Combinatorial Delivery of Docetaxel- and Erlotinib-Loaded Functionalized Nanostructured Lipid Carriers for the Treatment of Triple-Negative Breast Cancer Using Quality-by-Design Approach
Abstract
:1. Introduction
2. Materials
3. Cell Lines
4. Methods
4.1. Screening of Excipients
4.2. Compatibility Study
4.3. Preparation of DOC–ERL NLCs
4.4. Optimization of DOC–ERL NLCs
4.5. Preparation of FA–DOC–ERL NLCs
4.6. Characterization of NLCs
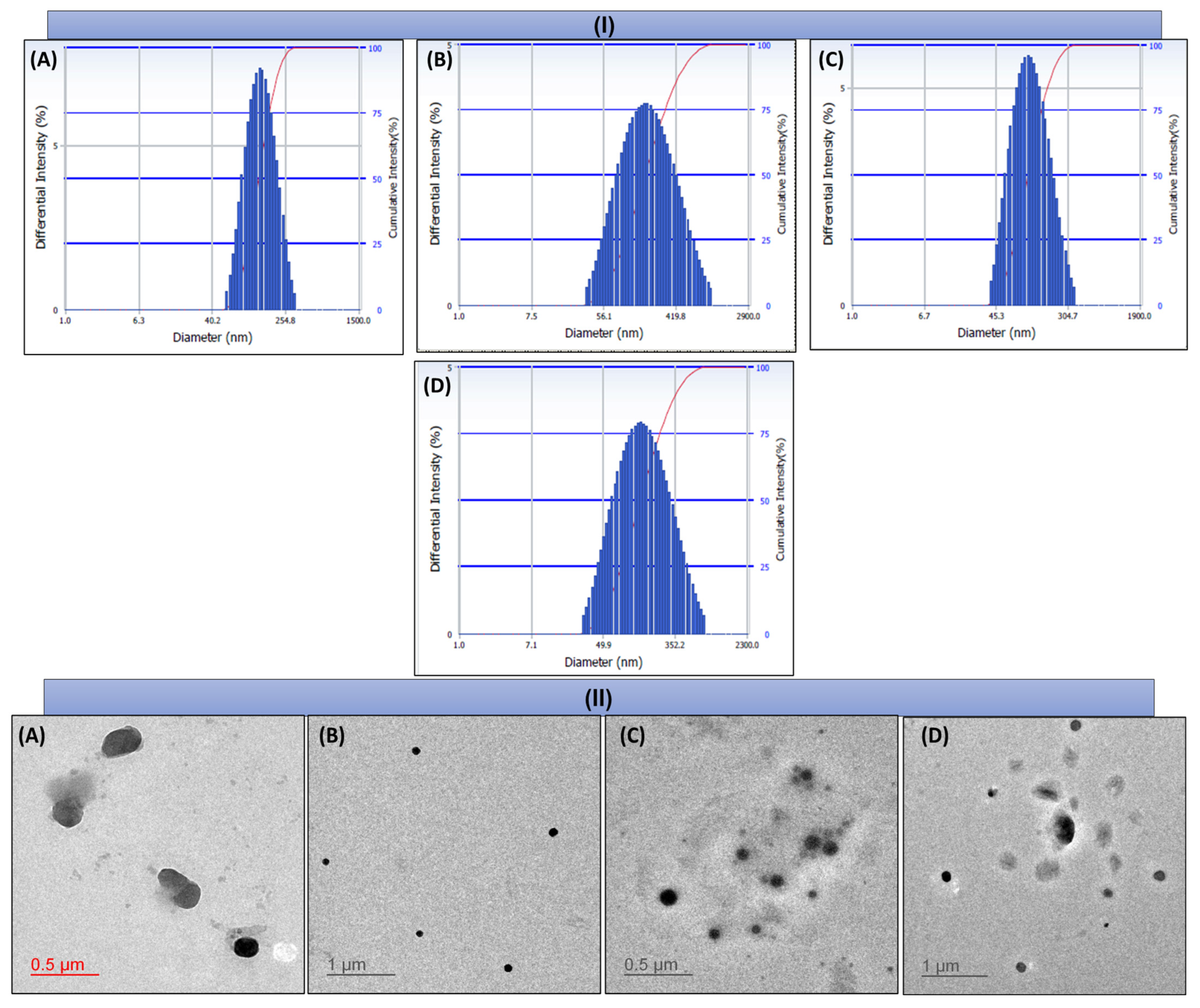
4.6.1. Particle Size (PS), Polydispersity Index (PDI), and Zeta Potential (ZP)
4.6.2. Entrapment Efficiency and Drug Loading
4.6.3. Morphology Analysis
4.6.4. Fourier Transform Infrared Spectroscopy–Attenuated Total Reflectance (FTIR-ATR)
4.7. Freeze-Drying Studies
4.8. In Vitro Drug Release
4.9. In Vitro Lipolysis Study
4.10. Stability Study
4.11. Cell Viability Assay
Determination of Combination Effect
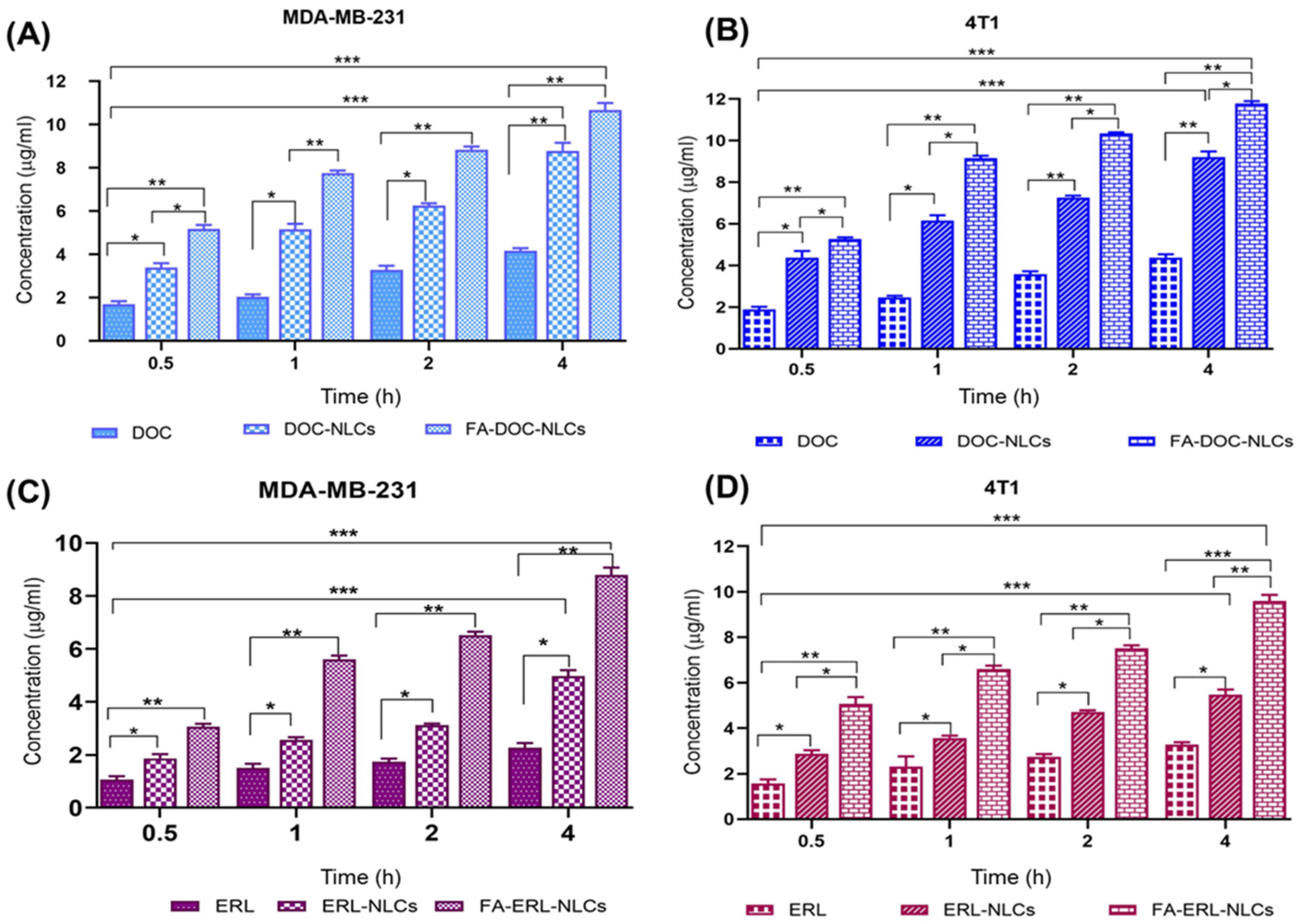
4.12. Quantitative and Qualitative Cellular Uptake Assay
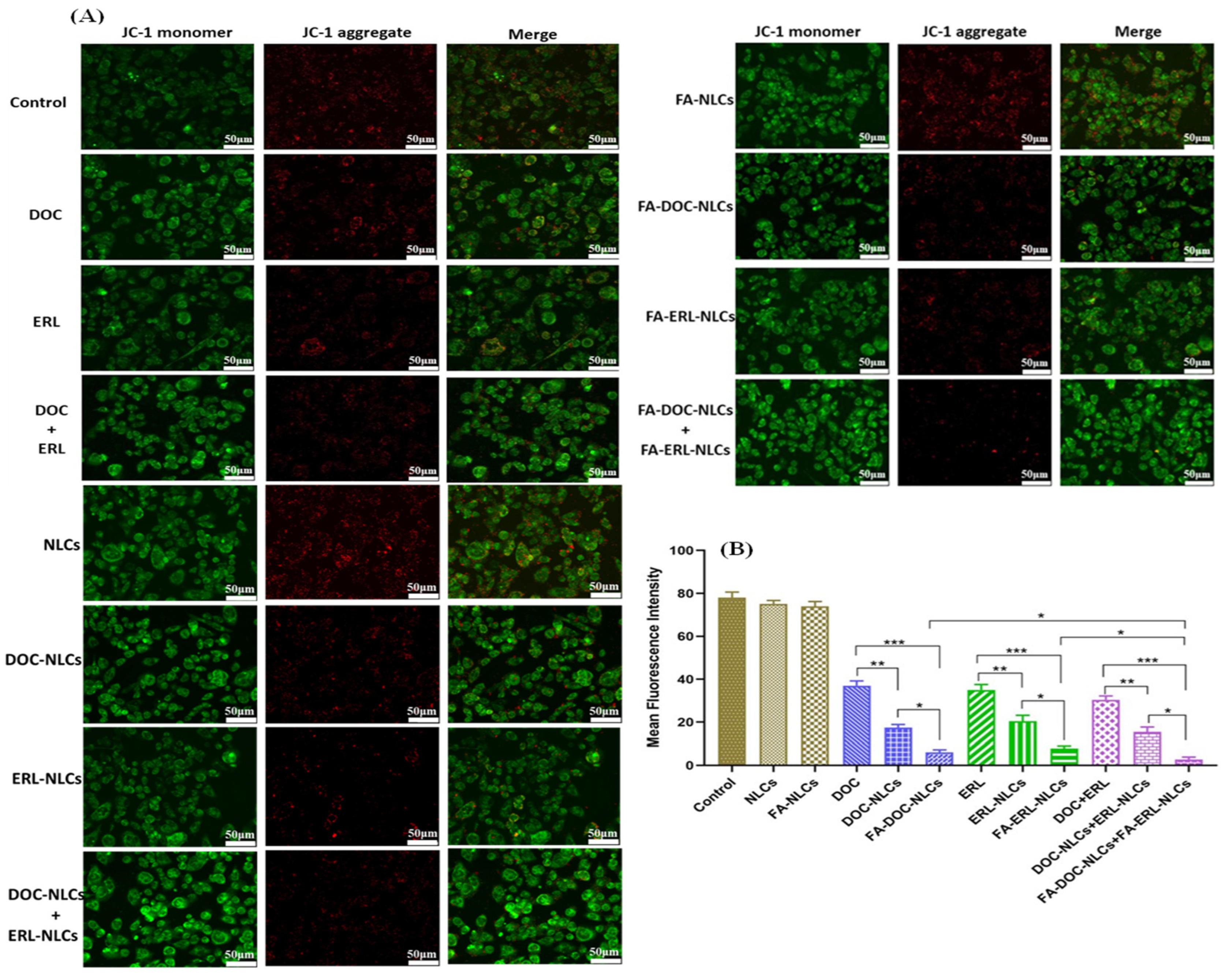
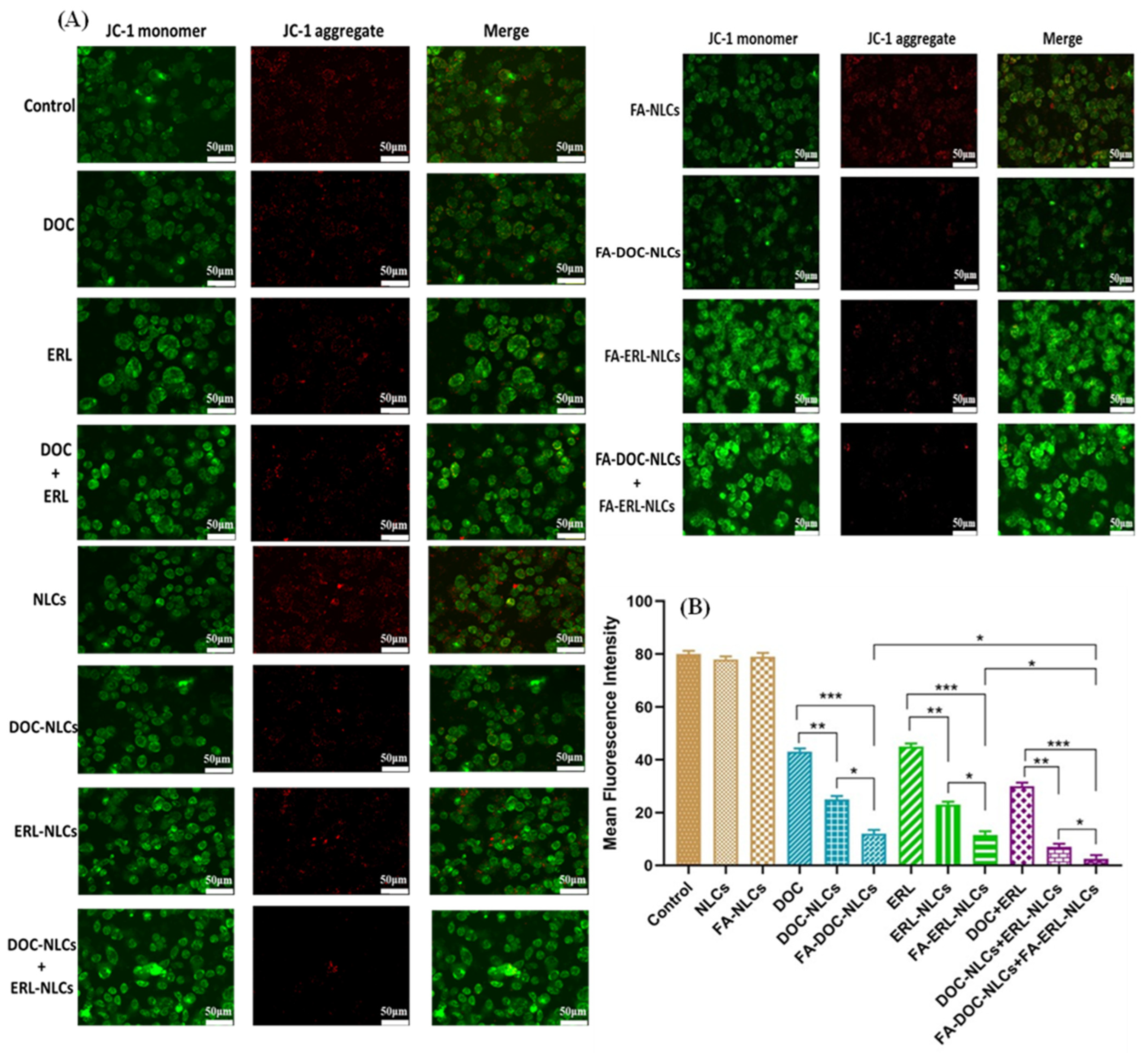
4.13. Mitochondrial Membrane Potential (MMP) Assay
4.14. Colony Formation Assay
4.15. Transwell Migration Assay
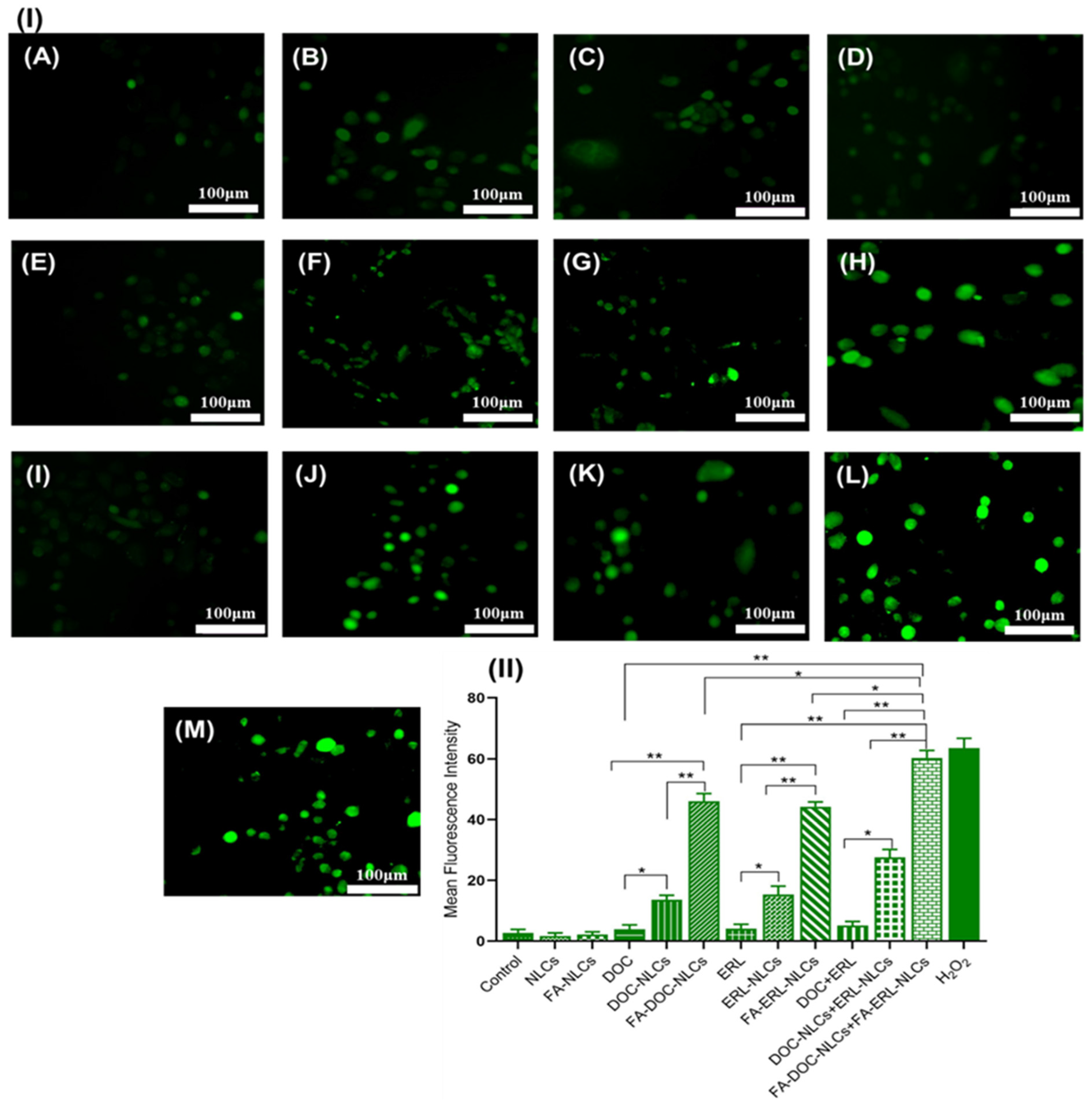
4.16. Reactive Oxygen Species (ROS) Assay
5. Statistical Analysis
6. Results
6.1. Screening of Excipients
6.2. Compatibility Study
6.3. Optimization of DOC–ERL NLCs
6.3.1. Plackett–Burman Design (PBD)
6.3.2. Box–Behnken Design (BBD)
6.4. Characterization of NLCs
6.5. Fourier-Transform Infrared Spectroscopy–Attenuated Total Reflectance (FTIR–ATR)
6.6. Freeze-Drying
6.7. In Vitro Drug Release
6.8. In Vitro Lipolysis
6.9. Stability Study
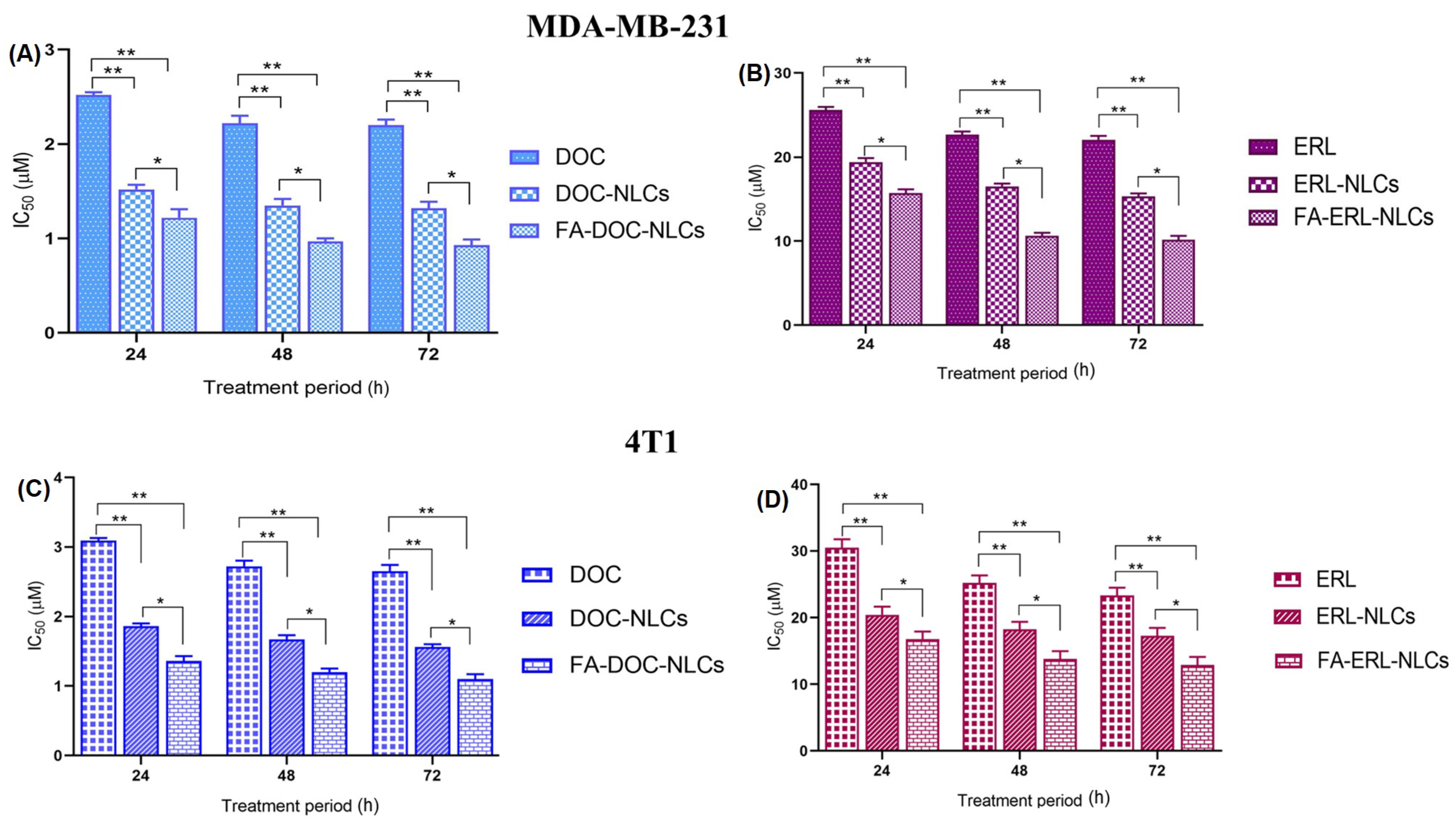
6.10. Cell Viability Assays
Determination of Combination Effect
6.11. Quantitative and Quantitative Cellular Uptake
6.12. Mitochondrial Membrane Potential (MMP) Assay
6.13. Colony Formation Assay
6.14. Transwell Migration Assay
6.15. Reactive Oxygen Species (ROS) Assay
7. Discussion
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.; Kumar, D.N.; Dehari, D.; Singh, S.; Kumar, P.; Bolla, P.K.; Kumar, D.; Agrawal, A.K. Emergence of Nanotechnology as a Powerful Cavalry against Triple-Negative Breast Cancer (TNBC). Pharmaceuticals 2022, 15, 542. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.V.; Ribeiro, L.N.M.; Moura, L.D.; Rodrigues da Silva, G.H.; Mitsutake, H.; Mendonca, T.C.; Geronimo, G.; Breitkreitz, M.C.; de Paula, E. Docetaxel Loaded in Copaiba Oil-Nanostructured Lipid Carriers as a Promising DDS for Breast Cancer Treatment. Molecules 2022, 27, 8838. [Google Scholar] [CrossRef] [PubMed]
- Newton, E.E.; Mueller, L.E.; Treadwell, S.M.; Morris, C.A.; Machado, H.L. Molecular Targets of Triple-Negative Breast Cancer: Where Do We Stand? Cancers 2022, 14, 482. [Google Scholar] [CrossRef] [PubMed]
- Corkery, B.; Crown, J.; Clynes, M.; O’Donovan, N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann. Oncol. 2009, 20, 862–867. [Google Scholar] [CrossRef] [PubMed]
- El Guerrab, A.; Bamdad, M.; Kwiatkowski, F.; Bignon, Y.-J.; Penault-Llorca, F.; Aubel, C. Anti-EGFR monoclonal antibodies and EGFR tyrosine kinase inhibitors as combination therapy for triple-negative breast cancer. Oncotarget 2016, 7, 73618–73637. [Google Scholar] [CrossRef] [PubMed]
- Lakkadwala, S.; Singh, J. Co-delivery of doxorubicin and erlotinib through liposomal nanoparticles for glioblastoma tumor regression using an in vitro brain tumor model. Colloids Surf. B Biointerfaces 2019, 173, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Xu, B.H.; Yuan, P.; Ma, F.; Wang, J.Y.; Ding, X.Y.; Zhang, P.; Li, Q.; Cai, R.G. Docetaxel-cisplatin might be superior to docetaxel-capecitabine in the first-line treatment of metastatic triple-negative breast cancer. Ann. Oncol. 2013, 24, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Ramos-Medina, R.; Bernat, R.; Garcia-Saenz, J.A.; Del Monte-Millan, M.; Alvarez, E.; Cebollero, M.; Moreno, F.; Gonzalez-Haba, E.; Bueno, O.; et al. Activity of docetaxel, carboplatin, and doxorubicin in patient-derived triple-negative breast cancer xenografts. Sci. Rep. 2021, 11, 7064. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhan, W.; Jia, M.; Zhao, Y.; Liu, Y.; Jha, R.K.; Zhou, L. Dual Loading of Nanoparticles with Doxorubicin and Icotinib for the Synergistic Suppression of Non-Small Cell Lung Cancer. Int. J. Med. Sci. 2020, 17, 390–402. [Google Scholar] [CrossRef]
- Granja, A.; Neves, A.R.; Sousa, C.T.; Pinheiro, M.; Reis, S. EGCG intestinal absorption and oral bioavailability enhancement using folic acid-functionalized nanostructured lipid carriers. Heliyon 2019, 5, e02020. [Google Scholar] [CrossRef] [PubMed]
- Paulmurugan, R.; Bhethanabotla, R.; Mishra, K.; Devulapally, R.; Foygel, K.; Sekar, T.V.; Ananta, J.S.; Massoud, T.F.; Joy, A. Folate Receptor-Targeted Polymeric Micellar Nanocarriers for Delivery of Orlistat as a Repurposed Drug against Triple-Negative Breast Cancer. Mol. Cancer Ther. 2016, 15, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Sakellari, G.I.; Zafeiri, I.; Batchelor, H.; Spyropoulos, F. Formulation design, production and characterisation of solid lipid nanoparticles (SLN) and nanostructured lipid carriers (NLC) for the encapsulation of a model hydrophobic active. Food Hydrocoll. Health 2021, 1, 100024. [Google Scholar] [CrossRef] [PubMed]
- Cirri, M.; Maestrini, L.; Maestrelli, F.; Mennini, N.; Mura, P.; Ghelardini, C.; Di Cesare Mannelli, L. Design, characterization and in vivo evaluation of nanostructured lipid carriers (NLC) as a new drug delivery system for hydrochlorothiazide oral administration in pediatric therapy. Drug Deliv. 2018, 25, 1910–1921. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, B.; Ali, J.; Baboota, S. Silymarin loaded nanostructured lipid carrier: From design and dermatokinetic study to mechanistic analysis of epidermal drug deposition enhancement. J. Mol. Liq. 2018, 255, 513–529. [Google Scholar] [CrossRef]
- Managuli, R.S.; Wang, J.T.; Faruqu, F.N.; Kushwah, V.; Raut, S.Y.; Shreya, A.B.; Al-Jamal, K.T.; Jain, S.; Mutalik, S. Asenapine maleate-loaded nanostructured lipid carriers: Optimization and in vitro, ex vivo and in vivo evaluations. Nanomedicine 2019, 14, 889–910. [Google Scholar] [CrossRef] [PubMed]
- Venishetty, V.K.; Komuravelli, R.; Kuncha, M.; Sistla, R.; Diwan, P.V. Increased brain uptake of docetaxel and ketoconazole loaded folate-grafted solid lipid nanoparticles. Nanomedicine 2013, 9, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.N.; Chaudhuri, A.; Dehari, D.; Shekher, A.; Gupta, S.C.; Majumdar, S.; Krishnamurthy, S.; Singh, S.; Kumar, D.; Agrawal, A.K. Combination Therapy Comprising Paclitaxel and 5-Fluorouracil by Using Folic Acid Functionalized Bovine Milk Exosomes Improves the Therapeutic Efficacy against Breast Cancer. Life 2022, 12, 1143. [Google Scholar] [CrossRef]
- Pereira, R.R.; Testi, M.; Rossi, F.; Silva Junior, J.O.C.; Ribeiro-Costa, R.M.; Bettini, R.; Santi, P.; Padula, C.; Sonvico, F. Ucuuba (Virola surinamensis) Fat-Based Nanostructured Lipid Carriers for Nail Drug Delivery of Ketoconazole: Development and Optimization Using Box-Behnken Design. Pharmaceutics 2019, 11, 284. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.K.; Urimi, D.; Harde, H.; Kushwah, V.; Jain, S. Folate appended chitosan nanoparticles augment the stability, bioavailability and efficacy of insulin in diabetic rats following oral administration. RSC Adv. 2015, 5, 105179–105193. [Google Scholar] [CrossRef]
- da Rocha, M.C.O.; da Silva, P.B.; Radicchi, M.A.; Andrade, B.Y.G.; de Oliveira, J.V.; Venus, T.; Merker, C.; Estrela-Lopis, I.; Longo, J.P.F.; Bao, S.N. Docetaxel-loaded solid lipid nanoparticles prevent tumor growth and lung metastasis of 4T1 murine mammary carcinoma cells. J. Nanobiotechnol. 2020, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Cunha, S.; Costa, C.P.; Loureiro, J.A.; Alves, J.; Peixoto, A.F.; Forbes, B.; Sousa Lobo, J.M.; Silva, A.C. Double Optimization of Rivastigmine-Loaded Nanostructured Lipid Carriers (NLC) for Nose-to-Brain Delivery Using the Quality by Design (QbD) Approach: Formulation Variables and Instrumental Parameters. Pharmaceutics 2020, 12, 599. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ahuja, A.; Ali, J.; Baboota, S. Curcumin-loaded lipid nanocarrier for improving bioavailability, stability and cytotoxicity against malignant glioma cells. Drug Deliv. 2016, 23, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Wijaya, W.; Zheng, H.; Zheng, T.; Su, S.; Patel, A.R.; Van der Meeren, P.; Huang, Q. Improved bioaccessibility of polymethoxyflavones loaded into high internal phase emulsions stabilized by biopolymeric complexes: A dynamic digestion study via TNO’s gastrointestinal model. Curr. Res. Food Sci. 2020, 2, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Shete, H.; Patravale, V. Long chain lipid based tamoxifen NLC. Part I: Preformulation studies, formulation development and physicochemical characterization. Int. J. Pharm. 2013, 454, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhao, L.; Su, X.; Zhang, P.; Ling, G. Repaglinide-loaded nanostructured lipid carriers with different particle sizes for improving oral absorption: Preparation, characterization, pharmacokinetics, and in situ intestinal perfusion. Drug Deliv. 2020, 27, 400–409. [Google Scholar] [CrossRef]
- Chaowanachan, T.; Krogstad, E.; Ball, C.; Woodrow, K.A. Drug synergy of tenofovir and nanoparticle-based antiretrovirals for HIV prophylaxis. PLoS ONE 2013, 8, e61416. [Google Scholar] [CrossRef]
- Novohradsky, V.; Zajac, J.; Vrana, O.; Kasparkova, J.; Brabec, V. Simultaneous delivery of olaparib and carboplatin in PEGylated liposomes imparts this drug combination hypersensitivity and selectivity for breast tumor cells. Oncotarget 2018, 9, 28456–28473. [Google Scholar] [CrossRef] [PubMed]
- Abdi Goushbolagh, N.; Farhood, B.; Astani, A.; Nikfarjam, A.; Kalantari, M.; Zare, M.H. Quantitative Cytotoxicity, Cellular Uptake and Radioprotection Effect of Cerium Oxide Nanoparticles in MRC-5 Normal Cells and MCF-7 Cancerous Cells. BioNanoScience 2018, 8, 769–777. [Google Scholar] [CrossRef]
- Kaithwas, V.; Dora, C.P.; Kushwah, V.; Jain, S. Nanostructured lipid carriers of olmesartan medoxomil with enhanced oral bioavailability. Colloids Surf. B Biointerfaces 2017, 154, 10–20. [Google Scholar] [CrossRef]
- Ducray, A.D.; Felser, A.; Zielinski, J.; Bittner, A.; Burgi, J.V.; Nuoffer, J.M.; Frenz, M.; Mevissen, M. Effects of silica nanoparticle exposure on mitochondrial function during neuronal differentiation. J. Nanobiotechnol. 2017, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Nordin, N.; Yeap, S.K.; Rahman, H.S.; Zamberi, N.R.; Abu, N.; Mohamad, N.E.; How, C.W.; Masarudin, M.J.; Abdullah, R.; Alitheen, N.B. In vitro cytotoxicity and anticancer effects of citral nanostructured lipid carrier on MDA MBA-231 human breast cancer cells. Sci. Rep. 2019, 9, 1614. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Pan, X.; Xie, F.; Lu, Y.; Zou, H.; Yin, C.; Zhang, Y.; Gao, J. Codelivery of doxorubicin and elacridar to target both liver cancer cells and stem cells by polylactide-co-glycolide/d-alpha-tocopherol polyethylene glycol 1000 succinate nanoparticles. Int. J. Nanomed. 2018, 13, 6855–6870. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Wu, Z.; Li, Z.; Liu, L. Celastrol Inhibits Migration and Invasion of Triple-Negative Breast Cancer Cells by Suppressing Interleukin-6 via Downregulating Nuclear Factor-kappaB (NF-kappaB). Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e922814. [Google Scholar] [CrossRef]
- Kumar, D.N.; Chaudhuri, A.; Kumar, D.; Singh, S.; Agrawal, A.K. Impact of the Drug Loading Method on the Drug Distribution and Biological Efficacy of Exosomes. AAPS PharmSciTech 2023, 24, 166. [Google Scholar] [CrossRef] [PubMed]
- Shilpi, D.; Kushwah, V.; Agrawal, A.K.; Jain, S. Improved Stability and Enhanced Oral Bioavailability of Atorvastatin Loaded Stearic Acid Modified Gelatin Nanoparticles. Pharm. Res. 2017, 34, 1505–1516. [Google Scholar] [CrossRef]
- Makoni, P.A.; Kasongo, K.W.; Walker, R.B. Preformulation studies of efavirenz with lipid excipients using thermal and spectroscopic techniques. Pharmazie 2020, 75, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Samy, A.M.; Afouna, M.I.; Dawaba, H.M.; Anwar, W. Screening Study for Formulation Variables in Preparation and Characterization of Candesartan Cilexetil Loaded Nanostructured Lipid Carriers. Univers. J. Pharm. Res. 2020, 4. [Google Scholar] [CrossRef]
- Chauhan, I.; Yasir, M.; Verma, M.; Singh, A.P. Nanostructured Lipid Carriers: A Groundbreaking Approach for Transdermal Drug Delivery. Adv. Pharm. Bull. 2020, 10, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Sharma, J.M.; Agrawal, A.K.; Mahajan, R.R. Surface stabilized efavirenz nanoparticles for oral bioavailability enhancement. J. Biomed. Nanotechnol. 2013, 9, 1862–1874. [Google Scholar] [CrossRef] [PubMed]
- Alam, T.; Khan, S.; Gaba, B.; Haider, M.F.; Baboota, S.; Ali, J. Adaptation of Quality by Design-Based Development of Isradipine Nanostructured-Lipid Carrier and Its Evaluation for In Vitro Gut Permeation and In Vivo Solubilization Fate. J. Pharm. Sci. 2018, 107, 2914–2926. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Roy, S.; Nath Bhaumik, K.; Kshetrapal, P.; Pillai, J. Comparative study of oral lipid nanoparticle formulations (LNFs) for chemical stabilization of antitubercular drugs: Physicochemical and cellular evaluation. Artif. Cells Nanomed. Biotechnol. 2018, 46, 540–558. [Google Scholar] [CrossRef] [PubMed]
- Antony, J. Screening Designs. In Design of Experiments for Engineers and Scientists; Elsevier: Amsterdam, The Netherlands, 2014; pp. 51–62. [Google Scholar]
- Sabzichi, M.; Mohammadian, J.; Mohammadi, M.; Jahanfar, F.; Movassagh Pour, A.A.; Hamishehkar, H.; Ostad-Rahimi, A. Vitamin D-Loaded Nanostructured Lipid Carrier (NLC): A New Strategy for Enhancing Efficacy of Doxorubicin in Breast Cancer Treatment. Nutr. Cancer 2017, 69, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Oshiro-Junior, J.A.; Sato, M.R.; Boni, F.I.; Santos, K.L.M.; de Oliveira, K.T.; de Freitas, L.M.; Fontana, C.R.; Nicholas, D.; McHale, A.; Callan, J.F.; et al. Phthalocyanine-loaded nanostructured lipid carriers functionalized with folic acid for photodynamic therapy. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 108, 110462. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.K.; Harde, H.; Thanki, K.; Jain, S. Improved stability and antidiabetic potential of insulin containing folic acid functionalized polymer stabilized multilayered liposomes following oral administration. Biomacromolecules 2014, 15, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kushwah, V.; Agrawal, A.K.; Jain, S. Insulin- and quercetin-loaded liquid crystalline nanoparticles: Implications on oral bioavailability, antidiabetic and antioxidant efficacy. Nanomedicine 2018, 13, 521–537. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Rehman, S.; Nabi, B.; Iqubal, A.; Nehal, N.; Fahmy, U.A.; Kotta, S.; Baboota, S.; Md, S.; Ali, J. Boosting the Brain Delivery of Atazanavir through Nanostructured Lipid Carrier-Based Approach for Mitigating NeuroAIDS. Pharmaceutics 2020, 12, 1059. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.V.; Adega, F.; Martins-Lopes, P.; Machado, R.; Lopes, C.M.; Lucio, M. pH-Responsive Hybrid Nanoassemblies for Cancer Treatment: Formulation Development, Optimization, and In Vitro Therapeutic Performance. Pharmaceutics 2023, 15, 326. [Google Scholar] [CrossRef] [PubMed]
- Houacine, C. Resveratrol Nanostructured Lipid Carrier for Targeted Delivery to Breast Cancer. Ph.D. Thesis, University of Central Lancashire, Preston, UK, 2018. [Google Scholar]
- De, A.; Roychowdhury, P.; Bhuyan, N.R.; Ko, Y.T.; Singh, S.K.; Dua, K.; Kuppusamy, G. Folic Acid Functionalized Diallyl Trisulfide-Solid Lipid Nanoparticles for Targeting Triple Negative Breast Cancer. Molecules 2023, 28, 1393. [Google Scholar] [CrossRef]
- Elmowafy, M.; Ibrahim, H.M.; Ahmed, M.A.; Shalaby, K.; Salama, A.; Hefesha, H. Atorvastatin-loaded nanostructured lipid carriers (NLCs): Strategy to overcome oral delivery drawbacks. Drug Deliv. 2017, 24, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Sahrayi, H.; Hosseini, E.; Karimifard, S.; Khayam, N.; Meybodi, S.M.; Amiri, S.; Bourbour, M.; Farasati Far, B.; Akbarzadeh, I.; Bhia, M.; et al. Co-Delivery of Letrozole and Cyclophosphamide via Folic Acid-Decorated Nanoniosomes for Breast Cancer Therapy: Synergic Effect, Augmentation of Cytotoxicity, and Apoptosis Gene Expression. Pharmaceuticals 2021, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Aditya, N.P.; Shim, M.; Lee, I.; Lee, Y.; Im, M.H.; Ko, S. Curcumin and genistein coloaded nanostructured lipid carriers: In vitro digestion and antiprostate cancer activity. J. Agric. Food Chem. 2013, 61, 1878–1883. [Google Scholar] [CrossRef] [PubMed]
- Thiruchenthooran, V.; Switalska, M.; Bonilla, L.; Espina, M.; Garcia, M.L.; Wietrzyk, J.; Sanchez-Lopez, E.; Gliszczynska, A. Novel Strategies against Cancer: Dexibuprofen-Loaded Nanostructured Lipid Carriers. Int. J. Mol. Sci. 2022, 23, 11310. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Kim, B.D.; Lee, T.H.; Kim, H.K.; Lyu, M.J.; Yoon, Y.I.; Goo, Y.T.; Kang, M.J.; Lee, S.; Choi, Y.W. Synergistic co-administration of docetaxel and curcumin to chemoresistant cancer cells using PEGylated and RIPL peptide-conjugated nanostructured lipid carriers. Cancer Nanotechnol. 2022, 13, 17. [Google Scholar] [CrossRef]
- Varshosaz, J.; Davoudi, M.A.; Rasoul-Amini, S. Docetaxel-loaded nanostructured lipid carriers functionalized with trastuzumab (Herceptin) for HER2-positive breast cancer cells. J. Liposome Res. 2017, 28, 285–295. [Google Scholar] [CrossRef]
- Pal, K.; Roy, S.; Parida, P.K.; Dutta, A.; Bardhan, S.; Das, S.; Jana, K.; Karmakar, P. Folic acid conjugated curcumin loaded biopolymeric gum acacia microsphere for triple negative breast cancer therapy in invitro and invivo model. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 95, 204–216. [Google Scholar] [CrossRef]
- Parvathaneni, V.; Shukla, S.K.; Gupta, V. Development and Characterization of Folic Acid-Conjugated Amodiaquine-Loaded Nanoparticles-Efficacy in Cancer Treatment. Pharmaceutics 2023, 15, 1001. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, X.; Yang, Z.; Tan, X.; Wang, J.; Chen, Y. Preparation and characterization of folic acid functionalized bioactive glass for targeted delivery and sustained release of methotrexate. J. Biomed. Mater. Res. A 2019, 107, 319–329. [Google Scholar] [CrossRef]
- Mishra, S.; Manna, K.; Kayal, U.; Saha, M.; Chatterjee, S.; Chandra, D.; Hara, M.; Datta, S.; Bhaumik, A.; Das Saha, K. Folic acid-conjugated magnetic mesoporous silica nanoparticles loaded with quercetin: A theranostic approach for cancer management. RSC Adv. 2020, 10, 23148–23164. [Google Scholar] [CrossRef]










| Plackett–Burman Design (PBD) | ||||
|---|---|---|---|---|
| Factors | Levels | |||
| Level 1 | Level 2 | |||
| BM (%w/v) | 1 | 2 | ||
| Surfactant (%w/v) | 1 | 2 | ||
| Mixing speed (rpm) | 250 | 750 | ||
| Homogenization time (min) | 1 | 5 | ||
| Homogenization speed (rpm) | 1000 | 5000 | ||
| Sonication time (min) | 2 | 5 | ||
| Sonication amplitude (%) | 40 | 80 | ||
| Responses | Constraints | |||
| Size (nm) | <200 | |||
| PDI | <0.35 | |||
| Box–Behnken Design (BBD) | ||||
| Factors | Levels | |||
| Low (−1) | Intermediate (0) | High (+1) | ||
| BM (%w/v) | 1 | 1.5 | 2 | |
| Surfactant (%w/v) | 1 | 1.5 | 2 | |
| Sonication time (min) | 2 | 3.5 | 5 | |
| Responses | Constraints | |||
| Size (nm) | <200 | |||
| PDI | <0.35 | |||
| Entrapment efficiency (%EE) | Maximum | |||
| Drug loading (%DL) | Maximum | |||
| S. No. | Conc. BM (%w/v) | Conc. Surf (%w/v) | Sonication Time (min) | Particle Size (nm) | PDI | EE (%) | DL (%) |
|---|---|---|---|---|---|---|---|
| 1 | 2 | 1.5 | 2 | 189.42 ± 2.42 | 0.278 ± 0.003 | 84.12 ± 1.24 | 3.62 ± 0.02 |
| 2 | 2 | 2 | 3.5 | 158.11 ± 1.32 | 0.167 ± 0.001 | 87.76 ± 2.11 | 3.39 ± 0.04 |
| 3 | 1.5 | 1.5 | 3.5 | 156.46 ± 1.36 | 0.211 ± 0.002 | 81.15 ± 1.67 | 4.14 ± 0.02 |
| 4 | 2 | 1 | 3.5 | 245.62 ± 2.77 | 0.344 ± 0.003 | 85.54 ± 1.48 | 4.08 ± 0.03 |
| 5 | 1.5 | 1 | 2 | 300.34 ± 3.17 | 0.329 ± 0.002 | 76.75 ± 1.35 | 4.54 ± 0.07 |
| 6 | 2 | 1.5 | 5 | 141.67 ± 4.16 | 0.156 ± 0.006 | 84.42 ± 1.27 | 3.62 ± 0.05 |
| 7 | 1 | 1.5 | 5 | 132.81 ± 2.49 | 0.125 ± 0.005 | 78.63 ± 2.03 | 3.89 ± 0.08 |
| 8 | 1.5 | 1.5 | 3.5 | 178.42 ± 2.16 | 0.232 ± 00.004 | 80.34 ± 2.16 | 4.12 ± 0.04 |
| 9 | 1.5 | 2 | 2 | 200.36 ± 2.37 | 0.254 ± 0.001 | 84.84 ± 1.86 | 3.64 ± 0.01 |
| 10 | 1 | 1.5 | 2 | 180.21 ± 1.65 | 0.134 ± 0.002 | 76.31 ± 1.53 | 3.34 ± 0.05 |
| 11 | 1 | 2 | 3.5 | 123.80 ± 2.10 | 0.175 ± 0.004 | 78.68 ± 1.41 | 4.09 ± 0.01 |
| 12 | 1 | 1 | 3.5 | 231.57 ± 3.08 | 0.245 ± 0.007 | 85.54 ± 1.37 | 3.12 ± 0.03 |
| 13 | 1.5 | 1.5 | 3.5 | 164.32 ± 2.64 | 0.209 ± 0.008 | 82.34 ± 1.26 | 3.41 ± 0.01 |
| 14 | 1.5 | 1 | 5 | 268.16 ± 2.81 | 0.314 ± 0.003 | 75.53 ± 2.33 | 3.54 ± 0.02 |
| 15 | 1.5 | 2 | 5 | 146.23 ± 2.58 | 0.166 ± 0.002 | 84.03 ± 1.42 | 3.67 ± 0.06 |
| S. No. | Conc. BM (%w/v) | Conc. Surf (%w/v) | Sonication Time (min) | Particle Size (nm) | PDI | EE (%) | DL (%) |
|---|---|---|---|---|---|---|---|
| 1 | 2 | 1.5 | 5 | 210.23 ± 1.65 | 0.208 ± 0.002 | 94.84 ± 1.33 | 3.64 ± 0.02 |
| 2 | 2 | 1 | 3.5 | 289.42 ± 2.43 | 0.291 ± 0.004 | 95.39 ± 1.14 | 4.08 ± 0.05 |
| 3 | 1.5 | 1.5 | 3.5 | 132.11 ± 2.74 | 0.113 ± 0.002 | 90.94 ± 1.25 | 3.74 ± 0.04 |
| 4 | 1.5 | 1.5 | 3.5 | 135.27 ± 2.12 | 0.122 ± 0.005 | 89.20 ± 1.38 | 4.03 ± 0.03 |
| 5 | 2 | 2 | 3.5 | 237.82 ± 1.44 | 0.225 ± 0.003 | 95.76 ± 1.05 | 3.39 ± 0.01 |
| 6 | 1 | 1.5 | 2 | 130.35 ± 1.31 | 0.128 ± 0.006 | 86.30 ± 2.41 | 3.34 ± 0.04 |
| 7 | 2 | 1.5 | 2 | 311.19 ± 1.57 | 0.200 ± 0.001 | 94.10 ± 2.15 | 3.62 ± 0.02 |
| 8 | 1 | 1 | 3.5 | 150.74 ± 2.17 | 0.146 ± 0.002 | 84.13 ± 1.22 | 4.04 ± 0.06 |
| 9 | 1.5 | 2 | 2 | 120.57 ± 2.76 | 0.213 ± 0.003 | 95.53 ± 1.52 | 4.13 ± 0.03 |
| 10 | 1.5 | 1 | 5 | 145.64 ± 2.83 | 0.178 ± 0.005 | 85.76 ± 1.73 | 3.64 ± 0.07 |
| 11 | 1.5 | 2 | 5 | 168.37 ± 1.58 | 0.196 ± 0.002 | 94.03 ± 1.74 | 3.47 ± 0.03 |
| 12 | 1 | 1.5 | 5 | 138.60 ± 1.22 | 0.109 ± 0.004 | 88.68 ± 1.29 | 3.43 ± 0.05 |
| 13 | 1.5 | 1 | 2 | 221.24 ± 1.40 | 0.225 ± 0.003 | 86.75 ± 1.14 | 4.35 ± 0.06 |
| 14 | 1.5 | 1.5 | 3.5 | 135.29 ± 2.47 | 0.132 ± 0.001 | 89.20 ± 1.20 | 4.01 ± 0.02 |
| 15 | 1 | 2 | 3.5 | 128.51 ± 2.50 | 0.233 ± 0.002 | 94.51 ± 2.11 | 4.09 ± 0.01 |
| Formulation | BM. Conc. (%w/v) | Surf. Conc. (%w/v) | Sonication Time (min) | Actual vs. Predicted | PS (nm) | PDI | %EE. | %DL |
|---|---|---|---|---|---|---|---|---|
| DOC NLCs | 1 | 2 | 3.5 | Predicted | 126.48 | 0.185 | 85.58 | 3.97 |
| Actual | 123.80 | 0.175 | 78.68 | 4.09 | ||||
| Prediction error | 2.12 | 5.41 | 8.06 | 2.93 | ||||
| ERL NLCs | 1.5 | 2 | 2 | Predicted | 123.89 | 0.201 | 93.92 | 4.33 |
| Actual | 120.57 | 0.213 | 95.53 | 4.13 | ||||
| Prediction error | 2.67 | 5.63 | 1.68 | 4.68 |
| Parameters | PS (nm) | PDI | ZP (mV) | EE (%) | DL (%) | Height (nm) in Z Axis |
|---|---|---|---|---|---|---|
| DOC NLCs | 123.80 ± 2.10 | 0.175 ± 0.004 | −15.25 ± 0.69 | 78.68 ± 1.41 | 4.09 ± 0.01 | 56.29 ± 1.32 |
| FA–DOC NLCs | 153.2 ± 2.52 | 0.241 ± 0.005 | −14.15 ± 0.45 | 80.56 ± 1.85 | 4.10 ± 0.05 | 38.76 ± 1.27 |
| ERL NLCs | 120.57 ± 2.76 | 0.213 ± 0.003 | −16.44 ± 0.63 | 95.53 ± 1.52 | 4.13 ± 0.03 | 87.18 ± 1.42 |
| FA–ERL NLCs | 147.40 ± 3.32 | 0.258 ± 0.001 | −15.86 ± 0.55 | 96.34 ± 1.55 | 4.20 ± 0.02 | 55.35 ± 1.38 |
| Combinations | IC50 Values (μM) at 24 h | |
|---|---|---|
| MDA-MB-231 | 4T1 | |
| DOC + ERL | 7.51 ± 0.87 | 8.42 ± 0.65 |
| DOC NLCs + ERL NLCs | 1.54 ± 0.27 | 1.79 ± 0.38 |
| FA–DOC NLCs + FA–ERL NLCs | 0.71 ± 0.06 | 0.81 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaudhuri, A.; Kumar, D.N.; Srivastava, S.K.; Kumar, D.; Patil, U.K.; Parmar, A.S.; Singh, S.; Agrawal, A.K. Combinatorial Delivery of Docetaxel- and Erlotinib-Loaded Functionalized Nanostructured Lipid Carriers for the Treatment of Triple-Negative Breast Cancer Using Quality-by-Design Approach. Pharmaceutics 2024, 16, 926. https://doi.org/10.3390/pharmaceutics16070926
Chaudhuri A, Kumar DN, Srivastava SK, Kumar D, Patil UK, Parmar AS, Singh S, Agrawal AK. Combinatorial Delivery of Docetaxel- and Erlotinib-Loaded Functionalized Nanostructured Lipid Carriers for the Treatment of Triple-Negative Breast Cancer Using Quality-by-Design Approach. Pharmaceutics. 2024; 16(7):926. https://doi.org/10.3390/pharmaceutics16070926
Chicago/Turabian StyleChaudhuri, Aiswarya, Dulla Naveen Kumar, Saurabh Kumar Srivastava, Dinesh Kumar, Umesh Kumar Patil, Avanish Singh Parmar, Sanjay Singh, and Ashish Kumar Agrawal. 2024. "Combinatorial Delivery of Docetaxel- and Erlotinib-Loaded Functionalized Nanostructured Lipid Carriers for the Treatment of Triple-Negative Breast Cancer Using Quality-by-Design Approach" Pharmaceutics 16, no. 7: 926. https://doi.org/10.3390/pharmaceutics16070926









