Optimizing mRNA-Loaded Lipid Nanoparticles as a Potential Tool for Protein-Replacement Therapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of LNP Formulations from Different Lipid Mixes (LM)
2.3. Determination of mRNA Encapsulation Efficiency (EE) by Ribogreen Assay
2.4. Particle Size, Zeta Potential (Z-Pot), and Polydispersity Index (PDI)
2.5. Cryogenic Transmission Electron Microscopy (Cryo-TEM)
2.6. Small-Angle X-ray Scattering (SAXS)
2.7. Evaluation of In Vitro Expression Levels
2.8. Isolation, Seeding, and LNP Treatment of PBMCs
2.9. Determination of Activation Markers in CD11c+ Cells from PBMCs
2.10. Cytokine Profile
2.11. Hemotoxicity Studies
2.12. Ethics Statement
2.13. In Vivo Biodistribution Assay
2.14. Statistical Analysis
3. Results and Discussion
3.1. Development and Physicochemical Characterization of LNP Formulations
3.2. Stability of LNPs
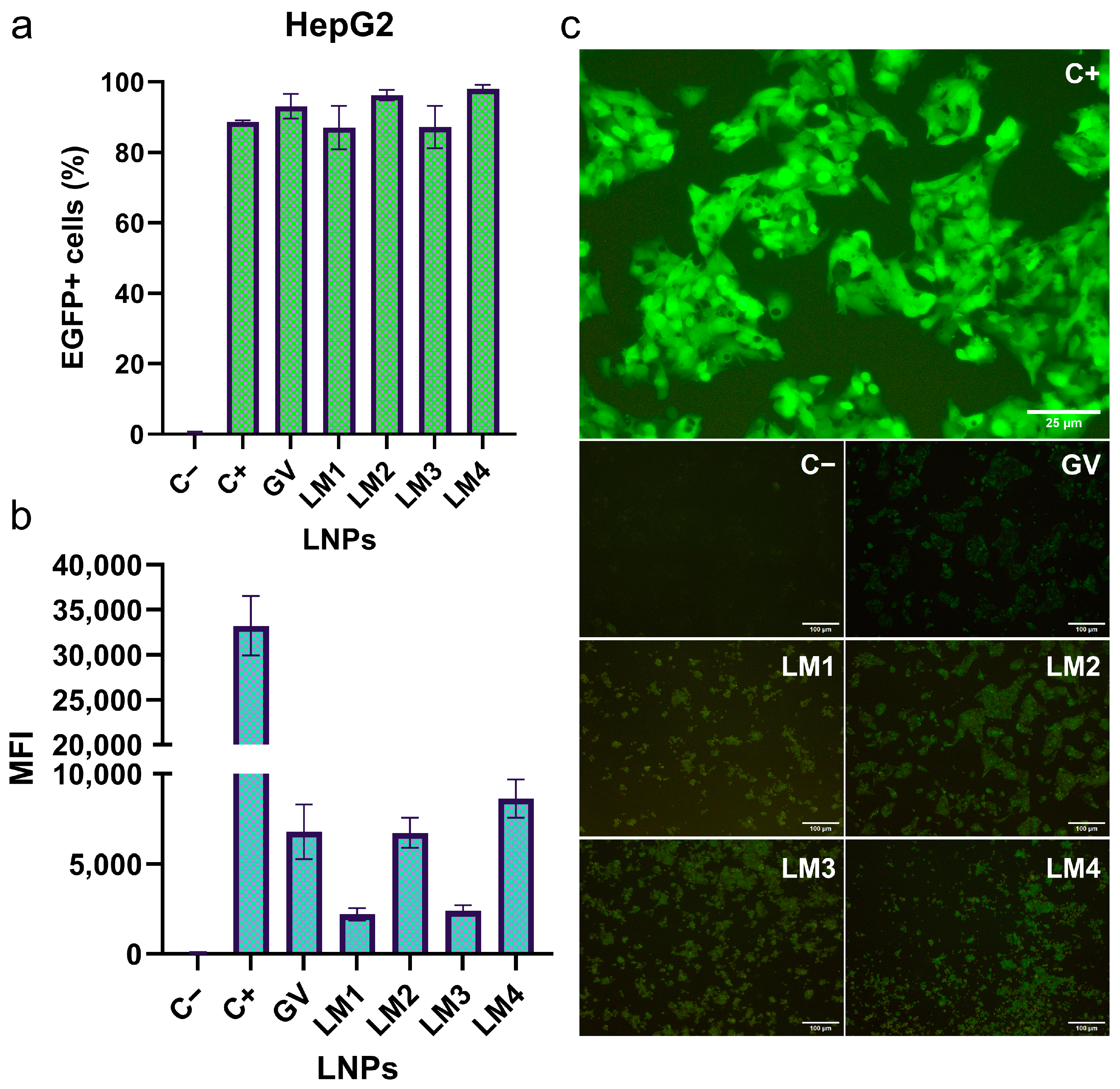
3.3. EGFP mRNA Transfection by LNP Formulations in HepG2 and Dendritic Cell Lines
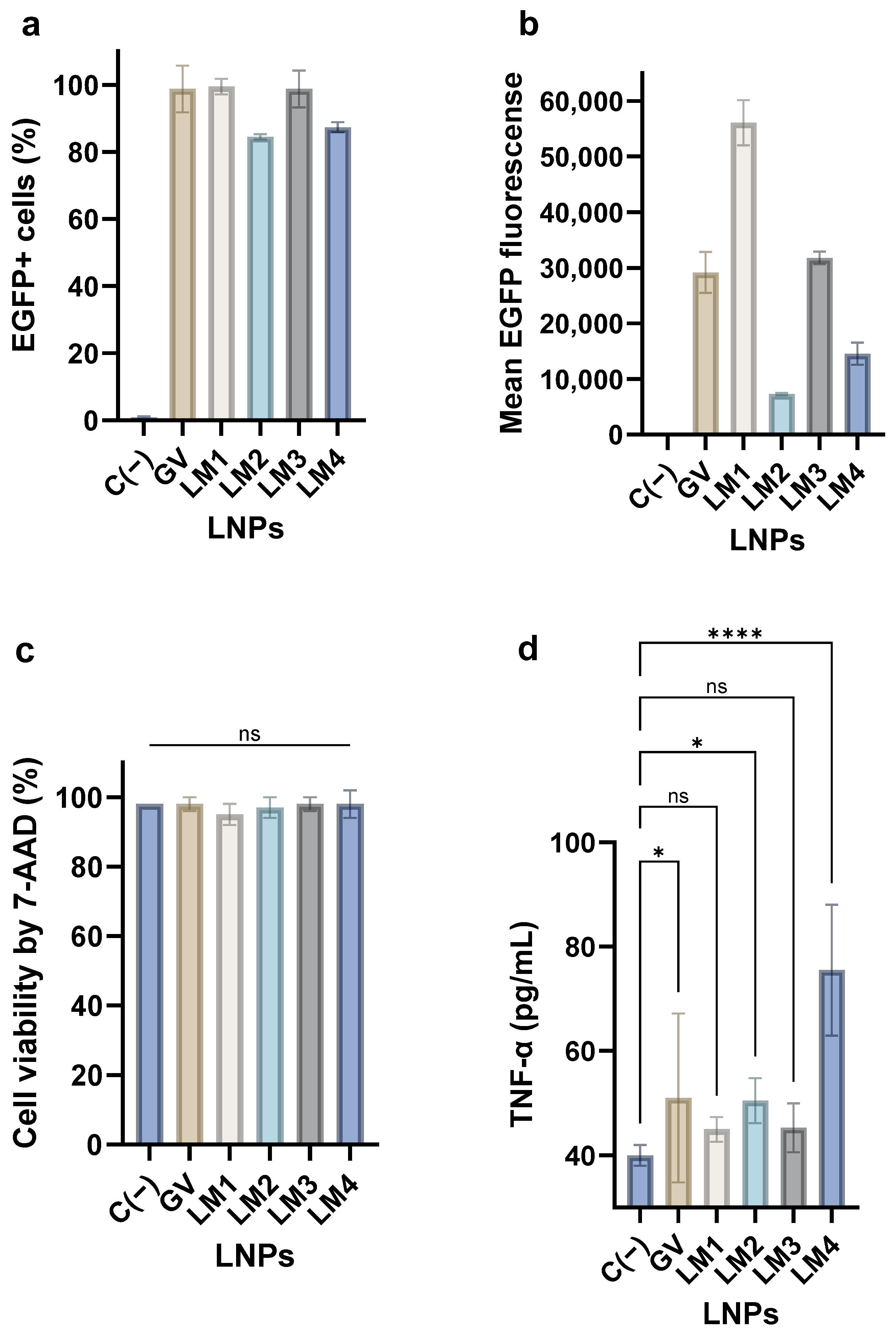
3.4. mRNA Transfection and Cytokine Secretion by PBMCs Treated with LNPs
3.5. Hemotoxicity of the LNPs
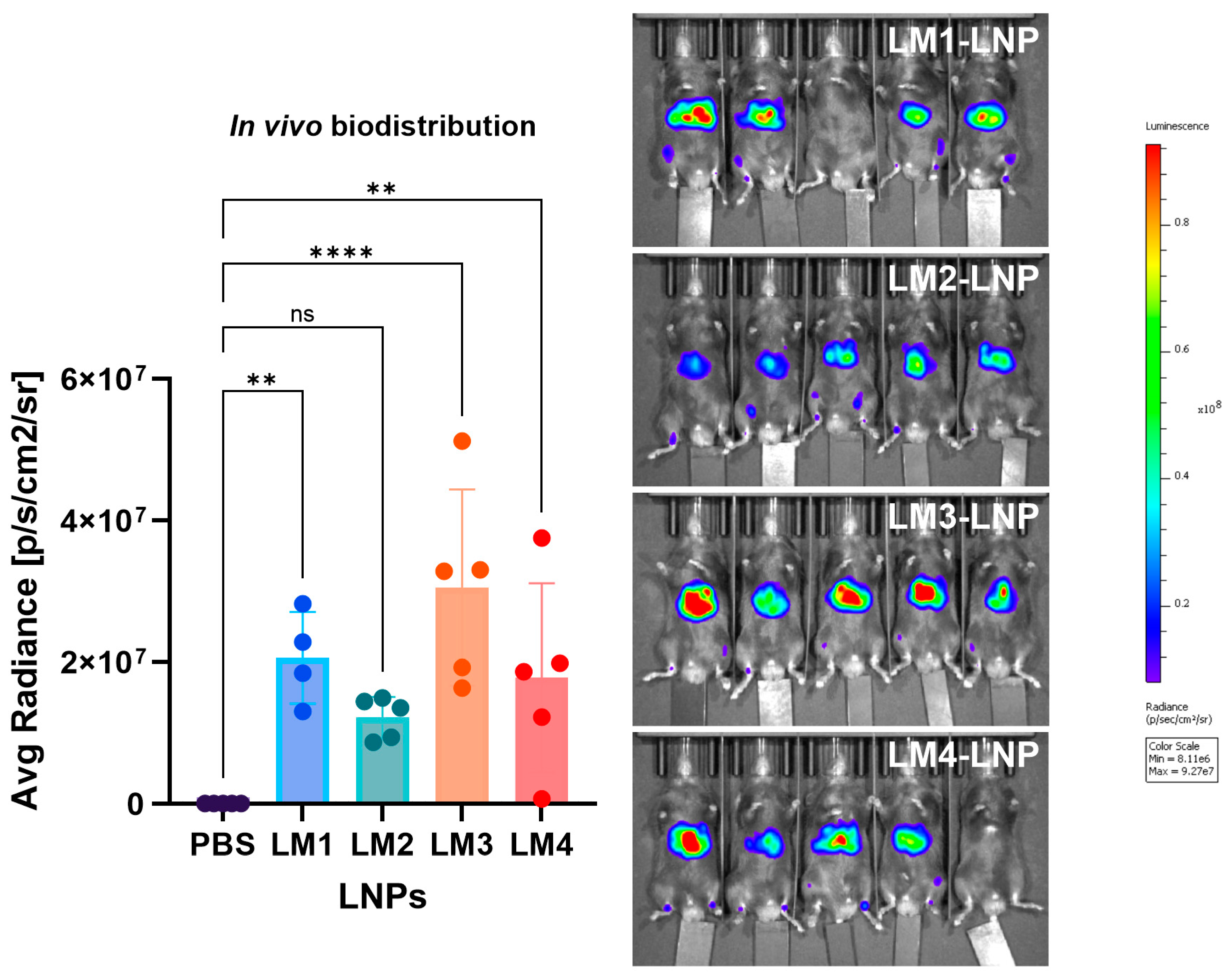
3.6. In Vivo Biodistribution
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reynolds, L.; Dewey, C.; Asfour, G.; Little, M. Vaccine efficacy against SARS-CoV-2 for Pfizer BioNTech, Moderna, and AstraZeneca vaccines: A systematic review. Front. Public Health 2023, 11, 1229716. [Google Scholar] [CrossRef] [PubMed]
- Alameh, M.-G.; Tombácz, I.; Bettini, E.; Lederer, K.; Ndeupen, S.; Sittplangkoon, C.; Wilmore, J.R.; Gaudette, B.T.; Soliman, O.Y.; Pine, M.; et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 2021, 54, 2877–2892.e7. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Jiang, A.Y.; Raji, I.; Atyeo, C.; Raimondo, T.M.; Gordon, A.G.R.; Rhym, L.H.; Samad, T.; MacIsaac, C.; Witten, J.; et al. Enhancing the immunogenicity of lipid-nanoparticle mRNA vaccines by adjuvanting the ionizable lipid and the mRNA. Nat. Biomed. Eng. 2023, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeong, M.; Park, J.; Jung, H.; Lee, H. Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics. Exp. Mol. Med. 2023, 55, 2085–2096. [Google Scholar] [CrossRef] [PubMed]
- Cacicedo, M.L.; Weinl-Tenbruck, C.; Frank, D.; Wirsching, S.; Straub, B.K.; Hauke, J.; Okun, J.G.; Horscroft, N.; Hennermann, J.B.; Zepp, F.; et al. mRNA-based therapy proves superior to the standard of care for treating hereditary tyrosinemia 1 in a mouse model. Mol. Ther. Methods Clin. Dev. 2022, 26, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Cacicedo, M.L.; Weinl-Tenbruck, C.; Frank, D.; Limeres, M.J.; Wirsching, S.; Hilbert, K.; Pasha Famian, M.A.; Horscroft, N.; Hennermann, J.B.; Zepp, F.; et al. Phenylalanine hydroxylase mRNA rescues the phenylketonuria phenotype in mice. Front. Bioeng. Biotechnol. 2022, 10, 993298. [Google Scholar] [CrossRef] [PubMed]
- Córdoba, K.M.; Jericó, D.; Sampedro, A.; Jiang, L.; Iraburu, M.J.; Martini, P.G.V.; Berraondo, P.; Avila, M.A.; Fontanellas, A. Chapter Two-Messenger RNA as a personalized therapy: The moment of truth for rare metabolic diseases. In International Review of Cell and Molecular Biology; Aranda, F., Berraondo, P., Galluzzi, L., Eds.; mRNA-Based Therapeutics; Academic Press: Cambridge, MA, USA, 2022; Volume 372, pp. 55–96. [Google Scholar] [CrossRef]
- Zhang, H.; Han, X.; Alameh, M.-G.; Shepherd, S.J.; Padilla, M.S.; Xue, L.; Butowska, K.; Weissman, D.; Mitchell, M.J. Rational design of anti-inflammatory lipid nanoparticles for mRNA delivery. J. Biomed. Mater. Res. A 2022, 110, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Friberg, D.; Bryant, J.; Shannon, W.; Whiteside, T.L. In vitro cytokine production by normal human peripheral blood mononuclear cells as a measure of immunocompetence or the state of activation. Clin. Diagn. Lab. Immunol. 1994, 1, 261–268. [Google Scholar] [CrossRef]
- Ashiotis, G.; Deschildre, A.; Nawaz, Z.; Wright, J.P.; Karkoulis, D.; Picca, F.E.; Kieffer, J. The fast azimuthal integration Python library: pyFAI. J. Appl. Crystallogr. 2015, 48 Pt 2, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.S. Analysis of small-angle scattering data from colloids and polymer solutions: Modeling and least-squares fitting. Adv. Colloid Interface Sci. 1997, 70, 171–210. [Google Scholar] [CrossRef]
- Sebastiani, F.; Yanez Arteta, M.; Lerche, M.; Porcar, L.; Lang, C.; Bragg, R.A.; Elmore, C.S.; Krishnamurthy, V.R.; Russell, R.A.; Darwish, T.; et al. Apolipoprotein E Binding Drives Structural and Compositional Rearrangement of mRNA-Containing Lipid Nanoparticles. ACS Nano 2021, 15, 6709–6722. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. Diffuse scattering from lamellar structures. Soft Matter 2022, 18, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Hald Albertsen, C.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Carreño, J.M.; Simon, V.; Dawson, K.; Krammer, F.; Kent, S.J. Impact of anti-PEG antibodies induced by SARS-CoV-2 mRNA vaccines. Nat. Rev. Immunol. 2023, 23, 135–136. [Google Scholar] [CrossRef] [PubMed]
- Ryals, R.C.; Patel, S.; Acosta, C.; McKinney, M.; Pennesi, M.E.; Sahay, G. The effects of PEGylation on LNP based mRNA delivery to the eye. PLoS ONE 2020, 15, e0241006. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.K.; Tam, Y.Y.C.; Chen, S.; Hafez, I.M.; Cullis, P.R. Microfluidic Mixing: A General Method for Encapsulating Macromolecules in Lipid Nanoparticle Systems. J. Phys. Chem. B 2015, 119, 8698–8706. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.; Sebastiani, F.; Arteta, M.Y.; Terry, A.; Fornell, A.; Russell, R.; Mahmoudi, N.; Nylander, T. Evolution of the structure of lipid nanoparticles for nucleic acid delivery: From in situ studies of formulation to colloidal stability. J. Colloid Interface Sci. 2024, 660, 66–76. [Google Scholar] [CrossRef]
- Maguire, C.M.; Rösslein, M.; Wick, P.; Prina-Mello, A. Characterisation of particles in solution—A perspective on light scattering and comparative technologies. Sci. Technol. Adv. Mater. 2018, 19, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; El-Mayta, R.; Murdoch, T.J.; Warzecha, C.C.; Billingsley, M.M.; Shepherd, S.J.; Gong, N.; Wang, L.; Wilson, J.M.; Lee, D.; et al. Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater. Sci. 2021, 9, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Eggesbø, J.B.; Hjermann, I.; Lund, P.K.; Joø, G.B.; Øvstebø, R.; Kierulf, P. LPS-induced release of IL-1β, IL-6, IL-8, TNF-α and sCD14 in whole blood and PBMC from persons with high or low levels of HDL-lipoprotein. Cytokine 1994, 6, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Anshita, D.; Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol. 2021, 101, 107598. [Google Scholar] [CrossRef] [PubMed]
- Bickel, M. The role of interleukin-8 in inflammation and mechanisms of regulation. J. Periodontol. 1993, 64, 456–460. [Google Scholar] [PubMed]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Toledo, C.; Gambaro, R.C.; Padula, G.; Vela, M.E.; Castro, G.R.; Chain, C.Y.; Islan, G.A. Binary Medical Nanofluids by Combination of Polymeric Eudragit Nanoparticles for Vehiculization of Tobramycin and Resveratrol: Antimicrobial, Hemotoxicity and Protein Corona Studies. J. Pharm. Sci. 2021, 110, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Chinnaiyan, S.K.; Karthikeyan, D.; Gadela, V.R. Development and Characterization of Metformin Loaded Pectin Nanoparticles for T2 Diabetes Mellitus. Pharm. Nanotechnol. 2018, 6, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Pateev, I.; Seregina, K.; Ivanov, R.; Reshetnikov, V. Biodistribution of RNA Vaccines and of Their Products: Evidence from Human and Animal Studies. Biomedicines 2024, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Yavuz, A.; Coiffier, C.; Garapon, C.; Gurcan, S.; Monge, C.; Exposito, J.-Y.; Arruda, D.C.; Verrier, B. DLin-MC3-Containing mRNA Lipid Nanoparticles Induce an Antibody Th2-Biased Immune Response Polarization in a Delivery Route-Dependent Manner in Mice. Pharmaceutics 2023, 15, 1009. [Google Scholar] [CrossRef] [PubMed]
- Ndeupen, S.; Qin, Z.; Jacobsen, S.; Bouteau, A.; Estanbouli, H.; Igyártó, B.Z. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 2021, 24, 103479. [Google Scholar] [CrossRef] [PubMed]
- Chander, N.; Basha, G.; Yan Cheng, M.H.; Witzigmann, D.; Cullis, P.R. Lipid nanoparticle mRNA systems containing high levels of sphingomyelin engender higher protein expression in hepatic and extra-hepatic tissues. Mol. Ther.-Methods Clin. Dev. 2023, 30, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, Z.; Jin, P.; Jiang, K.; Xu, Y.; Pan, F.; Tian, K.; Yuan, Z.; Liu, X.E.; Fu, J.; et al. Effects of PEG antibodies on in vivo performance of LNP-mRNA vaccines. Int. J. Pharm. 2024, 650, 123695. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Faggiano, S.; Mozzarelli, A. Enzyme Replacement Therapy for Genetic Disorders Associated with Enzyme Deficiency. Curr. Med. Chem. 2022, 29, 489–525. [Google Scholar] [CrossRef] [PubMed]










| Sample | Diameter (nm) | Multilayer (%) | N_av | χ2 |
|---|---|---|---|---|
| GV-LNP | 84.8 ± 0.1 | 72 | 2.3 ± 0.0 | 0.75 |
| LM1-LNP | 82.5 ± 0.2 | 40 | 1.2 ± 0.3 | 0.76 |
| LM2-LNP | 97.1 ± 0.2 | 55 | 2.2 ± 0.0 | 0.53 |
| LM3-LNP | 67.2 ± 0.2 | 0 | – | 0.93 |
| LM4-LNP | 57.4 ± 0.1 | 69 | 1.7 ± 0.1 | 0.74 |
| Hemolysis (%) | ||
|---|---|---|
| Formulation | Incubation Time (h) | |
| 1 | 24 | |
| LM1-LNP | 0.0 ± 0.0 | 1.0 ± 0.4 |
| LM2-LNP | 0.0 ± 0.0 | 1.9 ± 0.2 |
| LM3-LNP | 0.0 ± 0.0 | 0.2 ± 0.1 |
| LM4-LNP | 0.4 ± 0.6 | 1.0 ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gambaro, R.; Rivero Berti, I.; Limeres, M.J.; Huck-Iriart, C.; Svensson, M.; Fraude, S.; Pretsch, L.; Si, S.; Lieberwirth, I.; Gehring, S.; et al. Optimizing mRNA-Loaded Lipid Nanoparticles as a Potential Tool for Protein-Replacement Therapy. Pharmaceutics 2024, 16, 771. https://doi.org/10.3390/pharmaceutics16060771
Gambaro R, Rivero Berti I, Limeres MJ, Huck-Iriart C, Svensson M, Fraude S, Pretsch L, Si S, Lieberwirth I, Gehring S, et al. Optimizing mRNA-Loaded Lipid Nanoparticles as a Potential Tool for Protein-Replacement Therapy. Pharmaceutics. 2024; 16(6):771. https://doi.org/10.3390/pharmaceutics16060771
Chicago/Turabian StyleGambaro, Rocío, Ignacio Rivero Berti, María José Limeres, Cristián Huck-Iriart, Malin Svensson, Silvia Fraude, Leah Pretsch, Shutian Si, Ingo Lieberwirth, Stephan Gehring, and et al. 2024. "Optimizing mRNA-Loaded Lipid Nanoparticles as a Potential Tool for Protein-Replacement Therapy" Pharmaceutics 16, no. 6: 771. https://doi.org/10.3390/pharmaceutics16060771







