Identification, Synthesis, and In Vitro Activities of Antimicrobial Peptide from African Catfish against the Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish and Mucus Collection
2.2. Peptide Purification
2.3. Peptide Sequencing
2.4. Primary Structure Characterization
2.5. The 3-D Structure Prediction of the Antimicrobial Peptide
2.6. Peptide Synthesis
2.7. Antimicrobial Activity
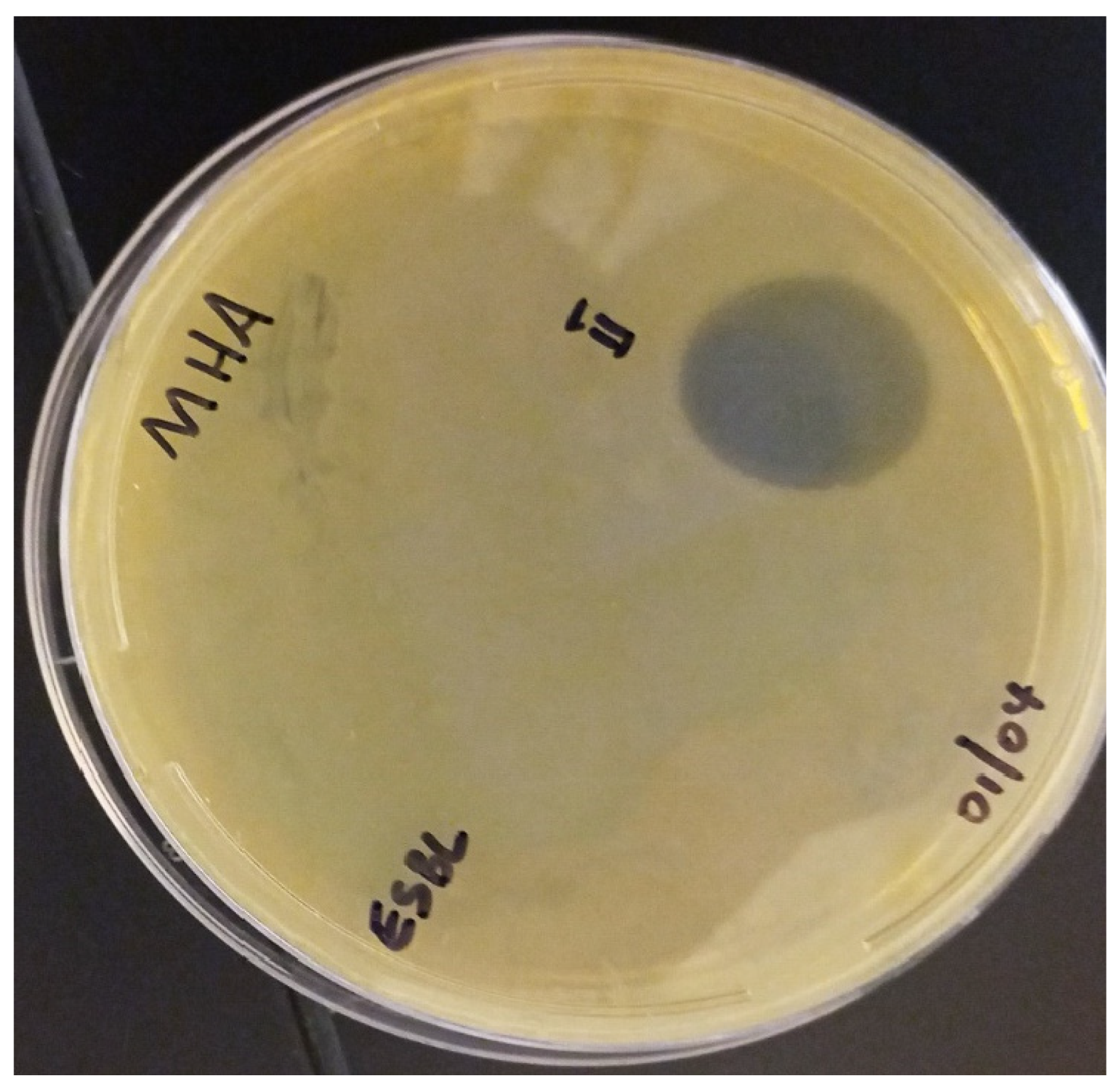
2.7.1. Spot-on-Lawn Method
2.7.2. Broth Dilution Method
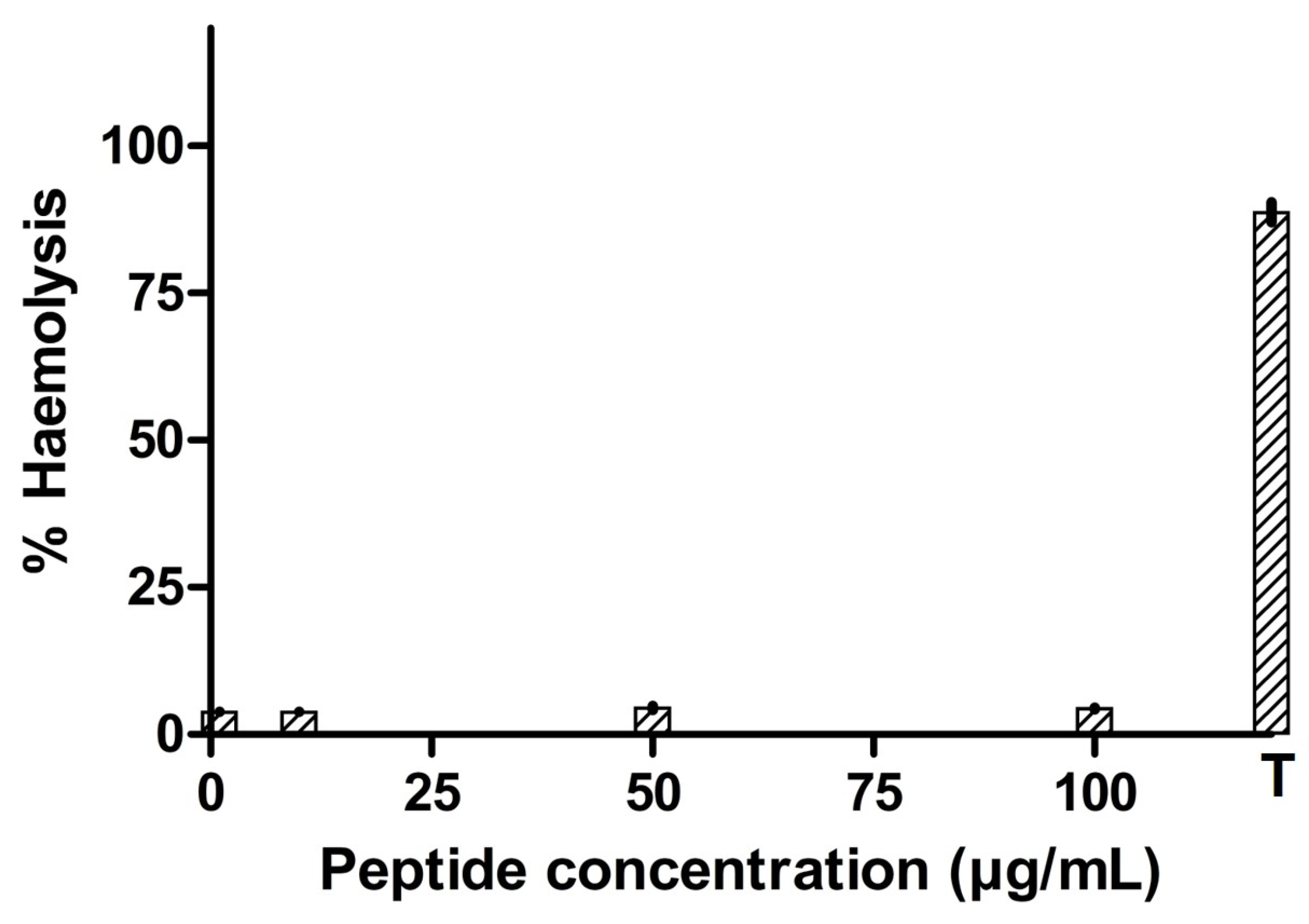
2.8. Hemotoxicity Profile of Antimicrobial Peptide
3. Results
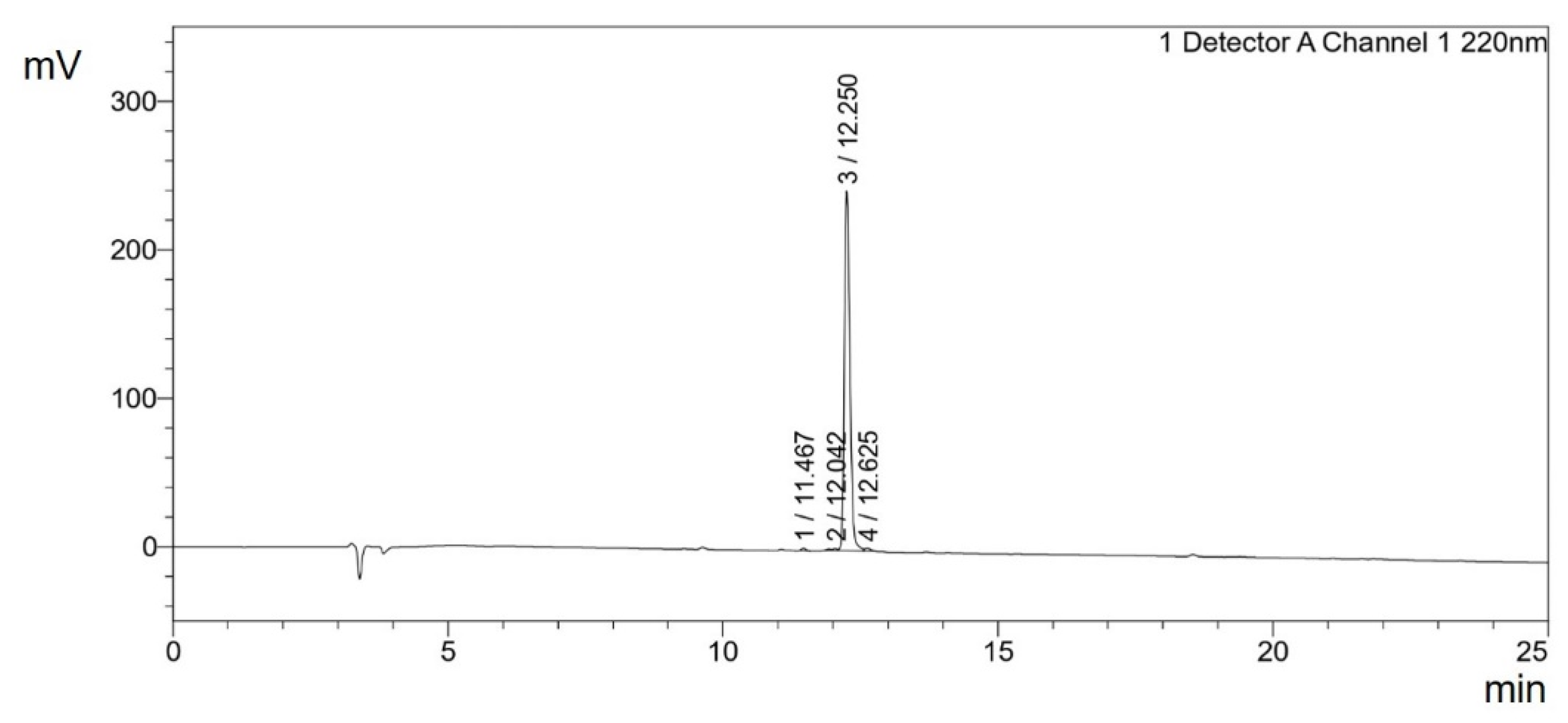
3.1. Solid Phase Extraction
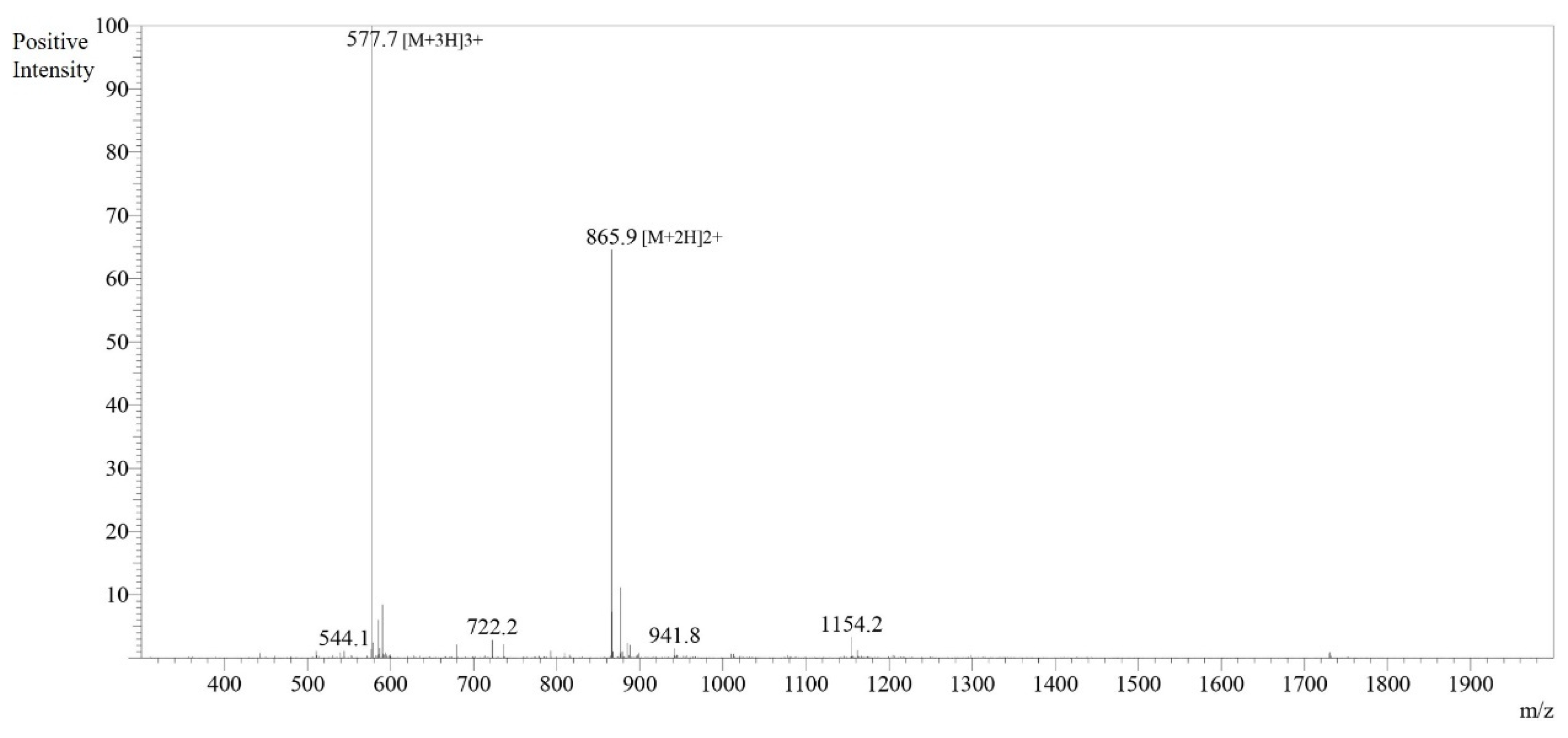
3.2. Peptide Identification and Characterization
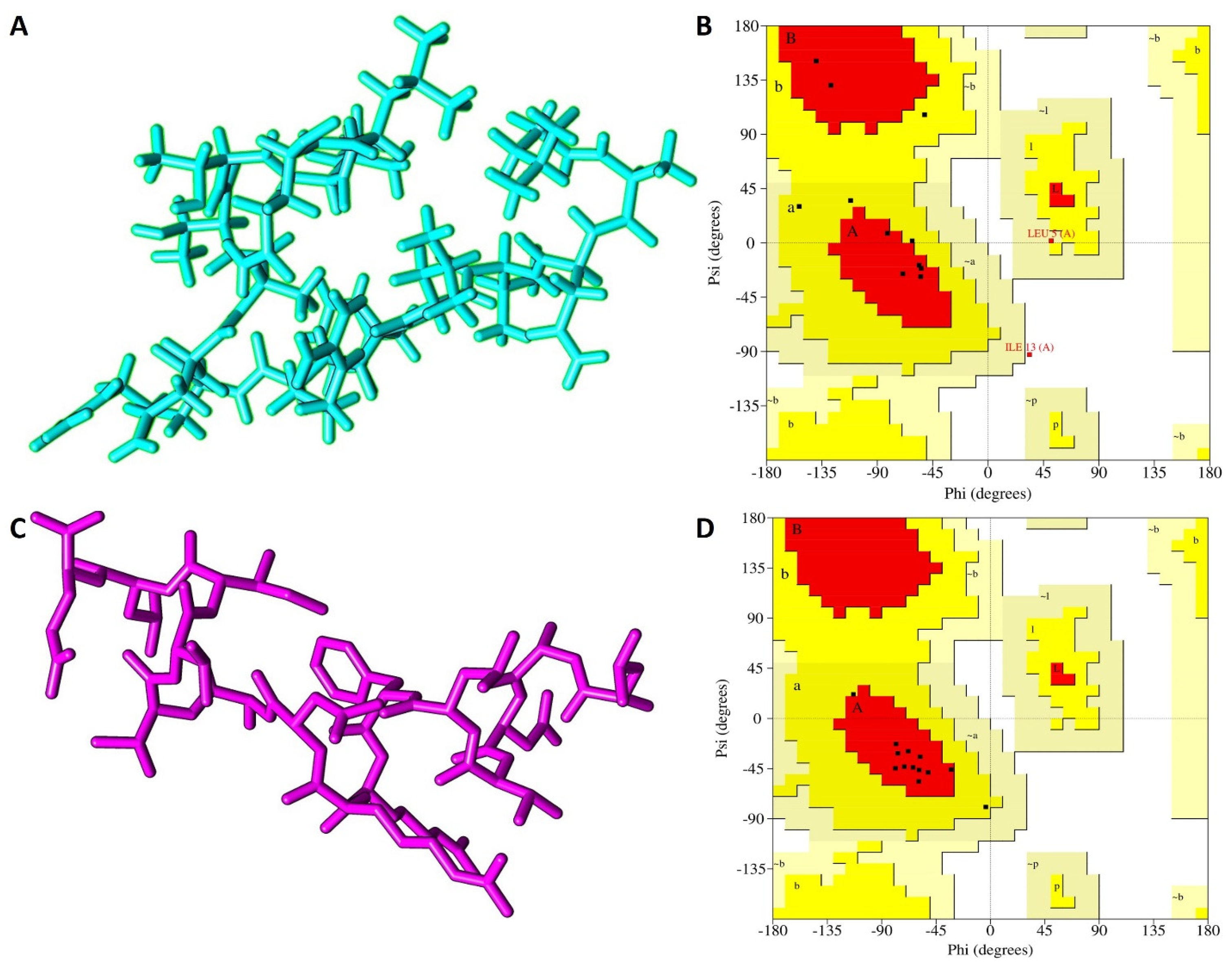
3.3. The 3-D Structure Modelling
3.4. Peptide Synthesis and In Vitro Antimicrobial Activity
3.5. Hemolytic Activity of the Antimicrobial Peptide
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- WHO. WHO Priority Pathogens List for R&D of New Antibiotics. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 18 April 2023).
- Paterson, D.L.; Bonomo, R.A. Clinical Update Extended-Spectrum Beta-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar]
- Luzzaro, F.; Mezzatesta, M.; Mugnaioli, C.; Perilli, M.; Stefani, S.; Amicosante, G.; Rossolini, G.M.; Toniolo, A. Trends in Production of Extended-Spectrum β-Lactamases among Enterobacteria of Medical Interest: Report of the Second Italian Nationwide Survey. J. Clin. Microbiol. 2006, 44, 1659–1664. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Benson, P.; Neuzil, K.M.; Grandjean, M.; Marino, J.L. Burden of Community-Onset Escherichia coli Bacteremia in Seniors. J. Infect. Dis. 2005, 191, 1523–1529. [Google Scholar] [CrossRef]
- Norrby, S.R.; Nord, C.E.; Finch, R. Lack of Development of New Antimicrobial Drugs: A Potential Serious Threat to Public Health. Lancet Infect. Dis. 2005, 5, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Musbau, S.; Yushau, M.; Dalha, W.T. Antibacterial Activities and Quantitative Phytochemical Screening of the Hexane and Ethanolic Oil Extracts of Syzygium aromaticum against ESBL Producing Bacteria Isolates. Eur. J. Med. Health Res. 2024, 2, 123–127. [Google Scholar] [CrossRef]
- Zai, M.J.; Cheesman, M.J.; Cock, I.E. Phytochemical Evaluation of Terminalia canescens DC. Radlk. Extracts with Antibacterial and Antibiotic Potentiation Activities against Selected β-Lactam Drug-Resistant Bacteria. Molecules 2024, 29, 1385. [Google Scholar] [CrossRef] [PubMed]
- Mogana, R.; Adhikari, A.; Tzar, M.N.; Ramliza, R.; Wiart, C. Antibacterial Activities of the Extracts, Fractions and Isolated Compounds from Canarium patentinervium Miq. Against Bacterial Clinical Isolates. BMC Complement. Med. Ther. 2020, 20, 55. [Google Scholar] [CrossRef]
- Hameed, A.S.H.; Karthikeyan, C.; Ahamed, A.P.; Thajuddin, N.; Alharbi, N.S.; Alharbi, S.A.; Ravi, G. In Vitro Antibacterial Activity of ZnO and Nd Doped ZnO Nanoparticles against ESBL Producing Escherichia coli and Klebsiella pneumoniae. Sci. Rep. 2016, 6, 24312. [Google Scholar] [CrossRef]
- Maruthupandy, M.; Rajivgandhi, G.; Muneeswaran, T.; Song, J.M.; Manoharan, N. Biologically Synthesized Zinc Oxide Nanoparticles as Nanoantibiotics against ESBLs Producing Gram Negative Bacteria. Microb. Pathog. 2018, 121, 224–231. [Google Scholar] [CrossRef]
- Togan, T.; Evren, E.; Çiftçi, Ö.; Narcı, H.; Özcan, M.; Arslan, H. The Antibacterial Effect of Propolis against Clinical Isolates. Sci.-Afr. J. Sci. Issues 2014, 2, 551–553. [Google Scholar]
- Skaradzińska, A.; Śliwka, P.; Kuźmińska-Bajor, M.; Skaradziński, G.; Rzasa, A.; Friese, A.; Roschanski, N.; Murugaiyan, J.; Roesler, U.H. The Efficacy of Isolated Bacteriophages from Pig Farms against ESBL/AmpC-Producing Escherichia coli from Pig and Turkey Farms. Front. Microbiol. 2017, 8, 530. [Google Scholar] [CrossRef]
- Erol, H.B.; Kaskatepe, B.; Ozturk, S.; Safi Oz, Z. The Comparison of Lytic Activity of Isolated Phage and Commercial Intesti Bacteriophage on ESBL Producer E. Coli and Determination of Ec_P6 Phage Efficacy with In Vivo Galleria mellonella Larvae Model. Microb. Pathog. 2022, 167, 105563. [Google Scholar] [CrossRef]
- Pienaar, U.D.V. The Freshwater Fished of the Kruger National Park. Koedoe 1968, 11, 1–82. [Google Scholar] [CrossRef]
- Okella, H.; Ajayi, C.O.; Ikiriza, H.; Mtewa, A.G.; Kaggwa, B.; Aber, J.A.; Kato, C.D.; Engeu, P.O. Bacterial Cell Envelope Lysis and Hemotoxicity of Peptides Previously Isolated from African Catfish, Clarias gariepinus. East Afr. Sci. 2022, 4, 93–100. [Google Scholar] [CrossRef]
- Okella, H.; Okello, E.; Mtewa, A.G.; Ikiriza, H.; Kaggwa, B.; Aber, J.; Ndekezi, C.; Nkamwesiga, J.; Ajayi, C.O.; Mugeni, I.M.; et al. ADMET Profiling and Molecular Docking of Potential Antimicrobial Peptides Previously Isolated from African Catfish, Clarias gariepinus. Front. Mol. Biosci. 2022, 9, 1039286. [Google Scholar] [CrossRef] [PubMed]
- Okella, H.; Ikiriza, H.; Ochwo, S.; Ajayi, C.O.; Ndekezi, C.; Nkamwesiga, J.; Kaggwa, B.; Aber, J.; Mtewa, A.G.; Koffi, T.K.; et al. Identification of Antimicrobial Peptides Isolated from the Skin Mucus of African Catfish, Clarias gariepinus (Burchell, 1822). Front. Microbiol. 2021, 12, 794631. [Google Scholar] [CrossRef] [PubMed]
- Hansen, I.K.Ø.; Lövdahl, T.; Simonovic, D.; Hansen, K.; Andersen, A.J.C.; Devold, H.; Richard, C.S.M.; Andersen, J.H.; Strøm, M.B.; Haug, T. Antimicrobial Activity of Small Synthetic Peptides Based on the Marine Peptide Turgencin a: Prediction of Antimicrobial Peptide Sequences in a Natural Peptide and Strategy for Optimization of Potency. Int. J. Mol. Sci. 2020, 21, 5460. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Rathinakumar, R.; Walkenhorst, W.F.; Wimley, W.C. Broad-Spectrum Antimicrobial Peptides by Rational Combinatorial Design and High-Throughput Screening: The Importance of Interfacial Activity. J. Am. Chem. Soc. 2009, 131, 7609–7617. [Google Scholar] [CrossRef]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides with Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 501450. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.O.; Molle, G.; Kemp, G.D.; Smith, V.J. Isolation and Characterisation of Oncorhyncin II, a Histone H1-Derived Antimicrobial Peptide from Skin Secretions of Rainbow Trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Bergsson, G.; Agerberth, B.; Jörnvall, H.; Gudmundsson, G.H. Isolation and Identification of Antimicrobial Components from the Epidermal Mucus of Atlantic Cod (Gadus Morhua). FEBS J. 2005, 272, 4960–4969. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Myxinidin, a Novel Antimicrobial Peptide from the Epidermal Mucus of Hagfish, Myxine glutinosa L. Mar. Biotechnol. 2009, 11, 748–757. [Google Scholar] [CrossRef]
- Alabssawy, A.N.; Abu-Elghait, M.; Azab, A.M.; Khalaf-Allah, H.M.M.; Ashry, A.S.; Ali, A.O.M.; Sabra, A.B.A.A.; Salem, S.S. Hindering the Biofilm of Microbial Pathogens and Cancer Cell Lines Development Using Silver Nanoparticles Synthesized by Epidermal Mucus Proteins from Clarias gariepinus. BMC Biotechnol. 2024, 24, 28. [Google Scholar] [CrossRef] [PubMed]
- Enan, G.; El-Essawy, A.A.; Uyttendaele, M.; Debevere, J. Antibacterial Activity of Lactobacillus plantarum UG1 Isolated from Dry Sausage: Characterization Production and Bactericidal Action of Plantaricin UG1. Int. J. Food Microbiol. 1996, 30, 189–215. [Google Scholar] [CrossRef] [PubMed]
- Song, H.P.; Yu, J.; Li, S.; Wei, Y.X.; Wang, W.X.; Xu, L.N.; Han, D. The Antibacterial Activity of Peptides from Porphyra Yezoensis. Adv. Fish. Sci. 2015, 36, 140–145. [Google Scholar]
- Rana, S.; Bajaj, R.; Mann, B. Characterization of Antimicrobial and Antioxidative Peptides Synthesized by L. Rhamnosus C6 Fermentation of Milk. Int. J. Pept. Res. Ther. 2018, 24, 309–321. [Google Scholar] [CrossRef]
- Schmid, F. Biological Macromolecules: UV-Visible Spectrophotometry; John Wiley & Sons Ltd.: Chichester, UK, 2001; Volume 1. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 4, D442–D450. [Google Scholar] [CrossRef]
- Moon, C.P.; Fleming, K.G. Side-Chain Hydrophobicity Scale Derived from Transmembrane Protein Folding into Lipid Bilayers. Proc. Natl. Acad. Sci. USA 2011, 108, 10174–10177. [Google Scholar] [CrossRef]
- Vishnepolsky, B.; Grigolava, M.; Managadze, G.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M.; Pirtskhalava, M. Comparative Analysis of Machine Learning Algorithms on the Microbial Strain-Specific AMP Prediction. Brief. Bioinform. 2022, 23, bbac233. [Google Scholar] [CrossRef] [PubMed]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An Updated de Novo Structure Prediction Server for Both Linear and Disulfide Bonded Cyclic Peptides. Nucleic Acids Res. 2012, 40, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Soomro, A.H.; Masud, T.; Sammi, S.; Rathore, H.A. Comparison of Different Methods for Detection of Antimicrobial Activity of Lactobacillus spp. Pak. J. Zool. 2007, 39, 265–268. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). M07: Clinical and Laboratory Standards Institute Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Clin. Lab. Stand. Inst. 2018, 38, 91. [Google Scholar]
- Teh, C.H.; Nazni, W.A.; Nurulhusna, A.H.; Norazah, A.; Lee, H.L. Determination of Antibacterial Activity and Minimum Inhibitory Concentration of Larval Extract of Fly via Resazurin-Based Turbidometric Assay. BMC Microbiol. 2017, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- De La Salud Bea, R.; Petraglia, A.F.; De Johnson, L.E.L. Synthesis, Antimicrobial Activity and Toxicity of Analogs of the Scorpion Venom BmKn Peptides. Toxicon 2015, 101, 79–84. [Google Scholar] [CrossRef]
- Je, J.Y.; Kim, S.Y.; Kim, S.K. Preparation and Antioxidative Activity of Hoki Frame Protein Hydrolysate Using Ultrafiltration Membranes. Eur. Food Res. Technol. 2005, 221, 157–162. [Google Scholar] [CrossRef]
- Sugiyama, N.; Araki, M.; Ishida, M.; Nagashima, Y.; Shiomi, K. Further Isolation and Characterization of Grammistins from the Skin Secretion of the Soapfish Grammistes sexlineatus. Toxicon 2005, 45, 595–601. [Google Scholar] [CrossRef]
- Su, Y. Isolation and Identification of Pelteobagrin, a Novel Antimicrobial Peptide from the Skin Mucus of Yellow Catfish (Pelteobagrus fulvidraco). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 158, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Kęska, P.; Stadnik, J. Antimicrobial Peptides of Meat Origin-An In Silico and In Vitro Analysis. Protein Pept. Lett. 2017, 24, 165–173. [Google Scholar] [PubMed]
- Bruston, F.; Lacombe, C.; Zimmermann, K.; Piesse, C.; El Amir, C. Structural Malleability of Plasticins: Preorganized Conformations in Solution and Relevance for Antimicrobial Activity. Biopolymers 2007, 86, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Kang, H.K.; Ko, S.J.; Hong, M.J.; Bang, J.K.; Seo, C.H.; Park, Y. Mechanisms Driving the Antibacterial and Antibiofilm Properties of Hp1404 and Its Analogue Peptides against Multidrug-Resistant Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 1763. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Burton, M.G.; Huang, Q.M.; Hossain, M.A.; Wade, J.D.; Clayton, A.H.A.; Gee, M.L. Long-Time-Scale Interaction Dynamics between a Model Antimicrobial Peptide and Giant Unilamellar Vesicles. Langmuir 2013, 29, 14613–14621. [Google Scholar] [CrossRef] [PubMed]
- Okella, H.; Georrge, J.J.; Ochwo, S.; Ndekezi, C.; Koffi, K.T.; Aber, J.; Ajayi, C.O.; Fofana, F.G.; Ikiriza, H.; Mtewa, A.G.; et al. New Putative Antimicrobial Candidates: In Silico Design of Fish-Derived Antibacterial Peptide-Motifs. Front. Bioeng. Biotechnol. 2020, 8, 604041. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Kong, X.; Hua, Y.; Chen, Y.; Zhang, C.; Chen, Y. Identification of Antibacterial Peptides Generated from Enzymatic Hydrolysis of Cottonseed Proteins. LWT 2020, 125, 109199. [Google Scholar] [CrossRef]
- Lüders, T.; Birkemo, G.A.; Nissen-Meyer, J.; Andersen, Ø.; Nes, I.F. Proline Conformation-Dependent Antimicrobial Activity of a Proline-Rich Histone H1 N-Terminal Peptide Fragment Isolated from the Skin Mucus of Atlantic Salmon. Antimicrob. Agents Chemother. 2005, 49, 2399–2406. [Google Scholar] [CrossRef]
- Cole, A.M.; Weis, P.; Diamond, G. Isolation and Characterization of Pleurocidin, an Antimicrobial Peptide in the Skin Secretions of Winter Flounder. J. Biol. Chem. 1997, 272, 12008–12013. [Google Scholar] [CrossRef]
- Boto, A.; De La Lastra, J.M.P.; González, C.C. The Road from Host-Defense Peptides to a New Generation of Antimicrobial Drugs. Molecules 2018, 23, 311. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, S.A.; Reinoso, C.; Morales, F.; Pilaquinga, F.; Morán-Marcillo, G.; Proaño-Bolaños, C.; Blasco-Zúñiga, A.; Rivera, M.; Meneses, L. Novel Antimicrobial Cruzioseptin Peptides Extracted from the Splendid Leaf Frog, Cruziohyla calcarifer. Amino Acids 2021, 53, 853–868. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Taheri, B.; Momenzadeh, N.; Salarinia, R.; Nabipour, I.; Farshadzadeh, Z.; Bargahi, A. Identification and Characterization of Novel Antimicrobial Peptide from Hippocampus comes by In Silico and Experimental Studies. Mar. Biotechnol. 2018, 20, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Fan, Y.; Zhao, W.; Ding, L.; Li, J.; Liu, J. Novel Angiotensin-Converting Enzyme Inhibitory Peptides Derived from Oncorhynchus mykiss Nebulin: Virtual Screening and In Silico Molecular Docking Study. J. Food Sci. 2018, 83, 2375–2383. [Google Scholar] [CrossRef] [PubMed]
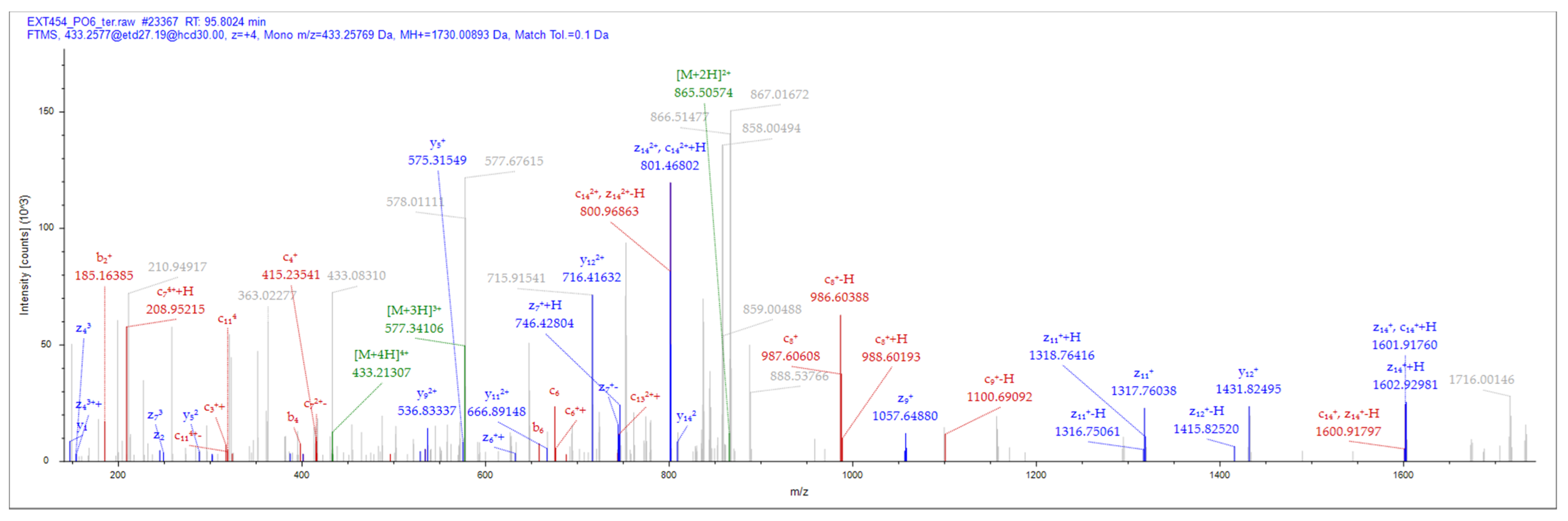
| Peptide ID | Sequence | Length | MW (Da) | Calculated Net Charge | pI | GRAVY | General Antimicrobial Activity |
|---|---|---|---|---|---|---|---|
| NACAP-I | VAPEEHPVLLTEAPLNPK | 18 | 1954.06 | −2 | 4.75 | −0.267 | Non-AMP |
| NACAP-II | LANVLFRRNATTILQ | 15 | 1730.01 | 2 | 12.00 | 0.373 | AMP |
| NACAP-III | TQAADLLGMVSRKSLWGQF | 19 | 2108.09 | 1 | 8.41 | 0.053 | Non-AMP |
| NACAP-IV | MLNLLHIKSGGCFLFLE | 17 | 1951.02 | 0 | 6.50 | 1.047 | Non-AMP |
| NACAP-V | KGVQINYRYFKGFFLES | 17 | 2096.20 | 2 | 9.52 | −0.359 | Non-AMP |
| NACAP-VI | GAGGGFGAGGGFG | 13 | 968.42 | 0 | 5.22 | 0.431 | AMP |
| NACAP-VII | NDDLIPPAQSIETTDMKTEVNC | 22 | 2434.11 | −4 | 3.77 | −0.673 | Non-AMP |
| NACAP-VIII | VSTMKSFMPGPCIHGDLP | 18 | 1948.90 | 0 | 6.71 | 0.172 | Non-AMP |
| Ramachandran Plot Parameters | I-TASSER | PEP-FOLD |
|---|---|---|
| Residues in most favored regions (%) | 53.8 | 84.6 |
| Residues in additional allowed regions (%) | 30.8 | 15.4 |
| Residues in generously allowed regions (%) | 7.7 | 0.0 |
| Residues in disallowed regions (%) | 7.7 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okella, H.; Odongo, S.; Vertommen, D.; Okello, E. Identification, Synthesis, and In Vitro Activities of Antimicrobial Peptide from African Catfish against the Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli. Pharmaceutics 2024, 16, 850. https://doi.org/10.3390/pharmaceutics16070850
Okella H, Odongo S, Vertommen D, Okello E. Identification, Synthesis, and In Vitro Activities of Antimicrobial Peptide from African Catfish against the Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli. Pharmaceutics. 2024; 16(7):850. https://doi.org/10.3390/pharmaceutics16070850
Chicago/Turabian StyleOkella, Hedmon, Steven Odongo, Didier Vertommen, and Emmanuel Okello. 2024. "Identification, Synthesis, and In Vitro Activities of Antimicrobial Peptide from African Catfish against the Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli" Pharmaceutics 16, no. 7: 850. https://doi.org/10.3390/pharmaceutics16070850





