Abstract
Novel antifungal drugs are urgently needed to treat candidiasis caused by the emerging fungal multidrug-resistant pathogen Candida auris. In this study, the most cost-effective drug repurposing technology was adopted to identify an appropriate option among the 1615 clinically approved drugs with anti-C. auris activity. High-throughput virtual screening of 1,3-beta-glucanosyltransferase inhibitors was conducted, followed by an analysis of the stability of 1,3-beta-glucanosyltransferase drug complexes and 1,3-beta-glucanosyltransferase–dutasteride metabolite interactions and the confirmation of their activity in biofilm formation and planktonic growth. The analysis identified dutasteride, a drug with no prior antifungal indications, as a potential medication for anti-auris activity in seven clinical C. auris isolates from Saudi Arabian patients. Dutasteride was effective at inhibiting biofilm formation by C. auris while also causing a significant reduction in planktonic growth. Dutasteride treatment resulted in disruption of the cell membrane, the lysis of cells, and crushed surfaces on C. auris, and significant (p-value = 0.0057) shrinkage in the length of C. auris was noted at 100,000×. In conclusion, the use of repurposed dutasteride with anti-C. auris potential can enable rapid recovery in patients with difficult-to-treat candidiasis caused by C. auris and reduce the transmission of nosocomial infection.
1. Introduction
After its discovery in Japan in 2009, Candida auris, a multidrug-resistant (MDR) fungus, was the first fungal human pathogen to be flagged globally as an imminent and immediate health risk [1,2,3,4,5,6]. It is a difficult pathogen to treat because of its strong innate and acquired resistance to known antifungal drugs [3]. Moreover, identifying C. auris using traditional microbiological methods is challenging [7]. As a result, the incidence and prevalence of C. auris have been increasing among immunocompromised patients and those under long-term hospitalization [1,3,8,9,10]. Owing to its high mortality rate, tendency to cause nosocomial invasive infections, and multidrug-resistant nature, C. auris poses a serious global public health concern [11,12,13,14]. C. auris has been reported to have a high rate of intrinsic resistance to antifungal treatments such as amphotericin B and fluconazole, but few countries have reported acquired echinocandin resistance [15,16]. In order to prevent a future epidemic, which is probable given the current status of C. auris, finding and identifying effective pharmaceutical components to overcome these drug-resistant pathogens must be prioritized [17]. A new antifungal medicine often takes 10 to 15 years to develop; thus, screening molecule libraries can provide a variety of solutions and expedite the identification of candidate drugs [18,19]. Among more than 4300 approved drugs, Cheng et al. reported six anti-C. auris compounds that had possible associations with 13 of them [18]. Their screening revealed the potential of amebicide iodoquinol and leishmanicide miltefosine as repositionable compounds, owing to their inhibition of C. auris growth [19].
The formation of biofilms (surface-adherent communities) is a major factor in C. auris pathogenesis as biofilms contribute to its drug resistance profile. C. auris isolates have been detected in several clinical locations, such as central venous catheters, wounds, and stents [20,21]. Biofilms play a crucial role in promoting the persistence and survival of C. auris in healthcare facilities as they allow the microorganism to survive on various surfaces for extended periods of time [22]. These factors are responsible for the emergence of C. auris as the first leading fungal pathogen to cause an international outbreak in healthcare institutions. Hence, the Centers for Disease Control and Prevention (CDC) released the “Antibiotic Resistance Threats in the United States” report in 2019, which classified C. auris as one of five “Urgent Threats” and advocated for assertive and expeditious measures against it [20]. As such, resistance to the limited number of antifungal treatments that are currently available will create various problems in the foreseeable future. Therefore, it is imperative to conduct thorough research on novel pharmaceuticals for combating infections induced by C. auris. A suitable strategic approach to acquiring novel antifungal drugs is to explore drug repurposing or repositioning [23,24,25]. For instance, anthelmintic niclosamide has been reported to have an inhibitory effect on C. auris biofilm formation. This drug was identified via a screening approach for inhibitors against C. albicans and has since been approved by the U.S. Food and Drug Administration (FDA) [26].
Drug repurposing studies are promising for various diseases [27,28,29,30] and have been conducted to identify several active drugs against C. auris, such as iodoquinol [20], Miltefosine [20,31], Robenidine [32], Salicylanilide oxyclozanide [33], Pyrvinium pamoate [34], Broxyquinoline [18], Tamoxifen citrate [34], Alexidine dihydrochloride [18,35], AC- 93253 iodide [18], Chloroxine [18], Clioquinol [18], Sertraline [36], Rolipram [34], Trifluoperazine [34], dihydrochloride [34], Thiethylperazine dimalate [34], ebselen [19,34], Disulfiram [37], (−)- MK 801 hydrogen maleate [34], Suloctidil [34], Ciclopirox ethanolamine [34], and Guanadrel sulfate [34]. Nanomaterial-based studies and drug repurposing approaches have led to the proposal of several alternative drugs with C. aruis growth inhibition activity. However, an effective treatment is still needed [30,38]. These compounds exhibit frequent activity against new targets that are currently unexplored in antifungal therapies and, consequently, in yeast adaptability. Thus, it is vital to improve infection control procedures by producing effective state-of-the-art disinfectants for managing C. auris and opportunistic infection [39]. The identification and development of novel and unique antifungal compounds are of paramount importance in the effective management of C. auris infections. While the results obtained using in silico screening approaches are not as precise as those collected from in vivo studies, the cost reduction justifies the efforts. Analyzing 1600 FDA-approved drugs through high-throughput virtual screening and experimental drug repurposing takes much less time compared with conducting in vivo studies for a similar number of drugs. Employing drug repurposing, a potential drug (dutasteride) was identified to inhibit 1,3-beta-glucanosyltransferase, a major cell-wall-associated protein in fungal organisms, including C. auris and Candida albicans, by retrieving a library of 1615 FDA-approved drugs from the ZINC database. The main objective of this research was to identify novel drugs against C. auris and C. albicans. This was achieved by performing a virtual screening of 1,3-beta-glucanosyltransferase inhibitors [40,41] to shortlist candidate drugs from the FDA-approved list based on docking scores. This was followed by analyzing the stability of proteins and protein–ligand complexes using molecular dynamics simulations. Then, the uppermost drug with the lowermost binding energy based on drug docking with 1,3-beta-glucanosyltransferase was further explored through an in vitro evaluation of its inhibitory activity against native isolates of C. auris and C. albicans. Its inhibitory effect was also evaluated by assessing fungal morphology under a scanning electron microscope after treatment with the candidate drug.
2. Materials and Methods
2.1. FDA-Approved Drugs and Virtual Screening
The ZINC 20 database [42] [Irwin and Shoichet Laboratories, University of California, USA] is a curated database of 230 million compounds, including FDA-approved drugs, for virtual screening. In addition, ZINC contains over 750 million purchasable compounds that can be searched for analogs in less than a minute. All of the FDA-approved drugs from the ZINC database [42] were obtained and underwent virtual screening to identify potential inhibitors of 1,3-beta-glucanosyltransferase (UniProt: A0A2H1A5Q4), which is one of the surface proteins of C. auris. The structure of 1,3-beta-glucanosyltransferase was constructed in PDB format as described earlier using SWISS-MODEL and PYMOL and validated through PROCHECK [43,44]. The FDA-approved drug molecules from the ZINC database and the PDB of 1,3-beta-glucanosyltransferase were imported to the PyRx workspace. PyRx 0.9.9 is a software program used for Computational Drug Discovery through Virtual Screening that enables the screening of compounds from libraries against potential drug targets. Virtual screening was performed individually for each FDA-approved drug molecule against 1,3-beta-glucanosyltransferase using the PyRx 0.9.9 virtual screening tool (https://pyrx.sourceforge.io/ accessed on 28 July 2023). “For computer time, this research used the resources of the Supercomputing Laboratory at King Abdullah University of Science & Technology (KAUST) in Thuwal, Saudi Arabia”.
To understand the behavior of the protein–ligand complex’s 1,3-beta-glucanosyltransferase from C. auris and dutasteride within a biological system’s aqueous environment, we employed molecular dynamics simulations. We simulated the docked conformation with the highest affinity for the target protein using Desmond software (version 4.1). The simulation used a TIP3P water model with a cubic box and a 10 Å side length. To maintain electrical neutrality, the system’s net charge was neutralized. We performed the simulations in the NPT ensemble, mimicking constant pressure (1 bar) and temperature (300 K) using the Berendsen coupling scheme, and acquired the final poses for the interacting 1,3-beta-glucanosyltransferase–dutasteride complex at the end. We ran a 100 ns production simulation for the protein–ligand complex after a 100 ns equilibration phase. The stability of the complex was confirmed by the root-mean-square deviation (RMSD) data of the 1,3-beta-glucanosyltransferase–dutasteride complex, dutasteride’s fit on 1,3-beta-glucanosyltransferase, and the development of hydrogen bonds, as well as hydrophobic and water bridges, during the interaction in the simulated 100 ns trajectories.
2.2. C. auris and C. albicans Strains
Clinical isolates of two organisms were used in this study including (1) C. auris (CA1, CA2, CA3, CA6, CA7, and CA8) and (2) C. albicans (CAL). C. auris isolates were previously collected from healthcare facilities in Saudi Arabia and identified as having multidrug-resistance mutations [38,45]. The C. auris and C. albicans (CAL) strains were reconfirmed using the 18S rRNA Gene and the ITSa and ITSb regions, as described earlier [38]. Multidrug-resistant C. auris CA1 and the clinical isolate C. albicans CAL were used for initial primary screening of the selected clinically approved drugs. Follow-up experiments were performed on the C. auris (CA1, CA2, CA3, CA6, CA7, and CA8) and C. albicans (CAL) strains. C. auris and C. albicans cultures were cultivated for 24 h by introducing cells into 10 mL of Sabouraud Dextrose Broth (SDB) medium in 150 mL flasks. The flasks were then placed in an orbital shaker operating at a speed of 150 to 180 revolutions per minute (rpm) and maintained at a temperature of 30 °C. After 18 h of incubation, 0.5 × 105 cells/mL were used for the planktonic assay and 1 × 106 cells/mL were suspended in RPMI medium for the biofilm assay (pH 6.9).
2.3. Primary Screening for Anti-C. auris Activity
Cell suspensions of C. auris and C. albicans were prepared and adjusted to a 0.5 McFarland standard to be used for the initial screening via the Kirby–Bauer disk diffusion method. By measuring the zone of inhibition, the antifungal susceptibility of DR1 [zinc000242548690 digoxin] and DR2 [zinc000003932831 dutasteride] to C. auris CA1 and C. albicans CAL was determined. Digoxin (0.250 mg tablet, Aspen Bad Oldesloe Gmbh, Bad Oldesloe, Germany) and dutasteride (0.5 mg capsules, GlaxoSmithKline, Poznan, Poland) were used in this study. Initially, 250 µg of dutasteride and 500 µg of digoxin were prepared separately in 1 mL of sterile distilled water as working stock solutions and stored at 4 °C. The antifungal susceptibility screening using the Kirby–Bauer disk diffusion method was performed in triplicate, and fluconazole was tested as a positive control for anti-C. auris and anti-C. albicans activities.
2.4. Determination of Anti-Candida Activity
The anti-C. auris and anti-C. albicans activities of the selected clinically approved drug dutasteride were analyzed in 96-well microtiter plates via the broth microdilution technique using dose–response assays, as described earlier, to inhibit planktonic growth and biofilm formation [19]. The screening was performed in quadruplicate.
2.5. Dose–Response Assays of Dutasteride-Mediated Inhibition of Planktonic Growth
The antifungal activity of medicines selected as positives through the initial screening was evaluated using dose–response tests, which measured their inhibition of the planktonic growth of five clinical strains of C. auris and one isolate of C. albicans. The starting concentration of the selected drugs was 32 μg/mL, and 0.0625 μg/mL was the final concentration achieved using sequential 2-fold dilutions performed across the 96-well microtiter plate rows. Additionally, positive (untreated) and negative controls (uninoculated SDB without Candida) were included, and all assays were performed in quadruplicate at each dose in the 96-well plate. After 24h in a shaking incubator at 35 °C, the cells in each well were homogenized, and a microtiter plate reader was used to determine the absorbance at 550 nm (OD550). A dose–response curve was constructed by converting the spectrophotometric measurements into normalized responses. The readings obtained from the positive and negative control wells were arbitrarily assigned growth values of 100% and 0%, respectively. The IC50, which represents the concentration of dutasteride required to inhibit 50% of planktonic growth, was determined by employing a linear equation (y = mx + n) to analyze the graph. Specifically, the y value corresponding to 50% inhibition was identified as the IC50 value using Microsoft Excel Spreadsheet Software (Microsoft Office Professional Plus 2016, Microsoft, Redmond, WA, USA), and the normalized values were fitted to a variable slope.
2.6. Assessment of Biofilm Inhibition Using Dutasteride
The biofilm biomass of the five C. auris clinical strains and one C. albicans isolate was assessed using the previously described method of crystal violet staining [19]. The dutasteride concentrations used for this assay ranged from 32 to 0.0625 μg/mL. Briefly, following biofilm formation, PBS was used to wash the plates once, and individual wells were fixed with 100 μL of methanol for 20 min. After removing the methanol, the plates were left to air dry. Then, 150 μL of 0.1% (wt/vol) crystal violet was used to stain the adherent biofilms for 10 min. After the removal of the crystal violet stain, the plates were left to dry and then washed (3 times) with 200 μL of distilled water. An inverted microscope (Olympus CKX41, Tokyo, Japan) was used to directly observe the stained samples on the 96-well plate. To estimate biomass, the crystal violet dye was dissolved by adding 100 μL of 33% glacial acetic acid to each well. The resulting solution was moved to a blank microtiter plate in order to measure the optical density at 550 nm (OD550). This measurement was used to quantify the degree of biofilm inhibition in comparison to the untreated control samples.
2.7. Assessment of Dutasteride-Treated C. auris and C. albicans Morphology Using SEM
The Kirby–Bauer disk diffusion method was utilized to assess the morphology of C. auris and C. albicans in SEM. After 24h of incubation with dutasteride, cells were collected from the following regions: the edge of the zone of inhibition by dutasteride against Candida [T1], the zone of inhibition by dutasteride against Candida [T2], and the usual growth region of Candida [T3]. The collected cells were transferred to separate tubes containing 500 µL of 3% glutaraldehyde, incubated for 16 h at 4 °C, and washed 3 times with distilled H2O for 10 min. After that, 1% osmium tetroxide (OsO4) was used to fix the samples for 16 h at 4 °C; then, the cells were washed with distilled water for 10 min followed by treatment with a series of acetone concentrations (30%, 50%, 70%, and 100%) for 10 min. Subsequently, the samples were placed in a solution of absolute acetone to facilitate the drying process. The sample was dried using a Leica EM CPD300 automated critical point drying apparatus, which utilizes a combination of acetone and carbon dioxide. The samples were sputter-coated with gold (Q150R ES machine), and double-sided carbon tape was used to mount them. C. auris and C. albicans cells were observed under a scanning electron microscope (Vega 3, TESCAN, Brno, Czech Republic) operated at 20 kV. The digitized photos were subjected to analysis using ImageJ 1.53t software [46].
3. Results
3.1. Virtual Screening for Anti-C. auris Activity
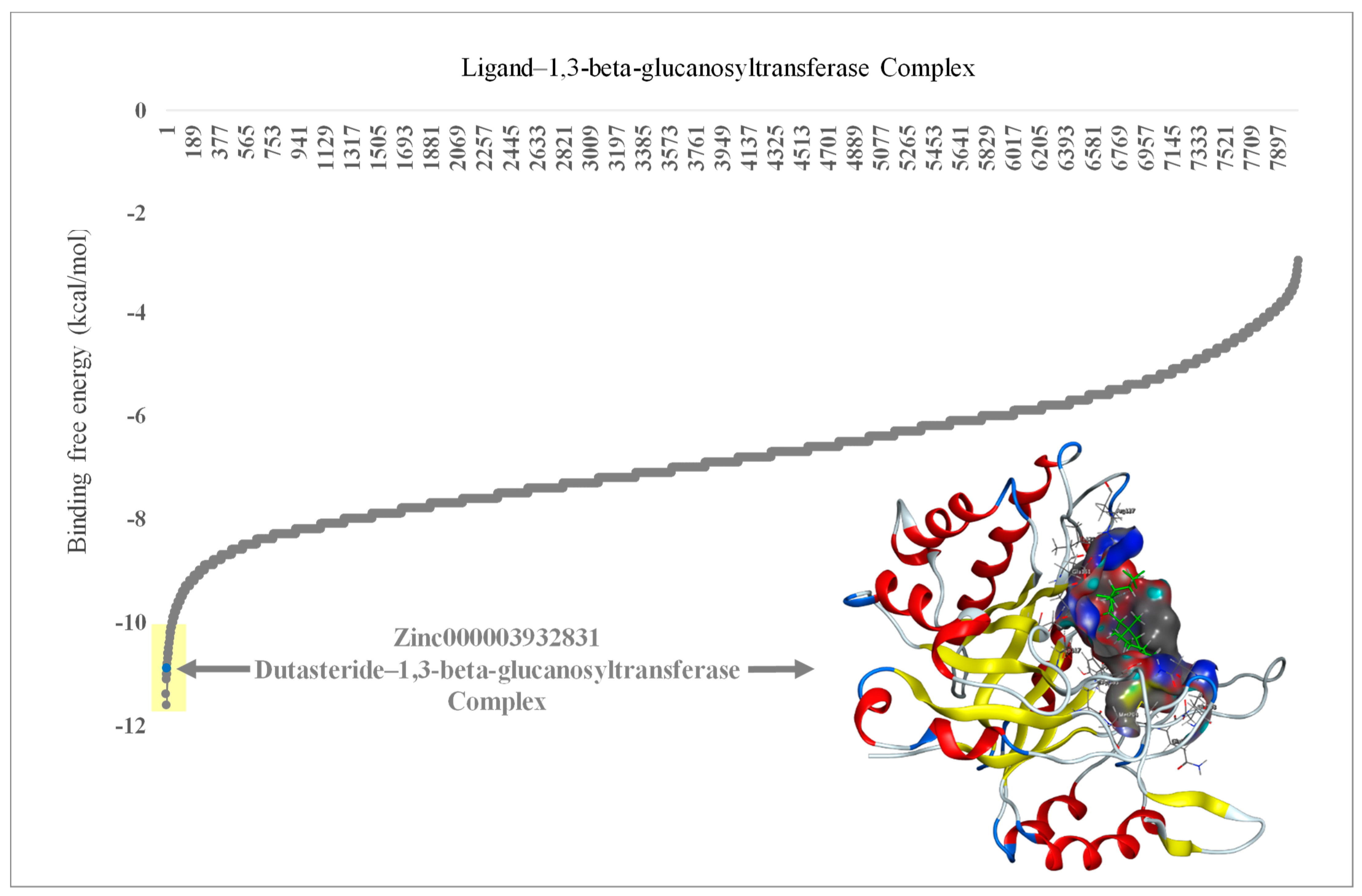
FDA-approved drugs (n = 1600) were obtained from the ZINC database and analyzed through virtual screening to detect inhibitors of 1,3-beta-glucanosyltransferase. The calculated binding affinities of the 1,3-beta-glucanosyltransferase-containing FDA-approved drugs were arranged from lowest to highest to identify the lowermost binding affinity (Figure 1). Ligands such as zinc000242548690 (digoxin), zinc000003932831 (dutasteride), zinc000052955754 (ergotamine), and zinc000203757351 (paritaprevir) were found to have a binding affinity of ≤−10 Kcal/mol. The two ligands [DR1: zinc000242548690 (digoxin) and DR2: zinc000003932831 (dutasteride)] with the lowermost binding affinity were selected for the in vitro evaluation of anti-C. auris and anti-C. albicans activity. The ligand DR2 (dutasteride) showed anti-C. auris and anti-C. albicans activity against the native fungal isolates, multidrug-resistant C. auris and C. albicans. However, DR1 showed no anti-Candida activity. Hence, DR2 was further explored by analyzing the stability of the 1,3-beta-glucanosyltransferase–DR2 complex through molecular dynamics (MD) simulations (Figure 2).
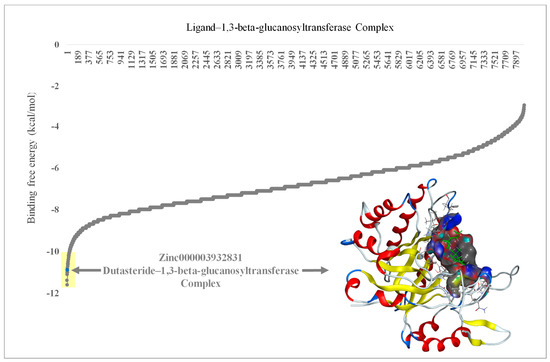
Figure 1.
Molecular docking results of 1,3-beta-glucanosyltransferase (UniProt: A0A2H1A5Q4) from C. auris and the FDA-approved drug complex, including energy minimization. The arrow indicates the lowest binding affinity of −11 Kcal/mol. Yellow shade indicates the lowest binding affinity of ≤−10 Kcal/mol in ligands zinc000242548690 (digoxin), zinc000003932831 (dutasteride), zinc000052955754 (ergotamine), and zinc000203757351 (paritaprevir). The embedded protein–ligand complex is the 1,3-beta-glucanosyltransferase–dutasteride complex.
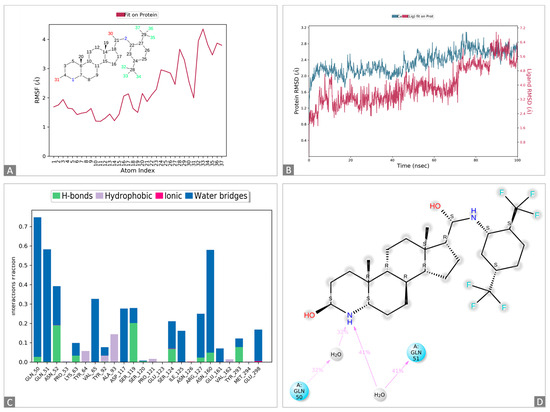
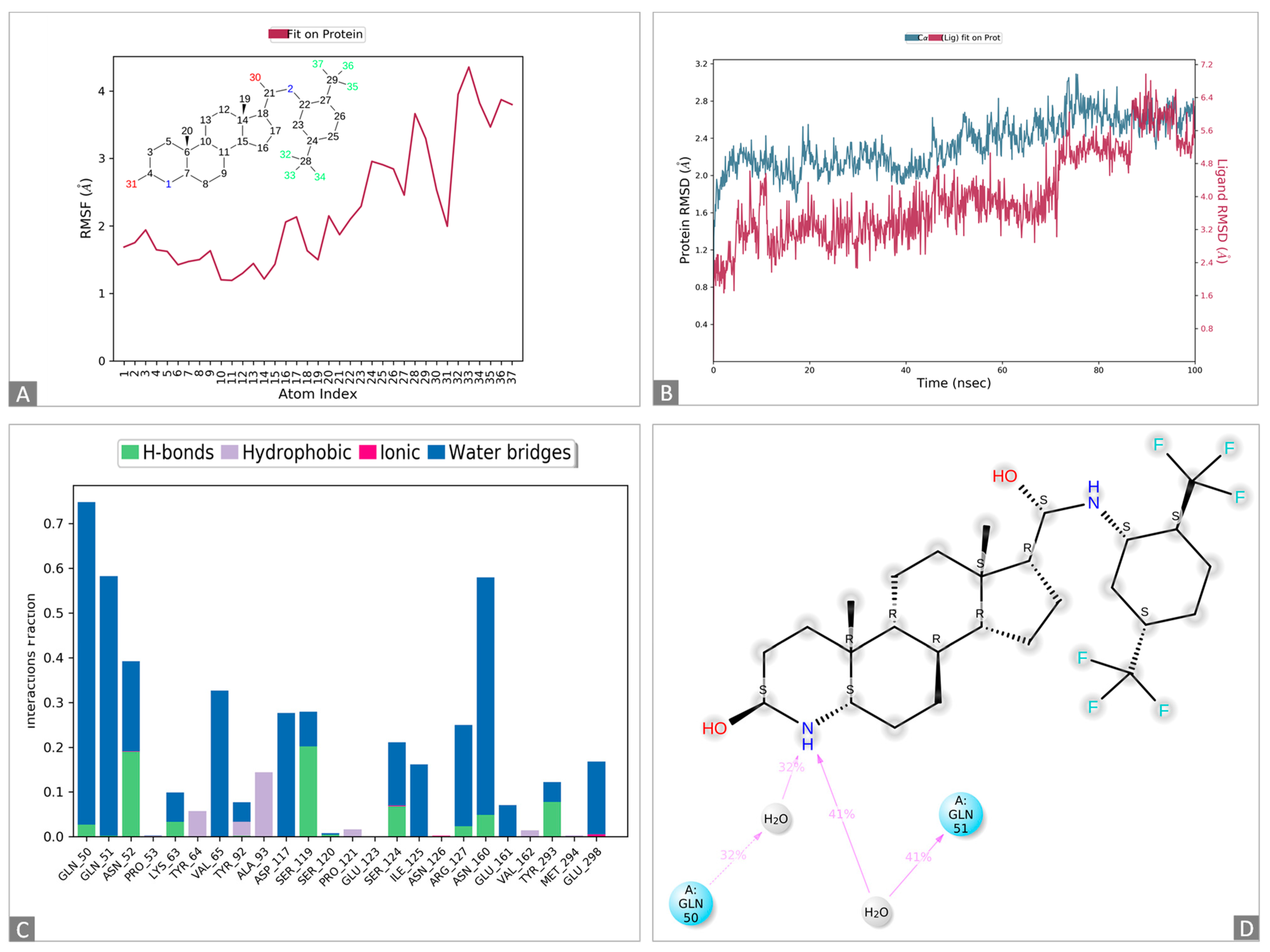
Figure 2.
Molecular dynamics simulations of the 1,3-beta-glucanosyltransferase (UniPort: A0A2H1A5Q4)-and-dutasteride complex. (A): Ligand dutasteride fluctuations with respect to 1,3-beta-glucanosyltransferase. (B): Protein–ligand root-mean-square deviation (RMSD) of 1,3-beta-glucanosyltransferase and dutasteride. (C): Types of 1,3-beta-glucanosyltransferase–dutasteride interactions or “contacts” of 1,3-beta-glucanosyltransferase and dutasteride. (D): Dutasteride atom interactions with 1,3-beta-glucanosyltransferase and dutasteride.
The docked complexes of 1,3-beta-glucanosyltransferase from C. auris and dutasteride were analyzed to assess their intermolecular interaction and stability with respect to time via a 100-nanosecond molecular dynamics simulation, and the final poses for the interacting 1,3-beta-glucanosyltransferase–dutasteride complex were acquired at the end (Figure 2 and Figure S1). The MD simulations of 1,3-beta-glucanosyltransferase–dutasteride confirmed the interaction in the docked complex. The root-mean-square deviation (RMSD) data of the 1,3-beta-glucanosyltransferase–dutasteride complex, dutasteride’s fit on 1,3-beta-glucanosyltransferase, and the development of hydrogen bonds, as well as hydrophobic and water bridges, during the interaction in the simulated 100 ns trajectories confirmed the stability of the complex. Metabolites of dutasteride, including 4′-hydroxydutasteride, 6-hydroxydutasteride, and 6,4′-dihydroxydutasteride, as well as a minor metabolite, 15-hydroxydutasteride, were tested for 1,3-beta-glucanosyltransferase–dutasteride metabolite interactions using virtual screening software. The interaction in the docked complex of the drug metabolite and 1,3-beta-glucanosyltransferase confirmed the high binding affinity of dutasteride metabolites, 6,4′-dihydroxydutasteride (binding affinity = −10.1), 6-hydroxydutasteride (binding affinity = −9.6), and 4′-hydroxydutasteride (binding affinity = −9.5) with the metabolites of dutasteride, except the minor metabolite, 15-hydroxydutasteride (binding affinity = −1.7).
3.2. Morphological Changes upon Dutasteride Treatment
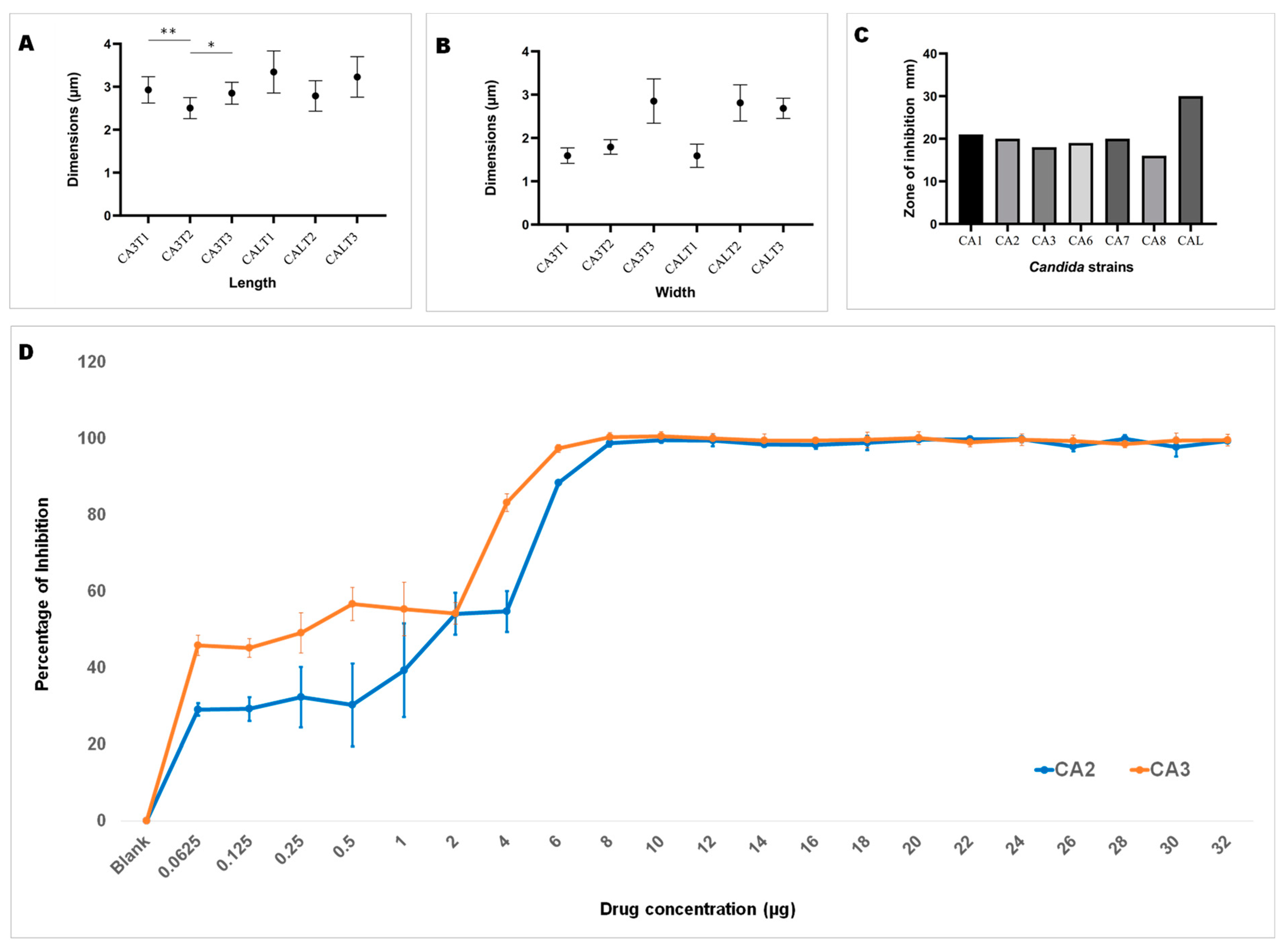
Upon ensuring the stability of the complex (1,3-beta-glucanosyltransferase–dutasteride), the morphological impacts of dutasteride on C. auris and C. albicans cells were analyzed using SEM. Antifungal susceptibility was assessed based on morphological changes in the structure (Figure 3) and dimensions (length, width) of Candida cells (Figure 4) after dutasteride treatment. One-way ANOVA in the horizontal dimension of C. auris cells (CA3T1: length 2.9294 ± 0.3072 µm; width = 1.5922 ± 0.1787 µm; CA3T2: length 2.5045 ± 0.2447 µm; width 1.7929 ± 0.1675 µm; CA3T3: length 2.8502 ± 0.2540 µm; width 2.8527 ± 0.5110 µm) revealed significant (F statistic = 6.2990; p-value = 0.0057) differences in length. One-way ANOVA in the horizontal dimension of C. albicans cells (CALT1: length 3.3452 ± 0.4906 µm; width 1.5892 ± 0.2680 µm; CALT2: length 2.7874 ± 0.3549 µm; width 2.8111 ± 0.4180 µm; CALT3: length 3.2296 ± 0.4716 µm; width 2.6848 ± 0.2331 µm) also showed significant (F statistic = 3.9722; p-value = 0.0307) differences in length (Figure 4). However, the SEM analysis of width after dutasteride treatment revealed insignificant changes in the Candida cells. The dutasteride treatment resulted in clear morphological changes indicated by cell membrane disruptions and lysed Candida cells. Furthermore, the SEM image confirmed the smooth phenotype of the healthy C. auris and C. albicans cells and the crushed phenotype of C. auris and C. albicans after dutasteride treatment. The positive control fluconazole was tested for anti-C. auris and anti-C. albicans activities, and both C. auris and C. albicans showed resistance to it.
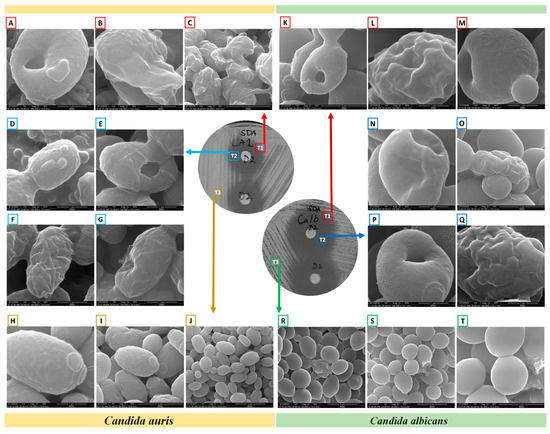
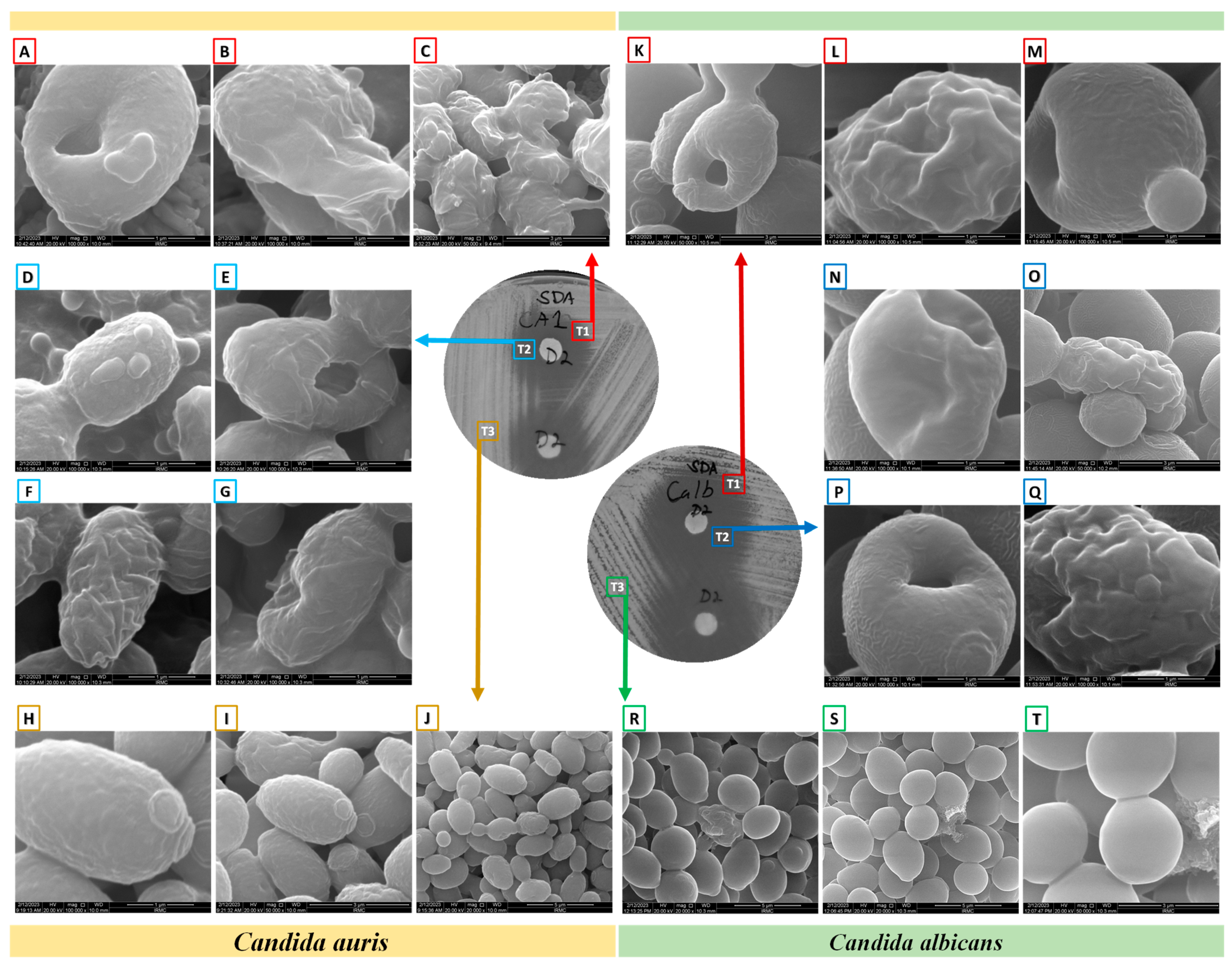
Figure 3.
Morphology of C. albicans and C. auris under a scanning electron microscope after determining their antifungal susceptibilities to dutasteride. (A–C,K–M): Morphologies of Candida from the edge of the zone of inhibition (ZOI) [T1]. (A–C): Changes in the morphologies of C. auris from the edge of the ZOI [T1] after treatment with dutasteride. (D–G,N–Q): Candida cells from the ZOI [T2]. (D–G): Changes in the morphologies of C. auris from the ZOI [T2] after treatment using dutasteride. (H–J): Structure of C. auris from the area of standard growth [T3]. (K–M): Changes in the morphologies of C. albicans from the edge of the ZOI [T1] after treatment using dutasteride. (N–Q): Changes in the morphologies of C. albicans from the ZOI [T2] after treatment using dutasteride. (R–T): Structure of C. albicans from the area of standard growth [T3].
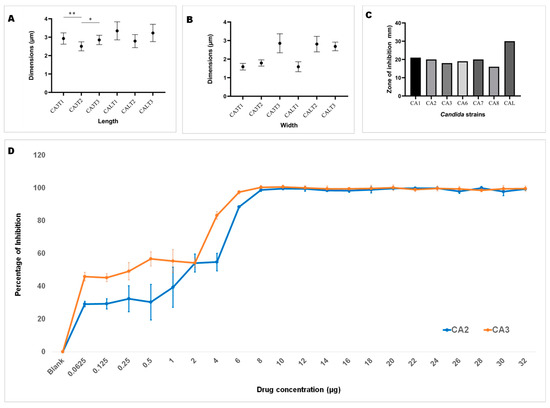
Figure 4.
Length and width of C. auris and C. albicans. (A): Length of C. auris (CA3) and C. albicans (CAL) cells after treatment using dutasteride (CA3T1, CA3T2, CALT1, and CALT2) and cells without treatment, i.e., the control (CA3T3 and CALT3). * Significant at a p-value of <0.05; ** significant at a p-value of <0.01. (B): Width of C. auris (CA3) and C. albicans (CAL) cells after treatment using dutasteride (CA3T1, CA3T2, CALT1, and CALT2) and Candida cells without treatment, i.e., the control (CA3T3 and CALT3). (C): Observation of the zone of inhibition resulting from dutasteride treatment against C. auris and C. albicans. (D): Inhibition of biofilm formation of C. auris CA2 and C. auris CA3 clinical isolates by dutasteride. Graphs illustrate the percentage of biofilm formation inhibition by dutasteride against C. auris CA2 and C. auris CA3. All assays of biofilm formation inhibition were executed in quadruplicate in a 96-well plate. Error bars in the graphs designate standard errors.
3.3. Dose–Response Assays of Dutasteride’s Inhibition of Planktonic Growth
Using the results of dutasteride dose–response curves (32 to 0.0625 µg/mL) to inhibit the planktonic growth of C. auris CA1, CA2, CA3, CA5, CA6, CA7, and CA8 and C. albicans CAL, we determined the 50% inhibitory concentration (IC50) (Table 1). The results revealed an IC50 of 8.6833 ± 1.832 µg/mL [CA1: 6.0097 µg; CA2: 6.5264 µg/mL; CA3: 10.3217 µg/mL; CA5: 9.5679 µg/mL; CA6: 10.1056 µg/mL; CA7: 10.2367 µg/mL; and CA8: 8.0151 µg/mL] for C. auris and an IC50 of 10.8116 µg/mL for C. albicans.

Table 1.
The planktonic and biofilm inhibition results.
3.4. Assessment of Biofilm Inhibition Using Dutasteride
Dutasteride was effective at inhibiting biofilm formation by C. auris and C. albicans (Figure 4D). Dutasteride revealed narrow variations in its inhibition of biofilm formation by the seven C. auris clinical isolates and one C. albicans clinical isolate. The formation of biofilm by C. auris CA8, CA7, and CA3 was efficiently inhibited by dutasteride compared with the inhibition of biofilm formation by C. albicans CAL (Figure 4D). The inhibiting ability of dutasteride against the formation of biofilm by C. auris CA1, CA2, CA6, and CA5 was reduced at lower concentrations of dutasteride. More than 98% of biofilm formation by the six tested clinical isolates, including C. auris CA3, CA5, CA6, CA7, and CA8 and C. albicans CAL, was inhibited by dutasteride at the highest concentration tested (Figure 4D). We determined the IC50 at which 50% biofilm formation was inhibited by dutasteride. The average biofilm IC50 for the tested clinical isolates of C. auris ranged from 4.840 to 11.632 µg/mL, with an average of 7.282 ± 2.799 µg/mL. The biofilm IC50 for the clinical isolate of C. albicans was 6.310 µg/mL (Table 1). The 1,3-beta-glucanosyltransferase–dutasteride complex showed lower binding free energy in the virtual screening results, which was confirmed by the MD simulations. Dutasteride’s activity in inhibiting the biofilm formation and planktonic growth of C. auris confirmed these predictions, indirectly indicating stability and a stronger interaction.
4. Discussion
Drug repurposing is a suitable method for finding new uses for drugs that have been previously researched and registered [47]. It can help avoid significant expenses related to the research and development of new drugs, and it also uses de-risked compounds to reduce attrition rates. As a result, this method has been used to repurpose medications against C. auris. The selection of appropriate target proteins is an important factor in virtual screening. This study considered Gibbs free energy and binding free energy by assessing negative values, which indicated whether a drug is predicted to bind to 1,3-beta–glucanosyltransferase in C. auris. We selected 1,3-beta-glucanosyltransferase, a major cell-wall-associated protein in genera like Candida [40,41]. The target 1,3-beta-glucanosyltransferase in C. auris was successfully inhibited by various FDA-approved drugs during virtual screening.
Through virtual screening, we found that a lower binding free energy indicates more stability and a stronger interaction, and we confirmed its potential association with higher drug efficacy using MD simulations and its activity in biofilm formation and planktonic growth. Specific interactions, including hydrogen bonding and other interactions between 1,3-beta-glucanosyltransferase and dutasteride, revealed improved binding. The conformational changes in MD simulations revealed how 1,3-beta-glucanosyltransferase might shift upon dutasteride binding, which signifies that dutasteride might work effectively when 1,3-beta-glucanosyltransferase adopts a specific conformation. Hydrogen bonding between the first amino group of dutasteride and the carbonyl group of glutamine (the 50th or 51st amino acid) can be crucial for understanding how dutasteride binds to 1,3-beta-glucanosyltransferase and designing new drugs. The observations made through MD simulations in this study may be useful for identifying new drug targets using 1,3-beta–glucanosyltransferase in C. auris and can predict off-target effects in the future by virtually screening existing drugs followed by wet-lab studies against various protein targets.
The four top hits [zinc000242548690 (digoxin C41H64O14), zinc000003932831 (dutasteride C27H30F6N2O2), zinc000052955754 (Ergotamine C33H35N5O5), and zinc000203757351 (Paritaprevir C40H43N7O7S)] are unrelated drugs, which were initially selected based on virtual screening for the inhibitors of 1,3-beta-glucanosyltransferase from C. auris. Digoxin (C41H64O14) is a cardiac glycoside used to treat moderate heart failure. Dutasteride is an antiandrogenic compound that inhibits 5-alpha reductase and is used to treat benign prostatic hyperplasia in adult males. Cluster headaches and migraines are treated with ergotamine, which is an alpha-1-selective adrenergic agonist vasoconstrictor. A direct-acting antiviral drug called paritaprevir is combined with other antiviral drugs to treat infections caused by the hepatitis C virus [48]. In the present study, we used clinical isolates of C. auris from Saudi Arabia, previously confirmed by our team as having various drug-resistance mutations, and all the strains were reconfirmed using molecular techniques [38,45]. We used the multidrug-resistant clinical isolate C. auris CA1 for the initial primary screening in parallel with the clinical isolate C. albicans CAL for inhibiting the activity of the top-rated (highlighted in yellow in Figure 1) clinically approved drugs from the virtual screening. As the major aim of this research was to identify FDA-approved drugs with anti-C. auris activity for use in drug repurposing, we focused on the top two drugs, i.e., zinc000242548690 (digoxin) and zinc000003932831 (dutasteride), from the highest predications. Furthermore, no antifungal drugs from the tested groups were in the top-rated category, with binding affinities of ≤−10 Kcal/mol. Here, the ability of C. auris to live on hospital surfaces and places other than the human body should be considered, and appropriate environmental decontamination and surface disinfection methods should be developed for hospitals and research labs to prevent the emergence of nosocomial pathogens like C. auris [19,49,50]. Even though zinc000242548690 digoxin (DR1) was rated highest in the prediction of binding affinity, it showed no anti-Candida activity, which was in line with earlier observations of C. albicans and Saccharomyces cerevisiae [51]. One hypothesis is that the cell wall or plasma membrane of C. albicans and C. auris prevented digoxin from entering the cytoplasm [51,52]. Digoxin, a cardiac glycoside, is a medication for heart conditions such as congestive heart failure and certain cardiac arrhythmias and has a narrow therapeutic window [53]. Our study once again confirms this, as despite a positive virtual screening result for binding, no wet-lab studies confirmed digoxin activity against C. auris cells. Digoxin is a toxic substance with a well-known cardiotoxic effect. Despite digoxin’s long-standing approval, it is important to acknowledge its potent side effects. If overdosed, the consequences can be severe [53]. This study confirmed a lack of anti-C. auris activity in fluconazole, as reported earlier by various studies [19].
zinc000003932831 dutasteride (DR2) was rated the second highest in the prediction of binding affinity, with anti-Candida activity confirmed in the primary screening. To our knowledge, this is the first report providing evidence for the anti-C. auris activity of dutasteride. A randomized, double-blind, placebo-controlled interventional trial observed reduced viral shedding in males with mild COVID-19 treated with dutasteride and other drugs [54]. More confirmative studies are needed to ensure that dutasteride will eventually be a more effective anti-C. auris treatment than the existing drugs or will have more promising antifungal potential. The primary screening of dutasteride on C. auris was extended to confirm its anti-C. auris activity through dose–response assays, over a range of concentrations (32 to 0.065 µg/mL) of dutasteride, against different clinical isolates of C. auris, and we calculated the IC50 against various clinical isolates of C. auris and C. albicans. Dutasteride at an 8.949 µg/mL concentration was able to inhibit 50% of the growth of clinical isolates of C. auris. The IC50 was used to identify the effective quantifying mechanism and revealed the half-maximum inhibitory concentration of dutasteride in inhibiting the growth of C. auris. It can also be used to determine appropriate dutasteride dosages for further testing and development studies. Regarding the inhibition of the planktonic growth of C. auris, the IC50 refers to the concentration of dutasteride needed to inhibit the growth of free-floating C. auris cells in broth culture. Biofilms are essential structural characteristics of C. auris attached to surfaces; the IC50 signifies the concentration of dutasteride required to disrupt an existing biofilm of C. auris and the formation of new biofilm by C. auris. The IC50 values observed for planktonic growth do not have a direct association with biofilm formation; however, they have a considerable influence on our understanding of the potential effectiveness of dutasteride.
The results of the electron microscopic studies clearly indicate that dutasteride is fungicidal. In order to identify potential drugs, screening libraries can provide a number of solutions. Through the Prestwick Chemical Library, a repurposing library, ebselen, which is an antioxidant, anti-inflammatory, and cytoprotective drug, was identified as a repositionable molecule and inhibited the growth of C. auris as well as biofilm formation [19]. A systemic review revealed studies about 12 repositionable drugs/compounds that inhibit the formation of C. auris biofilms, which included antiparasitic drugs (iodoquinol, Miltefosine, Niclosamide, Tri-Chloro-Salicyanilide), anti-inflammatory drugs (ebselen, AM-24 (2,4,6-triiodophenol)), anticancer drugs (Alexidine dihydrochloride), antidepressants (Sertraline), psychiatric agents (Disulfiram), a muscarinic receptor agonist (Tazomeline), a Farnesyl transferase inhibitor (Lonafarnib), and others (Provecta—rose bengal disodium) [18,30,31,32,33,35,36,37]. In addition, they revealed 22 repositionable drugs/compounds that inhibit planktonic growth in C. auris, which included antiparasitic drugs (iodoquinol, Miltefosine, Robenidine, Salicylanilide oxyclozanide, Pyrvinium pamoate, Broxyquinoline, Diiodohydroxyquinoline), anticancer drugs (Tamoxifen citrate, Alexidine dihydrochloride, AC-93253 iodide), antiseptics (Chloroxine, Clioquinol), antiemetics (Trifluoperazine: dihydrochloride, Thiethylperazine dimalate), and antidepressants (Sertraline, Rolipram), as well as antiviral drugs (Ribavirin), anti-inflammatory agents (ebselen), antifungal/antibacterial agents (Ciclopirox ethanolamine), anticonvulsants ((−)-MK 801 hydrogen maleate), antihypertensives (Guanadrel sulfate), antiplatelets (Suloctidil), and psychiatric agents (Disulfiram) [18,30,31,32,33,35,36,37]. A large screening study demonstrated 27 drugs that inhibited the growth of three different strains of C. auris with different geographical origins. These included antibacterial (Alexidine dihydrochloride, Benzethonium chloride, Chloroxine, Dequalinium dichloride, Methyl benzethonium chloride), antimalarial (Artemisinin), antibacterial/antifungal (Ciclopirox ethanolamine), antiamebic/antibacterial (Clioquinol), antipruritic (Dimethisoquin hydrochloride), local anesthetic (Dyclonine hydrochloride), anti-inflammatory (ebselen), anti-fatigue (Fipexide hydrochloride), antihypertensive (Guanadrel sulfate), antiseptic (Hexachlorophene, Thonzonium bromide), antipsychotic (Methiothepin maleate, Zotepine), anticonvulsant (MK 801 hydrogen maleate), anthelmintic (Pyrvinium pamoate), antiamebic/antipsychotic (Prochlorperazine dimaleate), antineoplastic (Tamoxifen citrate), antiemetic (Thiethylperazine dimalate, Trifluoperazine dihydrochloride), antidepressant (Rolipram, Sertraline), and antiplatelet (Suloctidil) drugs [34]. In addition to these drugs, the present study considers dutasteride an effective anti-Candida agent with promising inhibition of the planktonic growth of C. auris and C. albicans while also inhibiting biofilm formation by both Candida species. A smaller amount of dutasteride is needed to inhibit 50% of the biofilm formation in C. auris and C. albicans isolates, compared with 50% of planktonic growth for the strains C. auris CA3, CA5, CA6, CA7, and CA8 and C. albicans. However, it is different for the strains C. auris CA1 and CA2. The biofilm-inhibiting activity of dutasteride indicates the competency of the drug to fight the high-level resistance characteristics of C. auris and C. albicans [19,55,56,57,58]. On all of the tested clinical isolates of C. auris and C. albicans, in addition to biofilm and planktonic growth inhibition by dutasteride, we demonstrated dutasteride-induced morphological changes like cell disruption, a crushed phenotype structure, and a significant reduction in length.
In 2002, clinical research demonstrated the effectiveness of dutasteride at a dosage of 0.5 mg per day. The research findings indicated that dutasteride medication resulted in a smaller increase in adverse sexual events compared with a placebo. Nevertheless, its extended usage for a period exceeding 4 years did not result in an escalation of the adverse effects that were identified throughout the initial 2-year investigation. Hence, dutasteride not only enhances urinary symptoms and flow rate but is also linked to noteworthy enhancements in benign prostatic hyperplasia Impact Index scores, indicating benefits to quality of life for men with benign prostatic hyperplasia. During a 2-year period, a minimum of 1% of patients who received either dutasteride or a placebo experienced side effects, including impotence, ejaculation problems, decreased libido, and gynecomastia. Nevertheless, when dutasteride is used daily for a maximum of 2 years, its tolerability profile is similar to that of a placebo, with the exception of slightly higher occurrences of gynecomastia, impotence, and lower libido. Remarkably, the occurrence of the majority of drug-induced sexual side effects declined in individuals who were administered dutasteride consistently over the course of 48 months. Overall, long-term treatment with dutasteride does not result in an increased occurrence of negative outcomes, and patients generally adhere well to the treatment [59,60]. It is important to take these data into account when evaluating the future development of high dosages of dutasteride to treat individuals with C. auris infection. The liver extensively metabolizes dutasteride, primarily excreting it in the feces, and trace amounts are excreted in urine [61]. Researchers have identified C. auris in urinary tract infections worldwide. Therefore, we can consider dutasteride as a possible substitute if C. auris infection persists or recurs; however, the need for higher concentrations should not be ignored [62]. These observations confirm the need for further animal model studies and clinical trials to investigate dutasteride as a potential anti-Candida drug to treat C. auris and C. albicans infections.
5. Conclusions
The present study used high-throughput virtual screening of drug repurposing to significantly reduce costs and save time in identifying novel and effective anti-Candida drugs against the nosocomial pathogen C. auris, which exhibits multidrug resistance and is an emerging pathogen of increasing concern. The drug repurposing of dutasteride, which has potential antifungal efficiency (specifically with anti-C. auris and anti-C. albicans activity), could enable the rapid recovery of patients infected with multidrug-resistant C. auris. Furthermore, detailed investigations on dutasteride’s non-antifungal off-target effects using animal model studies are needed before it can be considered a repositionable clinical agent to reduce nosocomial infections and alleviate difficult-to-treat candidiasis and molds.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/pharmaceutics16060810/s1, Figure S1. Molecular dynamics simulations of the 1,3-beta-glucanosyltransferase (UniPort: A0A2H1A5Q4)-and-dutasteride complex. A: Structure (2D) of dutasteride (DrugBank: DB01126); B: root-mean-square fluctuation (RMSF) in protein (P-RMSF). D: Ligand dutasteride fluctuations with respect to 1,3-beta-glucanosyltransferase. C: Protein secondary structure element (P-SSE) timeline. D: 1,3-beta-glucanosyltransferase–dutasteride contact timeline. E: A dutasteride torsion plot of the conformational changes in every rotatable bond in dutasteride in the simulation (0.00–100.00 nanoseconds) trajectory. The histogram displays the conformational strain dutasteride undergoes to maintain a 1,3-beta-glucanosyltransferase-bound conformation. The colors in the inserted structure of dutasteride correspond to the rotatable bonds of dutasteride as well as the dial plot and bar plots.
Author Contributions
Conceptualization, J.F.B., N.B.A., P.S., R.A. (Reem AlJindan), D.A., S.A. (Sarah Almofty), T.S.D. and S.A. (Sayed AbdulAzeez); methodology, J.F.B., N.B.A., P.S., J.S.J., R.A. (Rahaf Alquwaie), E.A., N.F.A., R.A. (Razan Aldahhan), R.A. (Reem AlJindan), D.A., S.A. (Sarah Almofty), T.S.D. and S.A. (Sayed AbdulAzeez); software, J.F.B., P.S., J.S.J., R.A. (Rahaf Alquwaie), N.F.A., R.A. (Razan Aldahhan), S.A. (Sayed AbdulAzeez); validation, J.F.B., N.B.A., P.S., E.A., R.A. (Reem AlJindan), D.A., S.A. (Sarah Almofty), S.A. (Sayed AbdulAzeez); formal analysis, J.F.B., N.B.A., J.S.J., E.A., N.F.A., R.A. (Razan Aldahhan), D.A., S.A. (Sarah Almofty), T.S.D. and S.A. (Sayed AbdulAzeez); investigation, J.F.B., N.B.A., P.S., R.A. (Rahaf Alquwaie), E.A., N.F.A., R.A. (Reem AlJindan), D.A., S.A. (Sarah Almofty) and S.A. (Sayed AbdulAzeez); data curation, J.F.B., N.B.A., R.A. (Rahaf Alquwaie) and S.A. (Sayed AbdulAzeez); project administration, J.F.B., N.B.A., P.S. and S.A. (Sayed AbdulAzeez); writing—original draft preparation, J.F.B., N.B.A., P.S., J.S.J., R.A. (Rahaf Alquwaie), E.A., N.F.A., R.A. (Razan Aldahhan), R.A. (Reem AlJindan), D.A., S.A. (Sarah Almofty), T.S.D. and S.A. (Sayed AbdulAzeez); writing—review and editing, J.F.B., N.B.A., P.S.; J.S.J., R.A. (Rahaf Alquwaie), E.A., N.F.A., R.A. (Razan Aldahhan), R.A. (Reem AlJindan), D.A., S.A. (Sarah Almofty); T.S.D. and S.A. (Sayed AbdulAzeez). All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was approved by the Institutional Review Board (IRB) at Imam Abdulrahman Bin Faisal University (IRB approval number: IRB-2022-13-462).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
For computer time, this research used the resources of the Supercomputing Laboratory at King Abdullah University of Science & Technology (KAUST) in Thuwal, Saudi Arabia (Shaheen Projects K1484 and K1600). We are grateful to Sultan Akthar, Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia, for his support. We also appreciate the technical assistance provided by Ranilo M. Tumbaga, Horace T. Pacifico, Jee E. Aquino, and Edwardson Evancelista.
Conflicts of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Borgio, J.F.; Rasdan, A.S.; Sonbol, B.; Alhamid, G.; Almandil, N.B.; AbdulAzeez, S. Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula. Biology 2021, 10, 1144. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Nobile, C.J.; Huang, G. Candida Auris Infections in China. Virulence 2022, 13, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Abbasi, A.F.; Prakash, S.; Mangat, J.; Hosein, Z.; Haider, N.; Chan, J. Candida Auris: An Overview of the Emerging Drug-Resistant Fungal Infection. Infect. Chemother. 2022, 54, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida Auris Sp. Nov., a Novel Ascomycetous Yeast Isolated from the External Ear Canal of an Inpatient in a Japanese Hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Khari, A.; Biswas, B.; Gangwar, G.; Thakur, A.; Puria, R. Candida Auris Biofilm: A Review on Model to Mechanism Conservation. Expert Rev. Anti Infect. Ther. 2023, 21, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Rosario-Colon, J.A.; Eberle, K. Novel Intravenous Immunoglobulin Therapy for the Prevention and Treatment of Candida auris and Candida albicans Disseminated Candidiasis. mSphere 2023, 8, e00584-22. [Google Scholar] [CrossRef] [PubMed]
- Watkins, R.R.; Gowen, R.; Lionakis, M.S.; Ghannoum, M. Update on the Pathogenesis, Virulence, and Treatment of Candida Auris. Pathog. Immun. 2022, 7, 46–65. [Google Scholar] [CrossRef]
- Bravo Ruiz, G.; Lorenz, A. What Do We Know about the Biology of the Emerging Fungal Pathogen of Humans Candida Auris? Microbiol. Res. 2021, 242, 126621. [Google Scholar] [CrossRef]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, Biology, Antifungal Resistance, and Virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef]
- Thatchanamoorthy, N.; Rukumani Devi, V.; Chandramathi, S.; Tay, S.T. Candida Auris: A Mini Review on Epidemiology in Healthcare Facilities in Asia. J. Fungi 2022, 8, 1126. [Google Scholar] [CrossRef]
- Peyclit, L.; Yousfi, H.; Rolain, J.-M.; Bittar, F. Drug Repurposing in Medical Mycology: Identification of Compounds as Potential Antifungals to Overcome the Emergence of Multidrug-Resistant Fungi. Pharmaceuticals 2021, 14, 488. [Google Scholar] [CrossRef]
- Lone, S.A.; Ahmad, A. Candida Auris—The Growing Menace to Global Health. Mycoses 2019, 62, 620–637. [Google Scholar] [CrossRef] [PubMed]
- Preda, M.; Chivu, R.D.; Ditu, L.M.; Popescu, O.; Manolescu, L.S.C. Pathogenesis, Prophylaxis, and Treatment of Candida Auris. Biomedicines 2024, 12, 561. [Google Scholar] [CrossRef] [PubMed]
- Lohikoski, R.; Oldberg, K.; Rasmussen, M. Bacteraemia Caused by Non-Faecalis and Non-Faecium Enterococcus Species—A Retrospective Study of Incidence, Focus of Infection, and Prognosis. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R. Candida Auris and Multidrug Resistance: Defining the New Normal. Fungal Genet. Biol. 2019, 131, 103243. [Google Scholar] [CrossRef] [PubMed]
- Kean, R.; Ramage, G. Combined Antifungal Resistance and Biofilm Tolerance: The Global Threat of Candida Auris. mSphere 2019, 4, e00458-19. [Google Scholar] [CrossRef] [PubMed]
- Allaw, F.; Kara Zahreddine, N.; Ibrahim, A.; Tannous, J.; Taleb, H.; Bizri, A.R.; Dbaibo, G.; Kanj, S.S. First Candida Auris Outbreak during a COVID-19 Pandemic in a Tertiary-Care Center in Lebanon. Pathogens 2021, 10, 157. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Roma, J.S.; Shen, M.; Mota Fernandes, C.; Tsang, P.S.; Forbes, H.E.; Boshoff, H.; Lazzarini, C.; Del Poeta, M.; Zheng, W.; et al. Identification of Antifungal Compounds against Multidrug-Resistant Candida Auris Utilizing a High-Throughput Drug-Repurposing Screen. Antimicrob. Agents Chemother. 2021, 65, e01305-20. [Google Scholar] [CrossRef] [PubMed]
- Wall, G.; Chaturvedi, A.K.; Wormley, F.L.; Wiederhold, N.P.; Patterson, H.P.; Patterson, T.F.; Lopez-Ribot, J.L. Screening a Repurposing Library for Inhibitors of Multidrug-Resistant Candida Auris Identifies Ebselen as a Repositionable Candidate for Antifungal Drug Development. Antimicrob. Agents Chemother. 2018, 62, e01084-18. [Google Scholar] [CrossRef]
- Wall, G.; Chen, E.; Hull, M.V.; Lopez-Ribot, J.L. Screening the CALIBR ReFRAME Library in Search for Inhibitors of Candida Auris Biofilm Formation. Front. Cell Infect. Microbiol. 2020, 10, 597931. [Google Scholar] [CrossRef]
- Romera, D.; Aguilera-Correa, J.J.; Gadea, I.; Viñuela-Sandoval, L.; García-Rodríguez, J.; Esteban, J. Candida Auris: A Comparison between Planktonic and Biofilm Susceptibility to Antifungal Drugs. J. Med. Microbiol. 2019, 68, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.S.N.; Walraven, C.J.; Lee, S.A. Candida Auris: Disinfectants and Implications for Infection Control. Front. Microbiol. 2018, 9, 726. [Google Scholar] [CrossRef] [PubMed]
- Ashburn, T.T.; Thor, K.B. Drug Repositioning: Identifying and Developing New Uses for Existing Drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding New Light on Drug Metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Han, B.; Kumar, P.; Liu, X.; Ma, X.; Wei, X.; Huang, L.; Guo, Y.; Han, L.; Zheng, C.; et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010, 38, D787–D791. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.; Burgain, A.; Chaillot, J.; Pic, É.; Khemiri, I.; Sellam, A. A Phenotypic Small-Molecule Screen Identifies Halogenated Salicylanilides as Inhibitors of Fungal Morphogenesis, Biofilm Formation and Host Cell Invasion. Sci. Rep. 2018, 8, 11559. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lee, H.; Rho, S.B.; Park, M.K.; Lee, C.H. Ethacrynic Acid: A Promising Candidate for Drug Repurposing as an Anticancer Agent. Int. J. Mol. Sci. 2023, 24, 6712. [Google Scholar] [CrossRef]
- Ribeiro, E.; Costa, B.; Vasques-Nóvoa, F.; Vale, N. In Vitro Drug Repurposing: Focus on Vasodilators. Cells 2023, 12, 671. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, E.; Vale, N. Repurposing of the Drug Tezosentan for Cancer Therapy. Curr. Issues Mol. Biol. 2023, 45, 5118–5131. [Google Scholar] [CrossRef]
- Izadi, A.; Aghaei Gharehbolagh, S.; Sadeghi, F.; Talebi, M.; Darmiani, K.; Zarrinnia, A.; Zarei, F.; Peymaeei, F.; Khojasteh, S.; Borman, A.M.; et al. Drug Repurposing against Candida Auris: A Systematic Review. Mycoses 2022, 65, 784–793. [Google Scholar] [CrossRef]
- Barreto, T.L.; Rossato, L.; de Freitas, A.L.D.; Meis, J.F.; Lopes, L.B.; Colombo, A.L.; Ishida, K. Miltefosine as an Alternative Strategy in the Treatment of the Emerging Fungus Candida Auris. Int. J. Antimicrob. Agents 2020, 56, 106049. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Jiang, T.; Zou, Y.; Wang, Y.; Zhou, J.; Li, J.; Liu, L.; Tan, J.; Wei, L.; Li, J.; et al. FDA Approved Drug Library Screening Identifies Robenidine as a Repositionable Antifungal. Front. Microbiol. 2020, 11, 996. [Google Scholar] [CrossRef] [PubMed]
- Pic, E.; Burgain, A.; Sellam, A. Repurposing the Anthelminthic Salicylanilide Oxyclozanide against Susceptible and Clinical Resistant Candida albicans Strains. Med. Mycol. 2019, 57, 387–390. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, H.C.; Monteiro, M.C.; Rossi, S.A.; Pemán, J.; Ruiz-Gaitán, A.; Mendes-Giannini, M.J.S.; Mellado, E.; Zaragoza, O. Identification of Off-Patent Compounds that Present Antifungal Activity against the Emerging Fungal Pathogen Candida Auris. Front. Cell. Infect. Microbiol. 2019, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Mamouei, Z.; Alqarihi, A.; Singh, S.; Xu, S.; Mansour, M.K.; Ibrahim, A.S.; Uppuluri, P. Alexidine Dihydrochloride Has Broad-Spectrum Activities against Diverse Fungal Pathogens. mSphere 2018, 3, e00539-18. [Google Scholar] [CrossRef] [PubMed]
- Gowri, M.; Jayashree, B.; Jeyakanthan, J.; Girija, E.K. Sertraline as a Promising Antifungal Agent: Inhibition of Growth and Biofilm of Candida Auris with Special Focus on the Mechanism of Action in Vitro. J. Appl. Microbiol. 2020, 128, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Qiao, D.; Han, Y.; Du, N.; Li, X.; Fan, Y.; Ge, X.; Zhang, H. Identification of Disulfiram as a Potential Antifungal Drug by Screening Small Molecular Libraries. J. Infect. Chemother. 2021, 27, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Aldossary, H.A.; Rehman, S.; Jermy, B.R.; AlJindan, R.; Aldayel, A.; AbdulAzeez, S.; Akhtar, S.; Khan, F.A.; Borgio, J.F.; Al-Suhaimi, E.A. Therapeutic Intervention for Various Hospital Setting Strains of Biofilm Forming Candida Auris with Multiple Drug Resistance Mutations Using Nanomaterial Ag-Silicalite-1 Zeolite. Pharmaceutics 2022, 14, 2251. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Le, T.; Liu, Z.; Wang, L.; Guo, H.; Yang, J.; Chen, Q.; Hu, J. Different Efficacies of Common Disinfection Methods against Candida Auris and Other Candida Species. J. Infect. Public. Health 2020, 13, 730–736. [Google Scholar] [CrossRef]
- Okada, N.; Tatsuno, L.; Hanski, E.; Caparon, M.; Sasakawa, C. A 1,3-Beta-Glucanosyltransferase Isolated from the Cell Wall of Aspergillus Fumigatus Is a Homologue of the Yeast Bgl2p. Microbiology 1998, 144 Pt 11, 3079–3086. [Google Scholar] [CrossRef]
- Zamith-Miranda, D.; Amatuzzi, R.F.; Munhoz da Rocha, I.F.; Martins, S.T.; Lucena, A.C.R.; Vieira, A.Z.; Trentin, G.; Almeida, F.; Rodrigues, M.L.; Nakayasu, E.S.; et al. Transcriptional and Translational Landscape of Candida Auris in Response to Caspofungin. Comput. Struct. Biotechnol. J. 2021, 19, 5264–5277. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Tang, K.G.; Young, J.; Dandarchuluun, C.; Wong, B.R.; Khurelbaatar, M.; Moroz, Y.S.; Mayfield, J.; Sayle, R.A. ZINC20-A Free Ultralarge-Scale Chemical Database for Ligand Discovery. J. Chem. Inf. Model. 2020, 60, 6065–6073. [Google Scholar] [CrossRef] [PubMed]
- Borgio, J.F.; Alsuwat, H.S.; Al Otaibi, W.M.; Ibrahim, A.M.; Almandil, N.B.; Al Asoom, L.I.; Salahuddin, M.; Kamaraj, B.; AbdulAzeez, S. State-of-the-Art Tools Unveil Potent Drug Targets amongst Clinically Approved Drugs to Inhibit Helicase in SARS-CoV-2. Arch. Med. Sci. 2020, 16, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural Summaries of PDB Entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- AlJindan, R.; AlEraky, D.M.; Mahmoud, N.; Abdalhamid, B.; Almustafa, M.; AbdulAzeez, S.; Borgio, J.F. Drug Resistance-Associated Mutations in ERG11 of Multidrug-Resistant Candida Auris in a Tertiary Care Hospital of Eastern Saudi Arabia. J. Fungi 2020, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.J. ImageJ for Microscopy. Biotechniques 2007, 43, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Bauman, J.E.; Bologa, C.G.; Buranda, T.; Chigaev, A.; Edwards, B.S.; Jarvik, J.W.; Gresham, H.D.; Haynes, M.K.; Hjelle, B.; et al. Drug Repurposing from an Academic Perspective. Drug Discov. Today Ther. Strateg. 2011, 8, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Abdolrasouli, A.; Armstrong-James, D.; Ryan, L.; Schelenz, S. In Vitro Efficacy of Disinfectants Utilised for Skin Decolonisation and Environmental Decontamination during a Hospital Outbreak with Candida Auris. Mycoses 2017, 60, 758–763. [Google Scholar] [CrossRef]
- Kean, R.; Sherry, L.; Townsend, E.; McKloud, E.; Short, B.; Akinbobola, A.; Mackay, W.G.; Williams, C.; Jones, B.L.; Ramage, G. Surface Disinfection Challenges for Candida Auris: An in-Vitro Study. J. Hosp. Infect. 2018, 98, 433–436. [Google Scholar] [CrossRef]
- de Moraes, D.C.; Tessis, A.C.; Rollin-Pinheiro, R.; Princival, J.L.; Villar, J.A.F.P.; Barbosa, L.A.; Barreto-Bergter, E.; Ferreira-Pereira, A. Digoxin Derivatives Sensitize a Saccharomyces Cerevisiae Mutant Strain to Fluconazole by Inhibiting Pdr5p. J. Fungi 2022, 8, 769. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, K.; Kohli, A.; Prasad, R. Drug Susceptibilities of Yeast Cells Are Affected by Membrane Lipid Composition. Antimicrob. Agents Chemother. 2002, 46, 3695–3705. [Google Scholar] [CrossRef] [PubMed]
- Patocka, J.; Nepovimova, E.; Wu, W.; Kuca, K. Digoxin: Pharmacology and Toxicology—A Review. Environ. Toxicol. Pharmacol. 2020, 79, 103400. [Google Scholar] [CrossRef] [PubMed]
- Cadegiani, F.A.; McCoy, J.; Gustavo Wambier, C.; Goren, A. Early Antiandrogen Therapy With Dutasteride Reduces Viral Shedding, Inflammatory Responses, and Time-to-Remission in Males With COVID-19: A Randomized, Double-Blind, Placebo-Controlled Interventional Trial (EAT-DUTA AndroCoV Trial-Biochemical). Cureus 2021, 13, e13047. [Google Scholar] [CrossRef] [PubMed]
- Pierce, C.G.; Srinivasan, A.; Uppuluri, P.; Ramasubramanian, A.K.; López-Ribot, J.L. Antifungal Therapy with an Emphasis on Biofilms. Curr. Opin. Pharmacol. 2013, 13, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Vande Walle, K.; Wickes, B.L.; López-Ribot, J.L. Standardized Method for in Vitro Antifungal Susceptibility Testing of Candida Albicans Biofilms. Antimicrob. Agents Chemother. 2001, 45, 2475–2479. [Google Scholar] [CrossRef] [PubMed]
- Larkin, E.; Hager, C.; Chandra, J.; Mukherjee, P.K.; Retuerto, M.; Salem, I.; Long, L.; Isham, N.; Kovanda, L.; Borroto-Esoda, K.; et al. The Emerging Pathogen Candida Auris: Growth Phenotype, Virulence Factors, Activity of Antifungals, and Effect of SCY-078, a Novel Glucan Synthesis Inhibitor, on Growth Morphology and Biofilm Formation. Antimicrob. Agents Chemother. 2017, 61, e02396-16. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-Forming Capability of Highly Virulent, Multidrug-Resistant Candida Auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Marihart, S.; Harik, M.; Djavan, B. Dutasteride: A Review of Current Data on a Novel Dual Inhibitor of 5alpha Reductase. Rev. Urol. 2005, 7, 203–210. [Google Scholar] [PubMed]
- Li, Y.; Ma, J.; Qin, X.-H.; Hu, C.-Y. The Efficacy and Safety of Dutasteride and Finasteride in Patients with Benign Prostatic Hyperplasia: A Systematic Review and Meta-Analysis. Transl. Androl. Urol. 2022, 11, 313–324. [Google Scholar] [CrossRef]
- Miller, J.; Tarter, T.H. Update on the Use of Dutasteride in the Management of Benign Prostatic Hypertrophy. Clin. Interv. Aging 2007, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Griffith, N.; Danziger, L. Candida Auris Urinary Tract Infections and Possible Treatment. Antibiotics 2020, 9, 898. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).