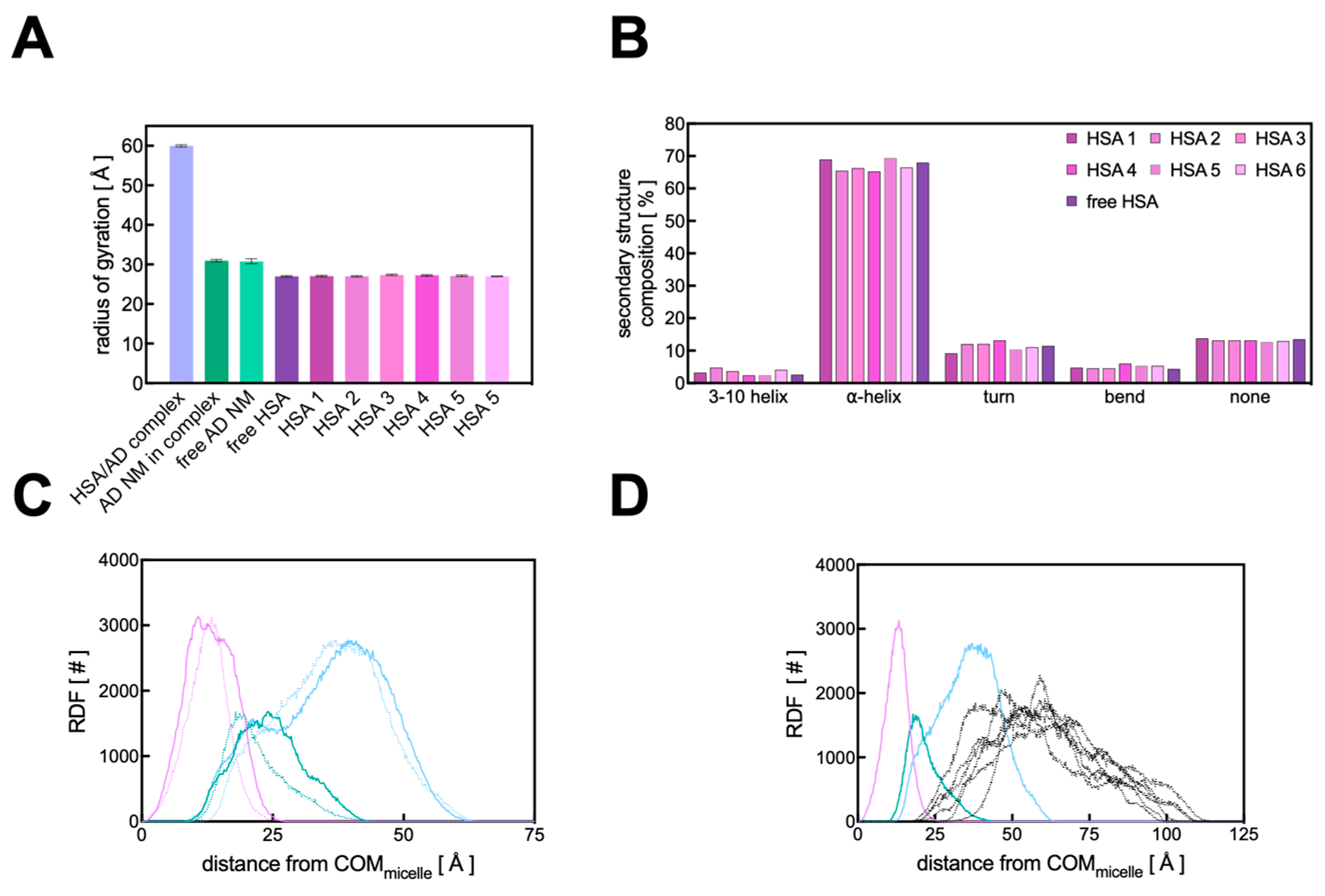
3.3. Computational Studies
3.3.1. Binding Analysis of the HSA/AD Complex
3.3.2. Binding Analysis of the HSA/AD Complex
4. Conclusions
- (i)
- mitigated carrier-induced toxicity, since an excessive binding affinity between any drug vector and HSA can perturb normal protein functions. A moderate binding pattern serves to alleviate such risks, thus increasing the safety of the drug delivery system;
- (ii)
- enhanced circulation stability, since moderate binding ensures a stable interaction between the drug carrier and HAS, and this, in turn, can enhance the circulation time of the drug delivery system in the bloodstream. This prolonged circulation time improves drug bioavailability and increases the likelihood of reaching target tissues;
- (iii)
- diminished immunogenicity since nanovector binding to HSA may mitigate the recognition of the drug delivery system as foreign by the immune system. This effect could potentially decrease immune responses, such as clearance by the reticuloendothelial system or the initiation of inflammatory reactions.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Astruc, D.; Boisselier, E.; Ornelas, C. Dendrimers designed for functions: From physical, photophysical, and supramolecular properties to applications in sensing, catalysis, molecular electronics, photonics, and nanomedicine. Chem. Rev. 2010, 110, 1857–1959. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Khanna, S.N. A Systematic Framework and Nanoperiodic Concept for Unifying Nanoscience: Hard/Soft Nanoelements, Superatoms, Meta-Atoms, New Emerging Properties, Periodic Property Patterns, and Predictive Mendeleev-like Nanoperiodic Tables. Chem. Rev. 2016, 116, 2705–2774. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Yañez, C.; Rodriguez, C.C. Dendrimers: Amazing Platforms for Bioactive Molecule Delivery Systems. Materials 2020, 13, 570. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ferreiro, M.; Abelairas, A.M.; Criado, A.; Gómez, I.J.; Mosquera, J. Dendrimers: Exploring Their Wide Structural Variety and Applications. Polymers 2023, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-P.; Ficker, M.; Christensen, J.B.; Trohopoulos, P.N.; Moghimi, S.M. Dendrimers in Medicine: Therapeutic Concepts and Pharmaceutical Challenges. Bioconjug. Chem. 2015, 26, 1198–1211. [Google Scholar] [CrossRef] [PubMed]
- Seidi, F.; Jenjob, R.; Crespy, D. Designing Smart Polymer Conjugates for Controlled Release of Payloads. Chem. Rev. 2018, 118, 3965–4036. [Google Scholar] [CrossRef] [PubMed]
- Saluja, V.; Mishra, Y.; Mishra, V.; Giri, N.; Nayak, P. Dendrimers based cancer nanotheranostics: An overview. Int. J. Pharm. 2021, 600, 120485. [Google Scholar] [CrossRef]
- Cruz, A.; Barbosa, J.; Antunes, P.; Bonifácio, V.D.B.; Pinto, S.N. A Glimpse into Dendrimers Integration in Cancer Imaging and Theranostics. Int. J. Mol. Sci. 2023, 24, 5430. [Google Scholar] [CrossRef] [PubMed]
- Tewabe, A.; Abate, A.; Tamrie, M.; Seyfu, A.; Siraj, E.A. Targeted Drug Delivery—From Magic Bullet to Nanomedicine: Principles, Challenges, and Future Perspectives. J. Multidiscip. Healthc. 2021, 14, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Tarach, P.; Janaszewska, A. Recent Advances in Preclinical Research Using PAMAM Dendrimers for Cancer Gene Therapy. Int. J. Mol. Sci. 2021, 22, 2912. [Google Scholar] [CrossRef]
- Kheraldine, H.; Rachid, O.; Habib, A.M.; Al Moustafa, A.-E.; Benter, I.F.; Akhtar, S. Emerging innate biological properties of nano-drug delivery systems: A focus on PAMAM dendrimers and their clinical potential. Adv. Drug Deliv. Rev. 2021, 178, 113908. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Chen, C.; Liu, J.; Liu, C.; Posocco, P.; Liu, X.; Cheng, Q.; Huo, S.; Liang, Z.; Fermeglia, M.; et al. Anticancer drug nanomicelles formed by self-assembling amphiphilic dendrimer to combat cancer drug resistance. Proc. Natl. Acad. Sci. USA 2015, 112, 2978–2983. [Google Scholar] [CrossRef] [PubMed]
- Russi, M.; Valeri, R.; Marson, D.; Danielli, C.; Felluga, F.; Tintaru, A.; Skoko, N.; Aulic, S.; Laurini, E.; Pricl, S. Some things old, new and borrowed: Delivery of dabrafenib and vemurafenib to melanoma cells via self-assembled nanomicelles based on an amphiphilic dendrimer. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2023, 180, 106311. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Landry, M.P.; Moore, A.; Coreas, R. The protein corona from nanomedicine to environmental science. Nat. Rev. Mater. Mar. 2023, 8, 422–438. [Google Scholar] [CrossRef] [PubMed]
- Bashiri, G.; Padilla, M.S.; Swingle, K.L.; Shepherd, S.J.; Mitchell, M.J.; Wang, K. Nanoparticle protein corona: From structure and function to therapeutic targeting. Lab. Chip. 2023, 23, 1432–1466. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, R.; Vallet-Regí, M. Hard and Soft Protein Corona of Nanomaterials: Analysis and Relevance. Nanomaterials 2021, 11, 888. [Google Scholar] [CrossRef]
- Kopac, T. Protein corona, understanding the nanoparticle-protein interactions and future perspectives: A critical review. Int. J. Biol. Macromol. 2021, 169, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Vroman, L.; Adams, A.L.; Fischer, G.C.; Munoz, P.C. Interaction of high molecular weight kininogen, factor XII, and fibrinogen in plasma at interfaces. Blood 1980, 55, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, Y.; Liang, H. Interactive association of drugs binding to human serum albumin. Int. J. Mol. Sci. 2014, 15, 3580–3595. [Google Scholar] [CrossRef] [PubMed]
- Sen, T.; Mandal, S.; Haldar, S.; Chattopadhyay, K.; Patra, A. Interaction of Gold Nanoparticle with Human Serum Albumin (HSA) Protein Using Surface Energy Transfer. J. Phys. Chem. C 2011, 115, 24037–24044. [Google Scholar] [CrossRef]
- Peng, Q.; Zhang, S.; Yang, Q.; Zhang, T.; Wei, X.-Q.; Jiang, L.; Zhang, C.-L.; Chen, Q.-M.; Zhang, Z.-R.; Lin, Y.-F. Preformed albumin corona, a protective coating for nanoparticles based drug delivery system. Biomaterials 2013, 34, 8521–8530. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Chen, C. The Crown and the Scepter: Roles of the Protein Corona in Nanomedicine. Adv. Mater. 2019, 31, 1805740. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.P.; Bobbala, S.; Karabin, N.B.; Frey, M.; Liu, Y.; Navidzadeh, J.O.; Stack, T.; Scott, E.A. Surface chemistry-mediated modulation of adsorbed albumin folding state specifies nanocarrier clearance by distinct macrophage subsets. Nat. Commun. 2021, 12, 648. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.W.; Laurini, E.; Posocco, P.; Pricl, S.; Smith, D.K. Chiral recognition at self-assembled multivalent (SAMul) nanoscale interfaces—Enantioselectivity in polyanion binding. Chem. Commun. 2016, 52, 10540–10543. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, A.C.; Bromfield, S.M.; Laurini, E.; Posocco, P.; Pricl, S.; Smith, D.K. Morphological control of self-assembled multivalent (SAMul) heparin binding in highly competitive media. Chem. Commun. 2017, 53, 6335–6338. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, K.A.; Laurini, E.; Pricl, S.; Smith, D.K. Enantiomeric and Diastereomeric Self-Assembled Multivalent Nanostructures: Understanding the Effects of Chirality on Binding to Polyanionic Heparin and DNA. Angew. Chem. Int. Ed. Engl. 2018, 57, 8530–8534. [Google Scholar] [CrossRef] [PubMed]
- Olesen, N.E.; Holm, R.; Westh, P. Determination of the aggregation number for micelles by isothermal titration calorimetry. Thermochim. Acta 2014, 588, 28–37. [Google Scholar] [CrossRef]
- Laurini, E.; Aulic, S.; Skoko, N.; Marson, D.; Fermeglia, M.; Pricl, S. ITC for Characterization of Self-Assembly Process of Cationic Dendrons for siRNA Delivery. Methods Mol. Biol. 2021, 2282, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Dhumal, D.; Maron, B.; Malach, E.; Lyu, Z.; Ding, L.; Marson, D.; Laurini, E.; Tintaru, A.; Ralahy, B.; Giorgio, S.; et al. Dynamic self-assembling supramolecular dendrimer nanosystems as potent antibacterial candidates against drug-resistant bacteria and biofilms. Nanoscale 2022, 14, 9286–9296. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.-Y. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39 (Suppl. 2), W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Miller, B.R.I.; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cisneros, G.A.; Cruzeiro, V.W.D.; et al. Amber 2023; University of California: San Francisco, CA, USA, 2023. [Google Scholar]
- Roe, D.R.; Cheatham, T.E.I. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Amber22.pdf. Available online: https://ambermd.org/doc12/Amber22.pdf (accessed on 13 March 2024).
- Bi, S.; Ding, L.; Tian, Y.; Song, D.; Zhou, X.; Liu, X.; Zhang, H. Investigation of the interaction between flavonoids and human serum albumin. J. Mol. Struct. 2004, 703, 37–45. [Google Scholar] [CrossRef]
- Chaves, O.A.; Oliveira, C.H.d.S.; Ferreira, R.C.; Ferreira, V.F.; Ferreira, S.B.; Serpa, C.; Cesarin-Sobrinho, D.; Silva, F.d.A.d.; Netto-Ferreira, J.C. Spectroscopic and in silico evaluation on the interactive behavior between substituted β-2,3-dihydrofuran naphthoquinones and human serum albumin. Chem. Phys. Impact 2024, 8, 100465. [Google Scholar] [CrossRef]
- Naveenraj, S.; Anandan, S. Binding of serum albumins with bioactive substances—Nanoparticles to drugs. J. Photochem. Photobiol. C Photochem. Rev. 2013, 14, 53–71. [Google Scholar] [CrossRef]
- Miles, A.J.; Janes, R.W.; Wallace, B.A. Tools and methods for circular dichroism spectroscopy of proteins: A tutorial review. Chem. Soc. Rev. 2021, 50, 8400–8413. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.; Heath, R.J. Structural and Biochemical Features of Human Serum Albumin Essential for Eukaryotic Cell Culture. Int. J. Mol. Sci. 2021, 22, 8411. [Google Scholar] [CrossRef] [PubMed]
- Hosainzadeh, A.; Gharanfoli, M.; Saberi, M.; Chamani, J. Probing the interaction of human serum albumin with bilirubin in the presence of aspirin by multi-spectroscopic, molecular modeling and zeta potential techniques: Insight on binary and ternary systems. J. Biomol. Struct. Dyn. 2012, 29, 1013–1050. [Google Scholar] [CrossRef] [PubMed]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggard, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Rocchia, W.; Alexov, E.; Honig, B. Extending the Applicability of the Nonlinear Poisson−Boltzmann Equation: Multiple Dielectric Constants and Multivalent Ions. J. Phys. Chem. B 2001, 105, 6507–6514. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Lyu, X.; Zhang, Q.; Liang, D.; Huang, Y. Interaction between human serum albumin and cholesterol-grafted polyglutamate as the potential carriers of protein drugs. Acta Pharm. Sin. B 2019, 9, 186–193. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, D.; Pan, Y.; Chen, B. Human Serum Albumin Based Nanodrug Delivery Systems: Recent Advances and Future Perspective. Polymers 2023, 15, 16. [Google Scholar] [CrossRef]
- Shen, Z.; Xiang, Y.; Vergara, S.; Chen, A.; Xiao, Z.; Santiago, U.; Jin, C.; Sang, Z.; Luo, J.; Chen, K.; et al. A resource of high-quality and versatile nanobodies for drug delivery. iScience 2021, 24, 103014. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavalieri, G.; Marson, D.; Giurgevich, N.; Valeri, R.; Felluga, F.; Laurini, E.; Pricl, S. Molecular Ballet: Investigating the Complex Interaction between Self-Assembling Dendrimers and Human Serum Albumin via Computational and Experimental Methods. Pharmaceutics 2024, 16, 533. https://doi.org/10.3390/pharmaceutics16040533
Cavalieri G, Marson D, Giurgevich N, Valeri R, Felluga F, Laurini E, Pricl S. Molecular Ballet: Investigating the Complex Interaction between Self-Assembling Dendrimers and Human Serum Albumin via Computational and Experimental Methods. Pharmaceutics. 2024; 16(4):533. https://doi.org/10.3390/pharmaceutics16040533
Chicago/Turabian StyleCavalieri, Gabriele, Domenico Marson, Nicoletta Giurgevich, Rachele Valeri, Fulvia Felluga, Erik Laurini, and Sabrina Pricl. 2024. "Molecular Ballet: Investigating the Complex Interaction between Self-Assembling Dendrimers and Human Serum Albumin via Computational and Experimental Methods" Pharmaceutics 16, no. 4: 533. https://doi.org/10.3390/pharmaceutics16040533
APA StyleCavalieri, G., Marson, D., Giurgevich, N., Valeri, R., Felluga, F., Laurini, E., & Pricl, S. (2024). Molecular Ballet: Investigating the Complex Interaction between Self-Assembling Dendrimers and Human Serum Albumin via Computational and Experimental Methods. Pharmaceutics, 16(4), 533. https://doi.org/10.3390/pharmaceutics16040533




