Dual-Omics Approach Unveils Novel Perspective on the Quality Control of Genetically Engineered Exosomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Expression Vectors for Genetic Engineering of Exosomes
2.3. Human Cell Culture and Transfection
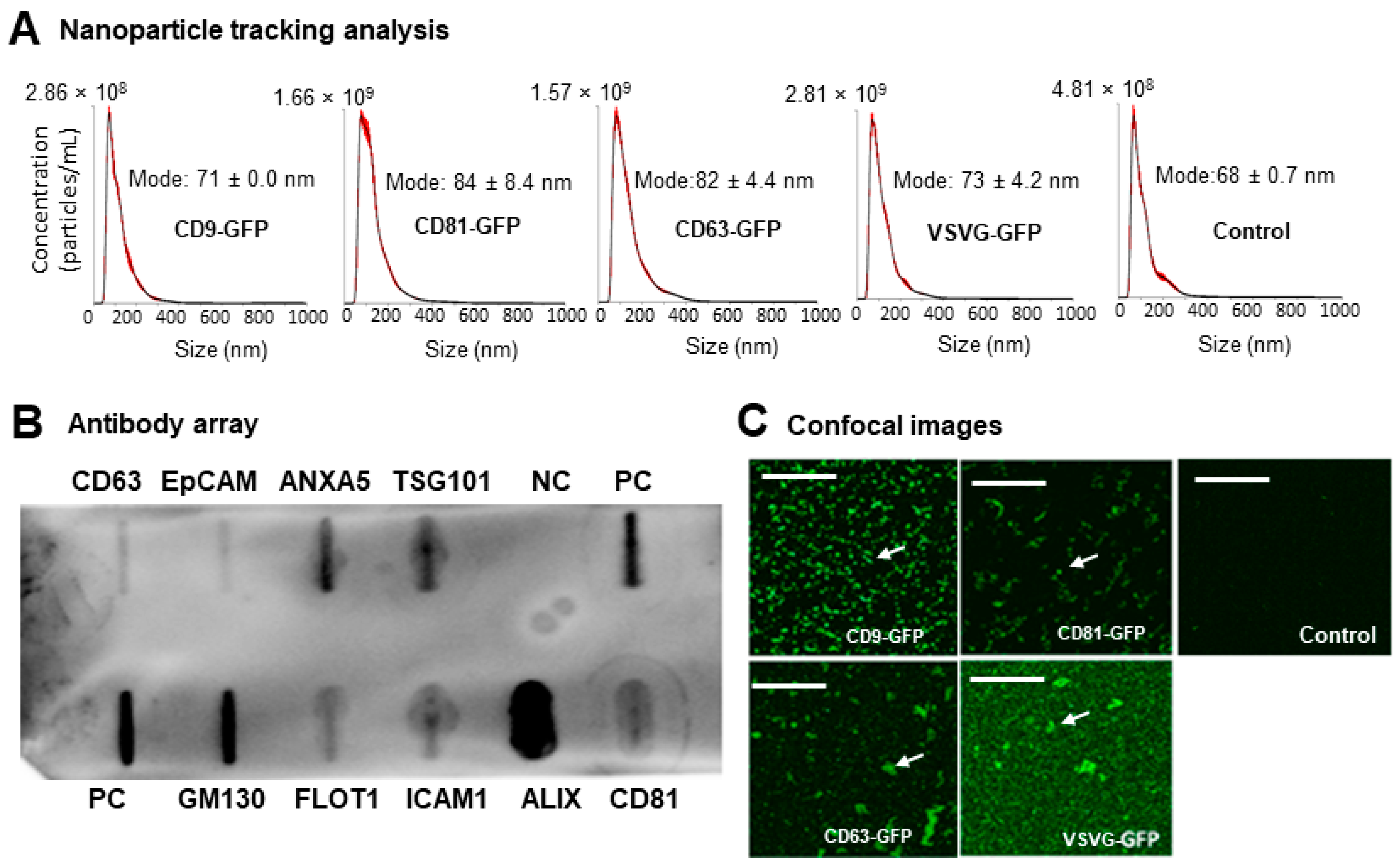
2.4. Molecular Tracking and Confocal Microscopy
2.5. Exosome Preparation
2.6. Nanoparticle Tracking Analysis (NTA)
2.7. Slot Blot Analysis
2.8. Next-Generation Sequencing (NGS)
2.9. Liquid Chromatography–Mass Spectrometry (LC-MS)
2.10. Dual-Omics Analysis and Protein Contaminant Detection
2.11. Pathway Analysis
3. Results
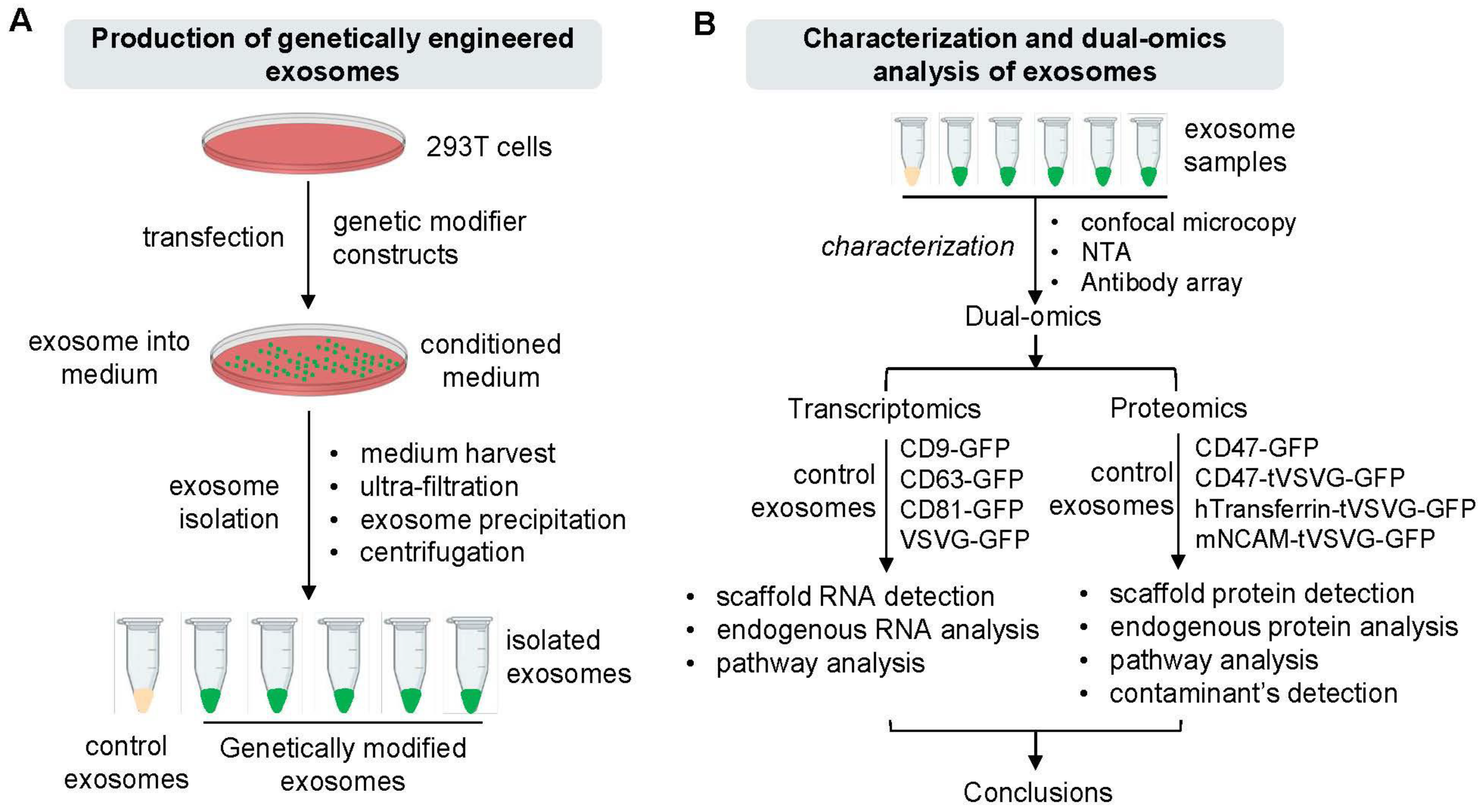
3.1. System Design and Genetic Engineering of Exosomes in Human 293T Cells
3.2. Preparation and Characterization of Genetically Engineered Exosomes
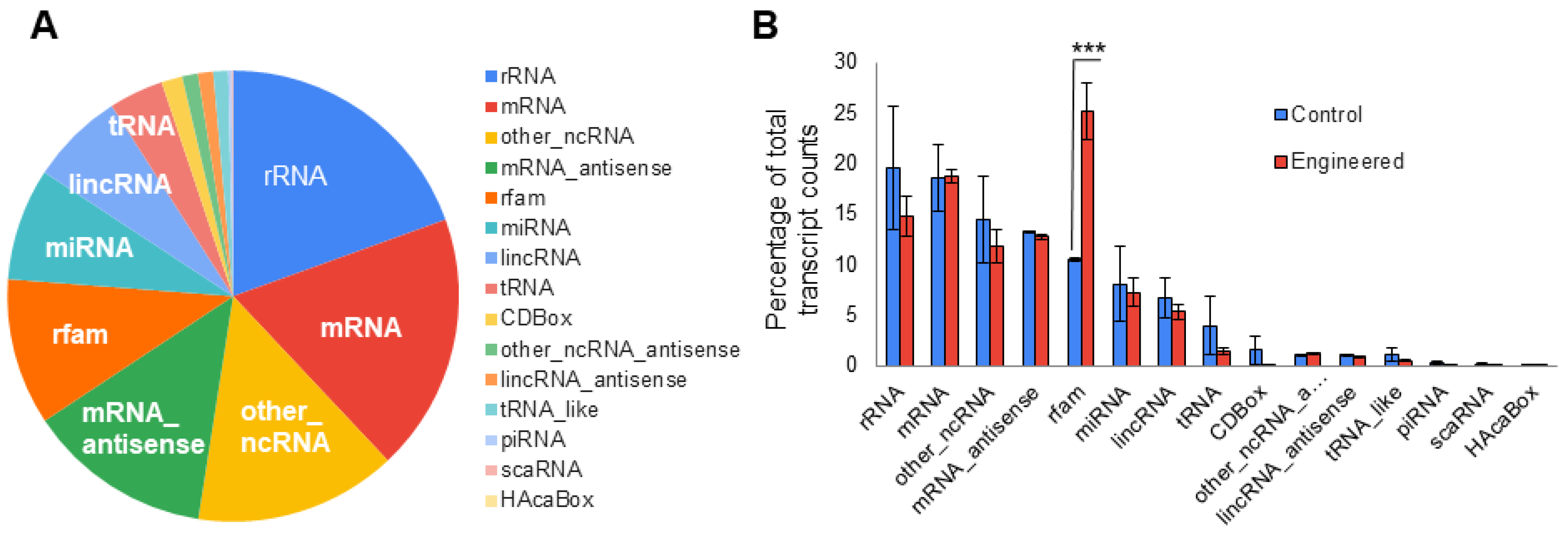
3.3. Dual-Omics Identifies Engineering-Induced Changes
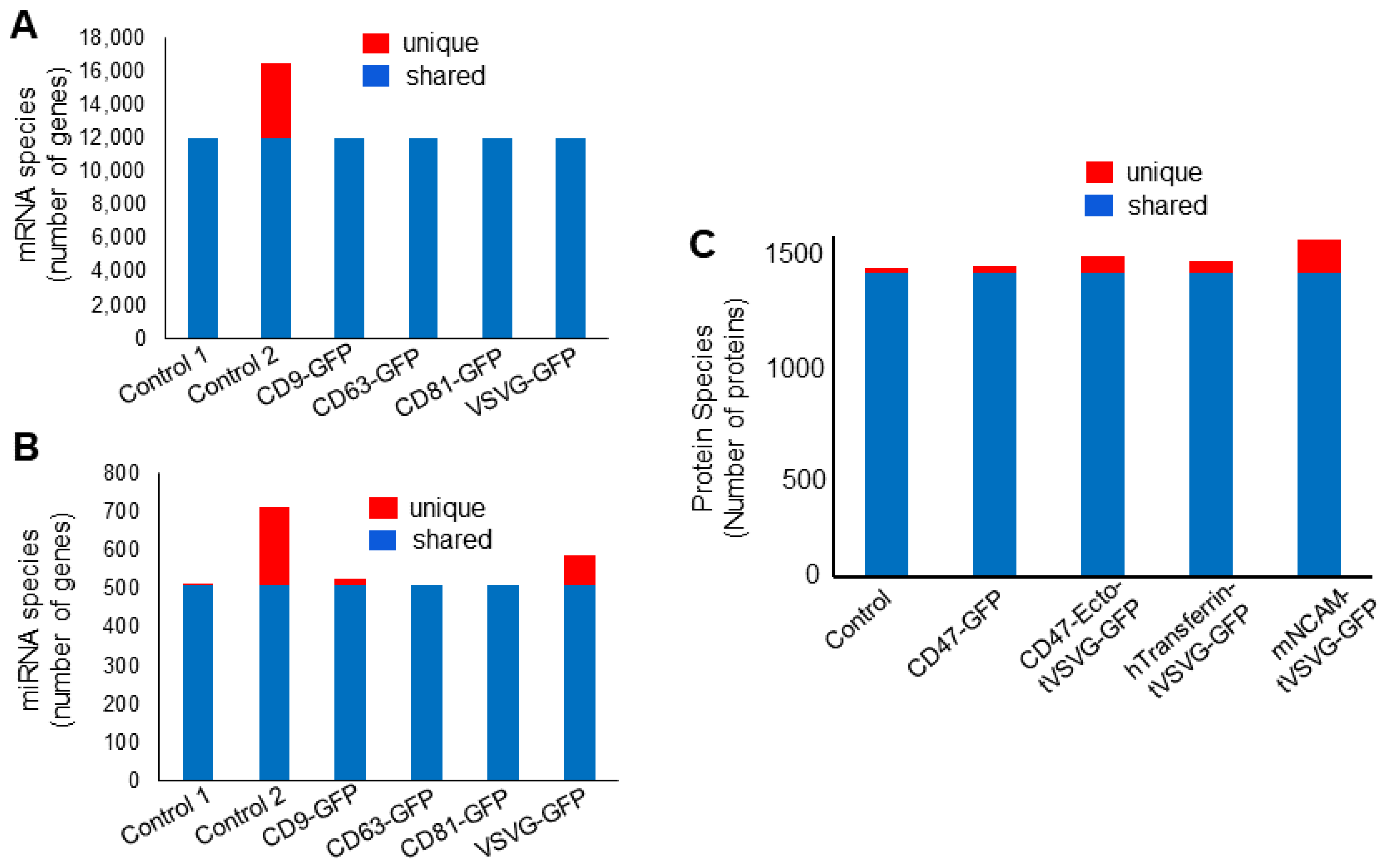
3.4. Dual-Omics Identifies a Diverse Ranges of Endogenous Cargos
3.5. Impact on Cellular Signaling and Metabolism by Endogenous Exosome Cargos
3.6. Proteomics Is Able to Identify Potential Protein Contaminants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hill, A.F. Therapeutically harnessing extracellular vesicles. Nat. Rev. Drug Discov. 2022, 21, 379–399. [Google Scholar] [CrossRef]
- Kar, R.; Dhar, R.; Mukherjee, S.; Nag, S.; Gorai, S.; Mukerjee, N.; Mukherjee, D.; Vatsa, R.; Chandrakanth Jadhav, M.; Ghosh, A.; et al. Exosome-Based Smart Drug Delivery Tool for Cancer Theranostics. ACS Biomater. Sci. Eng. 2023, 9, 577–594. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Ku, J.; Flojo, R.; Olson, C.; Bengford, D.; Marriott, G. Exosome- and extracellular vesicle-based approaches for the treatment of lysosomal storage disorders. Adv. Drug Deliv. Rev. 2022, 188, 114465. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Losordo, D.W. Exosomes and cardiac repair after myocardial infarction. Circ. Res. 2014, 114, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.T.; Xiong, Y.Y.; Ning, Y.; Tang, R.J.; Xu, J.Y.; Jiang, W.Y.; Li, X.S.; Zhang, L.L.; Chen, C.; Pan, Q.; et al. Nicorandil-Pretreated Mesenchymal Stem Cell-Derived Exosomes Facilitate Cardiac Repair after Myocardial Infarction via Promoting Macrophage M2 Polarization by Targeting miR-125a-5p/TRAF6/IRF5 Signaling Pathway. Int. J. Nanomed. 2024, 19, 2005–2024. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chopp, M. Exosome Therapy for Stroke. Stroke 2018, 49, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Schuldt, B.R.; Kalagara, R.; Chennareddy, S.; Odland, I.C.; Downes, M.H.; Reford, E.; Vicari, J.M.; Ali, M.; Bhimani, A.D.; Putrino, D.; et al. Exosome-Based Therapy for Ischemic Stroke: A Bibliometric Analysis of Current Trends and Future Directions. World Neurosurg. 2023, 171, e195–e205. [Google Scholar] [CrossRef]
- Duong, N.; Curley, K.; Brown, A.; Campanelli, A.; Do, M.A.; Levy, D.; Tantry, A.; Marriott, G.; Lu, B. Decoy exosomes as a novel biologic reagent to antagonize inflammation. Int. J. Nanomed. 2019, 14, 3413–3425. [Google Scholar] [CrossRef]
- Conceicao, M.; Forcina, L.; Wiklander, O.P.B.; Gupta, D.; Nordin, J.Z.; Vrellaku, B.; McClorey, G.; Mager, I.; Görgens, A.; Lundin, P.; et al. Engineered extracellular vesicle decoy receptor-mediated modulation of the IL6 trans-signalling pathway in muscle. Biomaterials 2021, 266, 120435. [Google Scholar] [CrossRef]
- Nasiri, Z.; Soleimanjahi, H.; Baheiraei, N.; Hashemi, S.M.; Pourkarim, M.R. The impact understanding of exosome therapy in COVID-19 and preparations for the future approaches in dealing with infectious diseases and inflammation. Sci. Rep. 2024, 14, 5724. [Google Scholar] [CrossRef] [PubMed]
- Do, M.A.; Levy, D.; Brown, A.; Marriott, G.; Lu, B. Targeted delivery of lysosomal enzymes to the endocytic compartment in human cells using engineered extracellular vesicles. Sci. Rep. 2019, 9, 17274. [Google Scholar] [CrossRef] [PubMed]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Li, H.; Yuan, Y.; Xie, Q.; Dong, Z. Exosomes: Potential targets for the diagnosis and treatment of neuropsychiatric disorders. J. Transl. Med. 2024, 22, 115. [Google Scholar] [CrossRef]
- Rehman, F.U.; Liu, Y.; Zheng, M.; Shi, B. Exosomes based strategies for brain drug delivery. Biomaterials 2023, 293, 121949. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Bisht, B.; Dutta, S.; Paul, M.K. Current advances in the use of exosomes, liposomes, and bioengineered hybrid nanovesicles in cancer detection and therapy. Acta Pharmacol. Sin. 2022, 43, 2759–2776. [Google Scholar] [CrossRef] [PubMed]
- De Leo, V.; Milano, F.; Agostiano, A.; Catucci, L. Recent Advancements in Polymer/Liposome Assembly for Drug Delivery: From Surface Modifications to Hybrid Vesicles. Polymers 2021, 13, 1027. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal. 2022, 20, 145. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Ryu, S.W.; Choi, H.; You, S.; Park, J.; Choi, C. Manufacturing Therapeutic Exosomes: From Bench to Industry. Mol. Cells 2022, 45, 284–290. [Google Scholar] [CrossRef]
- Zhang, K.; Cheng, K. Stem cell-derived exosome versus stem cell therapy. Nat. Rev. Bioeng. 2023, 1, 608–609. [Google Scholar] [CrossRef]
- van de Wakker, S.I.; Meijers, F.M.; Sluijter, J.P.G.; Vader, P. Extracellular Vesicle Heterogeneity and Its Impact for Regenerative Medicine Applications. Pharmacol. Rev. 2023, 75, 1043–1061. [Google Scholar] [CrossRef] [PubMed]
- Almeria, C.; Kress, S.; Weber, V.; Egger, D.; Kasper, C. Heterogeneity of mesenchymal stem cell-derived extracellular vesicles is highly impacted by the tissue/cell source and culture conditions. Cell Biosci. 2022, 12, 51. [Google Scholar] [CrossRef]
- Willms, E.; Johansson, H.J.; Mager, I.; Lee, Y.; Blomberg, K.E.; Sadik, M.; Alaarg, A.; Smith, C.I.; Lehtio, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.; Baek, R.; Pedersen, S.; Sondergaard, E.K.; Kristensen, S.R.; Varming, K. Extracellular Vesicle (EV) Array: Microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J. Extracell. Vesicles 2013, 2, 20920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Brown, A.; Johnson, B.; Diebold, D.; Asano, K.; Marriott, G.; Lu, B. Genetically Engineered Extracellular Vesicles Harboring Transmembrane Scaffolds Exhibit Differences in Their Size, Expression Levels of Specific Surface Markers and Cell-Uptake. Pharmaceutics 2022, 14, 2564. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrugger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Stickney, Z.; Losacco, J.; McDevitt, S.; Zhang, Z.; Lu, B. Development of exosome surface display technology in living human cells. Biochem. Biophys. Res. Commun. 2016, 472, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Losacco, J.; Stickney, Z.; Li, L.; Marriott, G.; Lu, B. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int. J. Nanomed. 2017, 12, 3153–3170. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Do, M.A.; Zhang, J.; Brown, A.; Lu, B. Orchestrating Extracellular Vesicle with Dual Reporters for Imaging and Capturing in Mammalian Cell Culture. Front. Mol. Biosci. 2021, 8, 680580. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, K. Peripheral Circulating Exosomal miRNAs Potentially Contribute to the Regulation of Molecular Signaling Networks in Aging. Int. J. Mol. Sci. 2020, 21, 1908. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Tanno, B.; De Stefano, I.; Giardullo, P.; Leonardi, S.; Merla, C.; Babini, G.; Tuncay Cagatay, S.; Mayah, A.; Kadhim, M.; et al. Micro-RNA and Proteomic Profiles of Plasma-Derived Exosomes from Irradiated Mice Reveal Molecular Changes Preventing Apoptosis in Neonatal Cerebellum. Int. J. Mol. Sci. 2022, 23, 2169. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, S.; McElroy, C.L.; Wang, B.; Jin, D.; Uteshev, V.V.; Jin, K. Circulating Pro-Inflammatory Exosomes Worsen Stroke Outcomes in Aging. Circ. Res. 2021, 129, e121–e140. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Chitti, S.V.; Gummadi, S.; Kang, T.; Shahi, S.; Marzan, A.L.; Nedeva, C.; Sanwlani, R.; Bramich, K.; Stewart, S.; Petrovska, M.; et al. Vesiclepedia 2024: An extracellular vesicles and extracellular particles repository. Nucleic Acids Res. 2024, 52, D1694–D1698. [Google Scholar] [CrossRef]
- Rodriguez, P.L.; Harada, T.; Christian, D.A.; Pantano, D.A.; Tsai, R.K.; Discher, D.E. Minimal “Self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science 2013, 339, 971–975. [Google Scholar] [CrossRef]
- Barclay, A.N.; Van den Berg, T.K. The interaction between signal regulatory protein alpha (SIRPalpha) and CD47: Structure, function, and therapeutic target. Annu. Rev. Immunol. 2014, 32, 25–50. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, K.B.; Burkhart, A.; Thomsen, L.B.; Andresen, T.L.; Moos, T. Targeting the transferrin receptor for brain drug delivery. Prog. Neurobiol. 2019, 181, 101665. [Google Scholar] [CrossRef]
- Pardridge, W.M.; Eisenberg, J.; Yang, J. Human blood-brain barrier transferrin receptor. Metabolism 1987, 36, 892–895. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Thery, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Lane, R.E.; Korbie, D.; Trau, M.; Hill, M.M. Purification Protocols for Extracellular Vesicles. Methods Mol. Biol. 2017, 1660, 111–130. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Wurdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, L.; Zeng, X.; Schwarz, H.; Nanda, H.S.; Peng, X.; Zhou, Y. Exosomes, a New Star for Targeted Delivery. Front. Cell Dev. Biol. 2021, 9, 751079. [Google Scholar] [CrossRef] [PubMed]


| Pathway | Control 1 | Control 2 | CD9 | CD63 | CD81 | VSVG |
|---|---|---|---|---|---|---|
| Cell Growth and Maintenance | 6.2 | 6.1 | 6.2 | 6 | 6 | 6.2 |
| Protein Metabolism | 7 | 6.9 | 7 | 7 | 7 | 6.9 |
| Energy Pathways | 7.4 | 7.5 | 7.4 | 7.4 | 7.4 | 7.7 |
| Transport | 7.6 *** | 7.7 *** | 7.6 *** | 7.5 *** | 7.5 *** | 7.7 *** |
| Metabolism | 7.7 | 8 | 7.7 | 7.7 | 7.7 | 7.8 |
| Nucleic Acid Metabolism | 17.6 *** | 17.5 *** | 17.6 *** | 17.7 *** | 17.8 *** | 17.7 *** |
| Cell Communication | 21.7 *** | 21.2 *** | 21.6 *** | 21.8 *** | 21.9 *** | 21.4 *** |
| Signal Transduction | 23.3 *** | 22.8 *** | 23.2 *** | 23.3 *** | 23.4 *** | 23.1 *** |
| Pathway | Control | CD47-GFP | CD47-tVSVG-GFP | hTransferrin-tVSVG-GFP | mNCAM-tVSVG-GFP |
|---|---|---|---|---|---|
| Transport | 8.4 | 8.3 | 8.2 *** | 8.1 | 8.8 |
| Cell Growth and Maintenance | 11.7 *** | 10.4 *** | 10.3 *** | 10.3 *** | 9.9 *** |
| Energy Pathways | 13.2 *** | 13.5 *** | 13.8 *** | 13.5 *** | 13 *** |
| Metabolism | 13.5 *** | 13.8 *** | 14 *** | 13.7 *** | 13.1 *** |
| Nucleic Acid Metabolism | 16.8 | 17.1 | 17.1 | 17.6 | 17.8 |
| Cell Communication | 17.5 | 17.1 | 16.4 | 16.6 | 16.5 |
| Protein Metabolism | 18.3 *** | 18.2 *** | 17.9 *** | 18.1 *** | 17.8 *** |
| Signal Transduction | 18.4 | 18.4 | 17.6 | 17.8 | 17.9 |
| Sample | Cow Plasma Proteins | Unknown Cow Proteins | Trypsin | Streptavidin |
|---|---|---|---|---|
| Control | 92 | 6 | Yes | Yes |
| CD47-GFP | 91 | 6 | Yes | Yes |
| CD47-Ecto-tVSVG | 93 | 6 | Yes | Yes |
| hTransferrin-tVSVG-GFP | 91 | 6 | Yes | Yes |
| mNCAM-tVSVG-GFP | 91 | 6 | Yes | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olson, C.; Ivanov, K.; Boyes, D.; Bengford, D.; Ku, J.; Flojo, R.; Zhang, P.; Lu, B. Dual-Omics Approach Unveils Novel Perspective on the Quality Control of Genetically Engineered Exosomes. Pharmaceutics 2024, 16, 824. https://doi.org/10.3390/pharmaceutics16060824
Olson C, Ivanov K, Boyes D, Bengford D, Ku J, Flojo R, Zhang P, Lu B. Dual-Omics Approach Unveils Novel Perspective on the Quality Control of Genetically Engineered Exosomes. Pharmaceutics. 2024; 16(6):824. https://doi.org/10.3390/pharmaceutics16060824
Chicago/Turabian StyleOlson, Christopher, Konstantin Ivanov, Darin Boyes, David Bengford, Joy Ku, Renceh Flojo, Pengyang Zhang, and Biao Lu. 2024. "Dual-Omics Approach Unveils Novel Perspective on the Quality Control of Genetically Engineered Exosomes" Pharmaceutics 16, no. 6: 824. https://doi.org/10.3390/pharmaceutics16060824
APA StyleOlson, C., Ivanov, K., Boyes, D., Bengford, D., Ku, J., Flojo, R., Zhang, P., & Lu, B. (2024). Dual-Omics Approach Unveils Novel Perspective on the Quality Control of Genetically Engineered Exosomes. Pharmaceutics, 16(6), 824. https://doi.org/10.3390/pharmaceutics16060824








