Extracellular Vesicles as Drug Delivery System for Cancer Therapy
Abstract
:1. Introduction
2. Discovery and Development of Extracellular Vesicles
2.1. Discovery of Extracellular Vesicles
2.2. Biogenesis and Classification of Extracellular Vesicles
2.3. Absorption/Uptake of Extracellular Vesicles
3. Strategies for Utilizing Extracellular Vesicles as a Drug Delivery System
3.1. Sources of Extracellular Vesicles for Drug Delivery
3.2. Characterization and Purification of Extracellular Vesicles
3.2.1. Characterization of Extracellular Vesicles
3.2.2. Purification of Extracellular Vesicles
3.3. General Methods for Drug Loading into Extracellular Vesicles
3.3.1. Pre-Loading Methods
3.3.2. Post-Loading Methods
| Cargo Loading Method | Therapeutic Cargo | Sources of EVs | Cancer Type | Function | Study Type | Year | Ref. |
|---|---|---|---|---|---|---|---|
| Pre-loding, coincubation | Phthalocyanine chloride tetrasulfonic acid (AlPcS4) | Gastric cancer MGC803 cells | Gastric cancer | Deconstruct exosome for releasing Dox and enable the photodynamics for combination therapy | In vitro and in vivo | 2021 | [148] |
| Pre-loding, coincubation | AIE-photosensitizer MBPN-TCyP | Dendritic cells | Breast cancer and colorectal cancer | Synergistic photodynamic immunotherapy elicits dramatic anti-tumor immune responses | In vitro and in vivo | 2022 | [149] |
| Pre-loding, coincuba-tion | MTX | Mouse hepatocarcinoma tumour cells H22 | Hepatocarcinoma | Inhibit ascites hepatocarcinoma growth without typical side effects | In vitro and in vivo | 2012 | [80] |
| Pre-loding, coincubation | Cisplatin/PTX | Human ovarian cancer tumour cells A2780 | Ovarian cancer | Inhibit human ovarian cancer growth without affecting liver and kidney functions of SCID mice | In vitro and in vivo | 2012 | [80] |
| Pre-loding, coincubation | MTX | Mouse fibroblast cells L929 | Glioblastoma | Facilitate extravasation across BBB and inhibit human brain tumor growth | In vitro and in vivo | 2018 | [150] |
| Pre-loding, coincubation | MTX | Primary malignant cells that are frequently accompanied by malignant pleural effusion (MPE) in their advanced stages | Lung cancer and colon cancer with MPE | Exhibit clinical activity in killing tumor cells and TAMs and induce antitumor immune responses | In vitro and in vivo | 2019 | [152] |
| Pre-loding, coincubation | ICG and PTX | HEK293T | Breast cancer | Increase the anticancer activity through combination of chemo/photothermal/photodynamic therapy | In vitro and in vivo | 2022 | [151] |
| Pre-loding, coincubation | PTX | BM-MSCs (SR4987) | Pancreatic adenocarcinoma | Exhibit strong antiproliferative activity on human pancreatic adenocarcinoma cells CFPAC-1 | In vitro | 2014 | [78] |
| Pre-loding, transfection | HGF siRNA | HEK293T | Gastric cancer | Suppress proliferation and migration of both cancer cells and vascular cells | In vitro and in vivo | 2018 | [154] |
| Pre-loding, electroporat | PTX/miR-16/Penicillin/MCP-1/Cas9-GFP | Differentiated human promyelocytic leukemia cells (dHL-60) and naïve HL-60 | Breast cancer cells (MCF-7)/Colon cancer cells (COLO205)/Jurkat cells | Dhl60 exhibit increased drug loading and production efficiency | In vitro | 2012 | [209] |
| Pre-loding, transduction and coincubation | TRAIL and Cabazitaxel (CTX) | MSCs | Oral squamous cell carcinoma | Synergistically inhibit the growth of cancer cells by inhibiting the activation of PI3K/Akt/mTOR pathway and inducing apoptosis | In vitro and in vivo | 2020 | [210] |
| Pre-loding, transduction | miR-379 | MSCs | Breast cancer | Inhibit the growth of breast cancer by downregulating cyclooxygenase (COX-2) | In vitro and in vivo | 2017 | [211] |
| Post-loding, coincubation | DOX | Brain endothelial cells | Brain cancer | Mediate drug delivery across the BBB and exert cytotoxic efficacy against brain cancer in zebrafish | In vitro and in vivo | 2015 | [158] |
| Post-loding, coincubation | Curcumin | Bovine milk | Multiple cancers (breast, lung and cervical cancer) | Enhance antiproliferative activity against multiple cancer cell lines (breast, lung, and cervical cancer) and e cervical tumor xenograft | In vitro and in vivo | 2017 | [159] |
| Post-loding, coincubation | Withaferin A (WFA)/Bilberry-derived anthocyanidins (Anthos)/Curcumin (Cur)/paclitaxel (PTX) and docetaxel (DOC) | Bovine milk | Lung cancer and breast cancer cells | Enhance anti-cancer and anti-inflammatory effects | In vitro | 2016 | [126] |
| Post-loding, coincubation for Ce6/electroporation for siRNA | Ce6/PD-L1 siRNA | NK cells | Hepatocellular carcinoma and Colon cancer | Effectively inhibit cancer progression by effective PDT or restoring the immunological surveillance function | In vitro and in vivo | 2022 | [160] |
| Post-loding, coincubation | Zinc Phthalocyanine | Metastatic murine melanoma cells (B16F10) | Colon cancer | Increase efficacy and selectivity of PDT | In vitro and in vivo | 2021 | [161] |
| Post-loading, coincubation | Zinc Phthalocyanine | M1/M2-like macrophages/B16F10/Milk | Colon cancer | Increase photodynamic therapy and promote immunological memory | In vitro and in vivo | 2022 | [162] |
| Post-loading, coincubation | DOX/Cholesterol-modified miRNA 159 | Human monocytes (THP-1) | Triple negative breast cancer (TNBC) | Co-delivering miR159 and Dox by targeted Exo for TNBC therapy | In vitro and in vivo | 2019 | [163] |
| Post-loading, coincubation | Cholesterol-modified miRNA 34a | HEK293T | Oral squamous cell carcinoma | Inhibition of oral squamous carcinoma HN6 cell proliferation, migration, and invasion by down regulating SATB2 expression | In vitro | 2022 | [172] |
| Post-loading, coincubation | DOX | RAW 264.7 cells pre-treated with hyaluronic acid (HA) and the β-blocker carvedilol (CV) | Breast cancer | Enhance the antitumor effects of DOX | In vitro and in vivo | 2022 | [164] |
| Post-loading, coincubation | DOX/Chemosensitizer lonidamine (LND) | Non-small cell lung carcinoma A549 cells | Non-small cell lung carcinoma | Synergistically increase anticancer activity | In vitro and in vivo | 2022 | [165] |
| Post-loading, calcium chloride transfection combined with heat shock/electroporation | miR-15a mimic/inhibitor | THP-1 cells | NA | Effectively enhance miRNA loading efficiency to EVs | In vitro | 2017 | [169] |
| Post-loading, transfection | miR-335 | Human hepatic stellate cell LX2 | Hepatocellular carcinoma | Inhibit hepatocellular carcinoma growth | In vitro and in vivo | 2018 | [167] |
| Post-loading, transfection | VEGF siRNA | Brain endothelial bEND.3 cells | Brain cancer | Mediate siRNA Delivery across the BBB to inhibit brain tumor growth | In vitro and in vivo | 2017 | [168] |
| Post-loading, saponin | DOX | Human GBM cell line U87 and U251 cells | Glioblastoma | Eliminate the original cargos of glioblastoma cell-derived small EVs for efficient drug delivery | In vitro and in vivo | 2022 | [212] |
| Post-loading, saponin/electroporation/extrusion/dialysis | Porphyrins | HMSCs/HUVECs/MDA-MB-231 cells | Breast cancer MDA-MB-231 cells | Induce a stronger phototoxic effect than free drug in a cancer cell model | In vitro | 2015 | [175] |
| Post-loading, saponin/sonication/extrusion/freeze-thaw cycles | Catalase | Raw 264.7 | Neuronal cells PC12 | Exhibit high loading efficiency, sustained release, and catalase preservation against proteases degradation and provide significant neuroprotective effects | In vitro and in vivo | 2015 | [174] |
| Post-loding, electroporation | DOX | HEK293F/B16F10 | Metastatic murine melanoma B16F10 cells | Optimized electroporation improves the loading of EVs with DOX | In vitro | 2022 | [179] |
| Post-loding, electroporation | ASOs/Cas9 mRNA and gRNA | Red blood cells (RBCs) | Leukemia/breast cancer | Exhibit highly robust microRNA inhibition and CRISPR–Cas9 genome editing | In vitro and in vivo | 2018 | [122] |
| Post-loding, electroporation for siRNA; Pre-loading, co-incubation for DOX | KRASG12D siRNA/DOX | Human umbilical cord mesenchymal stromal cells (UC-MSCs) | Pancreatic ductal adenocarcinoma (PDAC) | Co-delivery KRASG12D siRNA and DOX to PDAC cells to inhibit the cancer progression | In vitro | 2023 | [181] |
| Post-loding, electroporation | ITGB6 siRNAs | Prostate cancer cells (DU145 and PC3) | Prostate cancer | Delivery of siRNAs targeting the ITGB6 to inhibit adhesion and migration of recipient prostate cancer cells | In vitro | 2022 | [180] |
| Post-loding, sonication | HER2 siRNA | HEK293T/MCF-7 | Breast cancer | Knockdown of HER2, a therapeutic target that is overexpressed in numerous cancers | In vitro | 2016 | [185] |
| Post-loding, soni-cation | DOX | RAW 264.7 | TNBC | Significantly inhibit TNBC tumor growth | In vitro and in vivo | 2020 | [97] |
| Post-loding, sonication | PTX | RAW 264.7 macrophages | Lung cancer | Inhibit growth of pulmonary metastases and overcome MDR in Cancer cell | In vitro and in vivo | 2016 | [186] |
| Post-loding, sonication | Erastin/Rose Bengal | HEK293T | Hepatocellular carcinoma | Induce obvious ferroptosis in HCC with minimized toxicity in liver and kidney | In vitro and in vivo | 2021 | [187] |
| Post-loding, extrusion | PTX | Mesenchymal stem cells (MSCs) | Breast cancer | Exhibit therapeutically efficient for the treatment of breast cancer | In vitro and in vivo | 2018 | [193] |
| Post-loding, extrusion for DOX/electroporation for P-gp siRNA | DOX/P-gp siRNA | Normal ovarian epithelial Iose80 cells | Ovary cancer | Target delivery of chemotherapeutics to overcome drug resistance of ovarian cancer | In vitro and in vivo | 2023 | [194] |
| Post-loding, extrusion | DOX | HT1080/Hela | Fibrosarcoma | Tumor cell-derived exosomes preferentially targeted their cell of origin | In vitro and in vivo | 2020 | [195] |
| Post-loding, freeze-thaw cycles | Liposome | Mouse fibroblast sarcoma-derived CMS7-wt/CMS7-HE (HER2 overexpression)/Raw 264.7 | HeLa cells | Develop hybrid exosomes by fusing the membranes of exosomes with liposomes for loading therapeutic agents into exosomes | In vitro | 2016 | [199] |
| Post-loding, freeze-thaw cycles/extrusion,/sonication | DOX | Platelets | Breast cancer | Efficiently load DOX and kill breast cancer cells | In vitro | 2023 | [201] |
| Post-loding, freeze-thaw cycles | Folate-modified Liposomes with or without PTX | Mesenchymal stem cells (MSCs) | Colon carcinoma cell line CT26/Mouse melanoma cell line B16/Human ovarian cancer cell line A2780 | Increase therapeutic potential of PTX for cancer therapy | In vitro and in vivo | 2024 | [202] |
| Fused expression with tetraspanin CD63 | OVA | 293F cells | Immune cells | Significantly inhibit tumor growth by induce DNA vaccine-specific CD8+ T cell responses | In vitro and in vivo | 2017 | [206] |
| Fused expression with CD9 (CIBN and CRY interaction system) | Proteins: mCherry/luciferase/Bax/super repressor IκB (srIκB)/Cre recombinase | HEK293T | HeLa cells/Rat embryonic primary neurons/Neuronal cells | Significantly increase intracellular levels of cargo proteins and their function in recipient cells | In vitro and in vivo | 2016 | [207] |
| Fused expression with CD63 (FRB/FKBP heterodimerization system) | Proteins: Diphtheria toxin A (DTA) | DTA-resistant HT1080 cells | HT1080 cells | Efficient system enables to load any protein-based therapeutics into EVs | In vitro | 2023 | [208] |
4. Extracellular Vesicles Modification for Targeted Anti-Cancer Drug Delivery
4.1. Genetic Target Engineering
4.1.1. Genetic Target Engineering by Fusion Expression with LAMP-2B
4.1.2. Genetic Target Engineering by Fusion Expression with PDGFR TM Domain
4.1.3. Genetic Target Engineering by Fusion Expression with Lactadherin C1–C2 Domain
4.1.4. Genetic Target Engineering by Fusion Expression with the Tetraspanin Superfamily Proteins
4.1.5. Genetic Target Engineering by Fusion Expression with the CD47
4.1.6. Genetic Target Engineering by Fusion Expression with Glycosylphosphatidylinositol (GPI)-Anchor Signal Peptides
| Targeting Ligand | Transmembrane Protein on EVs | Therapeutic Cargo | Cargo Loading Method | Cell Sources of EVs | Cancer Type and Targets | Function | Study Type | Year | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| iRGD | LAMP-2B | DOX | Electroporation | immature dendritic cells | Breast cancer cells | Inhibit tumor growth without overt toxicity | In vitro and in vivo | 2014 | [99] |
| iRGD | LAMP-2B | DOX; GAPDH siRNA | Electroporation; Transfection | HEK293FT | Glioblastoma cells | Increase the drug internalization via across BBB | In vitro | 2022 | [218] |
| iRGD | LAMP-2B | KRAS siRNA | Transfection | HEK293T | Lung cancer cells | Target oncogenic KRAS | In vitro and in vivo | 2019 | [219] |
| iRGD | LAMP-2B | CPT1A siRNA | Transfection | HEK293T | Colon cancer cells | Target silencing CPT1A to inhibit FAO; reverse oxaliplatin resistance and inhibit tumour growth | In vitro and in vivo | 2021 | [220] |
| iRGD | LAMP-2B | BCL6 siRNA | Electroporation | immature dendritic cells | Diffuse large B-cell lymphoma cells (DLBCL) | Target silencing BLC6 to inhibit DLBCL tumor growth | In vitro and in vivo | 2022 | [221] |
| iRGD | LAMP-2B | miR-484 | Electroporation | HEK293T | Ovarian cancer cells; tumor vascular endothelial cells | Inhibit angiogenesis and sensitize the cancer to chemotherapy | In vitro and in vivo | 2022 | [217] |
| tLyP-1 | LAMP-2B | SOX2 siRNA | Electroporation | HEK293T | Lung cancer and cancer stem cells | Target silencing SOX2 expression of NSCLC cells and reducing the stemness of NSCLC stem cells | In vitro | 2020 | [224] |
| fragment of Interleukin 3 | LAMP-2B | Imatinib; BCR-ABL siRNA | Direct incubation; Transfection | HEK293T | Chronic myeloid leukemia (CML) cells | Target delivery of Imatinib or BCR-ABL siRNA to CML cells | In vitro and in vivo | 2017 | [227] |
| HER2-binding affibody zHER | LAMP-2B | 5-FU and miRNA-21 inhibitor | Electroporation | HEK293T | Her2 expressing colorectal cancer cells | Effectively reverse drug resistance and significantly enhanced the cytotoxicity in 5-FU-resistant colon cancer cells | In vitro and in vivo | 2020 | [183] |
| HER2-specific DARPin | LAMP-2B | TPD52 siRNA | Electroporation | HEK293T | HER2-positive breast cancer cells | Target silencing the TPD52 of Her2 positive cancer cells | In vitro | 2019 | [155] |
| HER2-specific DARPin | LAMP-2B | DOX | Electroporation | BM-MSCs | HER2-positive breast cancer cells | Specifically inhibit Her2 positive tumor growth | In vitro and in vivo | 2019 | [232] |
| HER2-specific DARPin | LAMP-2B | 99mTc | Chemical modification | HEK293T | HER2-positive ovarian cancer cells | In vivo HER2-positive tumor imaging | In vitro and in vivo | 2020 | [233] |
| GE11/EGF | PDGFR-TM | let-7a miRNA | Transfection | HEK293T | EGFR-positive breast cancer cells | Target delivery miRNAs to EGFR expressing cancer cells | In vitro and in vivo | 2013 | [237] |
| αCD3/αEGFR | PDGFR-TM | αCD3/αEGFR | Transfection | Expi293F cells | T cell and EGFR-expressing breast cancer cells | Induce cross-linking of T cells and EGFR-expressing breast cancer cells and elicit potent antitumor immunity. | In vitro and in vivo | 2018 | [240] |
| anti-HER2 scFv antibody (ML39) | Lactadherin C1-C2 domain | CNOB and HCHrR6 mRNA | Electroporation | HEK293 | HER2-overexpressing breast cancer cells | Delivery of functional exogenous mRNA to tumors | In vitro and in vivo | 2018 | [243] |
| anti-HER2 scFvs with different affinity | Lactadherin C1-C2 domain | CFSE | Chemical modification | HEK293 | HER2-overexpressing cancer cells | Monitor the target delivery of antiHER2-scFvs modified exosomes | In vitro and in vivo | 2018 | [239] |
| Two copies of the HER2 ligand | Lactadherin C1-C2 domain | HER2 miRNA | Transfection | HEK293 | HER2-overexpressing cancer cells | Specifically inhibit Her2 expressing tumor growth | In vitro and in vivo | 2020 | [244] |
| Anti-EGFR nanobodies | GPI-anchor Signal peptides | CellTracker Deep Red | Chemical modification | Neuro2A | EGFR-overexpressing cancer cells | Specifically target EGFR expression cells | In vitro | 2016 | [256] |
| Apo-A1 | CD63 | miRNA-26a | Electroporation | HEK293T | Hepatocellular Carcinoma (HepG2 | Inhibit tumor cell migration and proliferation | In vitro | 2018 | [252] |
| CDX peptide/CREKA | CD47 | PTEN mRNA | Cellular nanoporation biochip (CNP) | bone marrow-derived dendritic cells (BMDCs) | PTEN-deficient human U87 and mouse GL216 glioblastoma cells | Specifically inhibit PTEN-deficient glioblastoma | In vitro and in vivo | 2019 | [253] |
4.2. Chemical Modification of Extracellular Vesicles
4.2.1. Click Chemistry Method for Direct Modification
4.2.2. Hydrophobic Insertion Mediated Modification
| Targeting Ligand | Ligand Labeling Method | Therapeutic Cargo | Cargo Loading Method | Cell Sources of EVs | Cancer Type and Targets | Function | Study Type | Year | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| RGE | Copper catalyzed click chemistry | Curcumin and SPION | Electroporation | Mouse macrophage cell line Raw264.7 | Glioma | Simultaneous target imaging and therapy of glioma | In vitro and in vivo | 2018 | [258] |
| c(RGDyk) | Copper free click chemistry | Curcumin | Incubation | BM-MSCs | Integrin αvβ3 overexpressing cells (U87 glioblastoma cells and vascular endothelial cells) | Increase the drug internalization via across BBB and target delivery drugs to integrin αvβ3 overexpressing cells | In vitro | 2017 | [259] |
| RGD | DSPE-PEG-RGD | V2C Quantum Dots | Electroporation | MCF-7 cells | Integrin αvβ3-poitive breast cancer MCF-7 cells | Target delivery photothermal agents to integrin expressing cells | In vitro and in vivo | 2019 | [261] |
| Folic acid (FA) | DSPE-PEG-FA | Human hyaluronidase (PH20); DOX | Transfection; Electroporation | HEK293T | Folate receptor overexpressing cancer cells | Reduce hyaluronidase-induced metastasis and enhance target delivery of chemotherapy | In vitro and in vivo | 2021 | [262] |
| Folic acid (FA) | DSPE-PEG-FA | Erastin | Sonication | Human fetal lung fibroblasts HFL-1 | Folate receptor overexpressing cancer cells | Induce ferroptosis of folate receptor overexpression TNBC cells | In vitro | 2019 | [207] |
| Folic acid (FA) | DSPE-PEG-FA | DOX/P-gp siRNA | Extrusion; Electroporation | Normal ovarian epithelial Iose80 cells | Folate receptor overexpressing ovary cancer cells | Target delivery of chemotherapeutics to overcome drug resistance of ovarian can-cer | In vitro and in vivo | 2023 | [194] |
| Aminoethylanisamide (AA) | DSPE-PEG-AA | PTX | Sonication | Mouse macrophage cell line Raw264.7 | Sigma receptor overexpressing lung cancer cells | Improve drug circulation and inhibit lung cancer metastases | In vitro and in vivo | 2017 | [263] |
| AS1411 aptamer | Cholesterol-polypeptides | Let-7 miRNA/VEGF siRNA | Electroporation | BMDCs | Nucleolin overexpressing breast cancer cells | Target delivery siRNAs/miRNAs to nucleolin positive cancer cells | In vitro and in vivo | 2017 | [265] |
| AS1411 aptamer | Cholesterol-PEG2000 | PTX | Sonication | BMDCs | Nucleolin overexpressing breast cancer cells | Target delivery paclitaxel to nucleolin positive cancer cells | In vitro and in vivo | 2018 | [115] |
| PSMA RNA aptamer; EGFR RNA aptamer; Folic acid | Cholesterol-RNA nanoparticles | Survivin siRNA | Transfection | HEK293T | prostate cancer; breast cancer and colorectal cancer cells | Mediate RNA nanoparticles on EV memebrane | In vitro and in vivo | 2017 | [266] |
| LDL peptide | ApoA-I mimetic peptide | methotrexate, KLA (Lys-Leu-Ala) | Co-incubation | Mouse fibroblast L929 cells | LDLR overexpressing glioblastoma cells | Target treatment of LDLR overexpressing glioblastoma cells | In vitro and in vivo | 2018 | [150] |
| Aptamer sgc8 | Diacylipid-(PEG)2 | DOX | Electroporation | Immature dendritic cells (imDC) | Leukemia cells that overexpressed PTK7 | Target delivery of therapeutics to PTK7 overexpressing cancer cells | In vitro | 2019 | [259] |
5. Conclusion and Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Gao, J.; Karp, J.M.; Langer, R.; Joshi, N. The Future of Drug Delivery. Chem. Mater. 2023, 35, 359–363. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; Rodriguez-Torres, M.D.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Chen, Z.G.; Shin, D.M. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008, 7, 771–782. [Google Scholar] [CrossRef]
- Thi, T.T.H.; Suys, E.J.A.; Lee, J.S.; Nguyen, D.H.; Park, K.D.; Truong, N.P. Lipid-Based Nanoparticles in the Clinic and Clinical Trials: From Cancer Nanomedicine to COVID-19 Vaccines. Vaccines 2021, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H. Tumor-selective delivery of macromolecular drugs via the EPR effect: Background and future prospects. Bioconjugate Chem. 2010, 21, 797–802. [Google Scholar] [CrossRef]
- Bi, Y.; Hao, F.; Yan, G.; Teng, L.; Lee, R.J.; Xie, J. Actively Targeted Nanoparticles for Drug Delivery to Tumor. Curr. Drug Metab. 2016, 17, 763–782. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Trieu, V.; Yao, Z.; Louie, L.; Ci, S.; Yang, A.; Tao, C.; De, T.; Beals, B.; Dykes, D.; et al. Increased antitumor activity, intratumor paclitaxel concentrations, and endothelial cell transport of cremophor-free, albumin-bound paclitaxel, ABI-007, compared with cremophor-based paclitaxel. Clin. Cancer Res. 2006, 12, 1317–1324. [Google Scholar] [CrossRef]
- Huang, Y.W.; Cambre, M.; Lee, H.J. The Toxicity of Nanoparticles Depends on Multiple Molecular and Physicochemical Mechanisms. Int. J. Mol. Sci. 2017, 18, 2702. [Google Scholar] [CrossRef]
- Kai, W.; Xiaojun, X.; Ximing, P.; Zhenqing, H.; Qiqing, Z. Cytotoxic effects and the mechanism of three types of magnetic nanoparticles on human hepatoma BEL-7402 cells. Nanoscale Res. Lett. 2011, 6, 480. [Google Scholar] [CrossRef]
- Sukhanova, A.; Bozrova, S.; Sokolov, P.; Berestovoy, M.; Karaulov, A.; Nabiev, I. Dependence of Nanoparticle Toxicity on Their Physical and Chemical Properties. Nanoscale Res. Lett. 2018, 13, 44. [Google Scholar] [CrossRef]
- Su, H.; Wang, Y.; Gu, Y.; Bowman, L.; Zhao, J.; Ding, M. Potential applications and human biosafety of nanomaterials used in nanomedicine. J. Appl. Toxicol. JAT 2018, 38, 3–24. [Google Scholar] [CrossRef]
- Moller, A.; Lobb, R.J. The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer 2020, 20, 697–709. [Google Scholar] [CrossRef]
- Di Bella, M.A. Overview and Update on Extracellular Vesicles: Considerations on Exosomes and Their Application in Modern Medicine. Biology 2022, 11, 804. [Google Scholar] [CrossRef]
- Clayton, A.; Harris, C.L.; Court, J.; Mason, M.D.; Morgan, B.P. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur. J. Immunol. 2003, 33, 522–531. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, B.; Ocansey, D.K.W.; Xu, W.; Qian, H. Extracellular vesicles: A bright star of nanomedicine. Biomaterials 2021, 269, 120467. [Google Scholar] [CrossRef]
- Frolova, L.; Li, I.T.S. Targeting Capabilities of Native and Bioengineered Extracellular Vesicles for Drug Delivery. Bioengineering 2022, 9, 496. [Google Scholar] [CrossRef]
- Lenzini, S.; Bargi, R.; Chung, G.; Shin, J.W. Matrix mechanics and water permeation regulate extracellular vesicle transport. Nat. Nanotechnol. 2020, 15, 217–223. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, F.; Xu, W.; Qian, H. Engineered Extracellular Vesicles as a Targeted Delivery Platform for Precision Therapy. Tissue Eng. Regen. Med. 2023, 20, 157–175. [Google Scholar] [CrossRef]
- Sadeghi, S.; Tehrani, F.R.; Tahmasebi, S.; Shafiee, A.; Hashemi, S.M. Exosome engineering in cell therapy and drug delivery. Inflammopharmacology 2023, 31, 145–169. [Google Scholar] [CrossRef]
- Bahmani, L.; Ullah, M. Different sourced extracellular vesicles and their potential applications in clinical treatments. Cells 2022, 11, 1989. [Google Scholar] [CrossRef]
- Burnouf, T.; Agrahari, V.; Agrahari, V. Extracellular vesicles as nanomedicine: Hopes and hurdles in clinical translation. Int. J. Nanomed. 2019, 14, 8847–8859. [Google Scholar] [CrossRef]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [CrossRef]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Mathew, A.; Mason, A.B.; Teng, K. Exosome formation during maturation of mammalian and avian reticulocytes: Evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell. Physiol. 1991, 147, 27–36. [Google Scholar] [CrossRef]
- Lee, Y.J.; Jy, W.; Horstman, L.L.; Janania, J.; Reyes, Y.; Kelley, R.E.; Ahn, Y.S. Elevated platelet microparticles in transient ischemic attacks, lacunar infarcts, and multiinfarct dementias. Thromb. Res. 1993, 72, 295–304. [Google Scholar] [CrossRef]
- Singh, N.; Gemmell, C.H.; Daly, P.A.; Yeo, E.L. Elevated platelet-derived microparticle levels during unstable angina. Can. J. Cardiol. 1995, 11, 1015–1021. [Google Scholar]
- Jackson, K.K.; Mata, C.; Marcus, R.K. A rapid capillary-channeled polymer (C-CP) fiber spin-down tip approach for the isolation of plant-derived extracellular vesicles (PDEVs) from 20 common fruit and vegetable sources. Talanta 2023, 252, 123779. [Google Scholar] [CrossRef]
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef]
- Robinson, D.G.; Ding, Y.; Jiang, L. Unconventional protein secretion in plants: A critical assessment. Protoplasma 2016, 253, 31–43. [Google Scholar] [CrossRef]
- Liu, X.M.; Ma, L.; Schekman, R. Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. elife 2021, 10, e71982. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Sharma, S.; Masud, M.K.; Kaneti, Y.V.; Rewatkar, P.; Koradia, A.; Hossain, M.S.A.; Yamauchi, Y.; Popat, A.; Salomon, C. Extracellular Vesicle Nanoarchitectonics for Novel Drug Delivery Applications. Small 2021, 17, e2102220. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Ståhl, A.L.; Johansson, K.; Mossberg, M.; Kahn, R.; Karpman, D. Exosomes and microvesicles in normal physiology, pathophysiology, and renal diseases. Pediatr. Nephrol. 2019, 34, 11–30. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Du, S.; Guan, Y.; Xie, A.; Yan, Z.; Gao, S.; Li, W.; Rao, L.; Chen, X.; Chen, T. Extracellular vesicles: A rising star for therapeutics and drug delivery. J. Nanobiotechnol. 2023, 21, 231. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Zhang, Q.; Franklin, J.L.; Coffey, R.J. Extracellular vesicles and nanoparticles: Emerging complexities. Trends Cell Biol. 2023, 33, 667–681. [Google Scholar] [CrossRef]
- Matsui, T.; Osaki, F.; Hiragi, S.; Sakamaki, Y.; Fukuda, M. ALIX and ceramide differentially control polarized small extracellular vesicle release from epithelial cells. EMBO Rep. 2021, 22, e51475. [Google Scholar] [CrossRef]
- Shiri, F.; Feng, H.; Petersen, K.E.; Sant, H.; Bardi, G.T.; Schroeder, L.A.; Merchant, M.L.; Gale, B.K.; Hood, J.L. Separation of U87 glioblastoma cell-derived small and medium extracellular vesicles using elasto-inertial flow focusing (a spiral channel). Sci. Rep. 2022, 12, 6146. [Google Scholar] [CrossRef]
- Yan, J.; Fan, Y.J.; Bao, H.; Li, Y.G.; Zhang, S.M.; Yao, Q.P.; Huo, Y.L.; Jiang, Z.L.; Qi, Y.X.; Han, Y. Platelet-derived microvesicles regulate vascular smooth muscle cell energy metabolism via PRKAA after intimal injury. J. Cell Sci. 2022, 135, jcs259364. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, J.; Kadungure, T.; Beyene, J.; Zhang, H.; Lu, Q. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 2018, 9, 960. [Google Scholar] [CrossRef]
- Saliba, D.G.; Cespedes-Donoso, P.F.; Balint, S.; Compeer, E.B.; Korobchevskaya, K.; Valvo, S.; Mayya, V.; Kvalvaag, A.; Peng, Y.; Dong, T.; et al. Composition and structure of synaptic ectosomes exporting antigen receptor linked to functional CD40 ligand from helper T cells. elife 2019, 8, e47528. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.W.; Suwanpradid, J.; Kim, I.H.; Staats, H.F.; Haniffa, M.; MacLeod, A.S.; Abraham, S.N. Perivascular dendritic cells elicit anaphylaxis by relaying allergens to mast cells via microvesicles. Science 2018, 362, eaao0666. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Nevo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.J.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan-Chari, V.; Clancy, J.; Plou, C.; Romao, M.; Chavrier, P.; Raposo, G.; D’Souza-Schorey, C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 2009, 19, 1875–1885. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hill, A.F. Therapeutically harnessing extracellular vesicles. Nat. Rev. Drug Discov. 2022, 21, 379–399. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavik, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, I.; Terrasi, A.; Martelli, C.; Gaudioso, G.; Di Cristofori, A.; Storaci, A.M.; Formica, M.; Braidotti, P.; Todoerti, K.; Ferrero, S.; et al. A GBM-like V-ATPase signature directs cell-cell tumor signaling and reprogramming via large oncosomes. eBioMedicine 2019, 41, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Soriani, A.; Vulpis, E.; Cuollo, L.; Santoni, A.; Zingoni, A. Cancer extracellular vesicles as novel regulators of NK cell response. Cytokine Growth Factor Rev. 2020, 51, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Woith, E.; Fuhrmann, G.; Melzig, M.F. Extracellular Vesicles-Connecting Kingdoms. Int. J. Mol. Sci. 2019, 20, 5695. [Google Scholar] [CrossRef]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef]
- Liu, D.; Kou, X.; Chen, C.; Liu, S.; Liu, Y.; Yu, W.; Yu, T.; Yang, R.; Wang, R.; Zhou, Y.; et al. Circulating apoptotic bodies maintain mesenchymal stem cell homeostasis and ameliorate osteopenia via transferring multiple cellular factors. Cell Res. 2018, 28, 918–933. [Google Scholar] [CrossRef]
- Richardson, C.E.; Shen, K. Neurite Development and Repair in Worms and Flies. Annu. Rev. Neurosci. 2019, 42, 209–226. [Google Scholar] [CrossRef]
- Sisirak, V.; Sally, B.; D’Agati, V.; Martinez-Ortiz, W.; Ozcakar, Z.B.; David, J.; Rashidfarrokhi, A.; Yeste, A.; Panea, C.; Chida, A.S.; et al. Digestion of Chromatin in Apoptotic Cell Microparticles Prevents Autoimmunity. Cell 2016, 166, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.C.; Tan, S.S.; Yeo, R.W.Y.; Choo, A.B.H.; Reiner, A.T.; Su, Y.; Shen, Y.; Fu, Z.; Alexander, L.; Sze, S.K. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J. Extracell. Vesicles 2016, 5, 29828. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed]
- Prada, I.; Meldolesi, J. Binding and Fusion of Extracellular Vesicles to the Plasma Membrane of Their Cell Targets. Int. J. Mol. Sci. 2016, 17, 1296. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef]
- Edlich, A.; Volz, P.; Brodwolf, R.; Unbehauen, M.; Mundhenk, L.; Gruber, A.D.; Hedtrich, S.; Haag, R.; Alexiev, U.; Kleuser, B. Crosstalk between core-multishell nanocarriers for cutaneous drug delivery and antigen-presenting cells of the skin. Biomaterials 2018, 162, 60–70. [Google Scholar] [CrossRef]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef]
- Salunkhe, S.; Dheeraj; Basak, M.; Chitkara, D.; Mittal, A. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. J. Control. Release 2020, 326, 599–614. [Google Scholar]
- Ohnami, N.; Nakamura, A.; Miyado, M.; Sato, M.; Kawano, N.; Yoshida, K.; Harada, Y.; Takezawa, Y.; Kanai, S.; Ono, C.; et al. CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol. Open 2012, 1, 640–647. [Google Scholar] [CrossRef]
- Breakefield, X.O.; Frederickson, R.M.; Simpson, R.J. Gesicles: Microvesicle “cookies” for transient information transfer between cells. Mol. Ther. 2011, 19, 1574–1576. [Google Scholar] [CrossRef]
- Sedgwick, A.E.; Clancy, J.W.; Olivia Balmert, M.; D’Souza-Schorey, C. Extracellular microvesicles and invadopodia mediate non-overlapping modes of tumor cell invasion. Sci. Rep. 2015, 5, 14748. [Google Scholar] [CrossRef]
- Costa Verdera, H.; Gitz-Francois, J.J.; Schiffelers, R.M.; Vader, P. Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J. Control. Release 2017, 266, 100–108. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-‘t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Zhuang, J.; Ibarra, A.; Acosta, A.; Karns, A.P.; Aballi, J.; Nerenberg, M.; Sninsky, J.J.; Quake, S.R.; Toden, S. Survey of extracellular communication of systemic and organ-specific inflammatory responses through cell free messenger RNA profiling in mice. eBioMedicine 2022, 83, 104242. [Google Scholar] [CrossRef]
- Santavanond, J.P.; Rutter, S.F.; Atkin-Smith, G.K.; Poon, I.K.H. Apoptotic Bodies: Mechanism of Formation, Isolation and Functional Relevance. Subcell. Biochem. 2021, 97, 61–88. [Google Scholar]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Majidpoor, J.; Mortezaee, K. Extracellular vesicle-based drug delivery in cancer immunotherapy. Drug Deliv. Transl. Res. 2023, 13, 2790–2806. [Google Scholar] [CrossRef]
- Tang, K.; Zhang, Y.; Zhang, H.; Xu, P.; Liu, J.; Ma, J.; Lv, M.; Li, D.; Katirai, F.; Shen, G.X.; et al. Delivery of chemotherapeutic drugs in tumour cell-derived microparticles. Nat. Commun. 2012, 3, 1282. [Google Scholar] [CrossRef]
- Fonseka, P.; Liem, M.; Ozcitti, C.; Adda, C.G.; Ang, C.S.; Mathivanan, S. Exosomes from N-Myc amplified neuroblastoma cells induce migration and confer chemoresistance to non-N-Myc amplified cells: Implications of intra-tumour heterogeneity. J. Extracell. Vesicles 2019, 8, 1597614. [Google Scholar] [CrossRef]
- Wiklander, O.P.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mager, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef]
- Kang, M.; Jordan, V.; Blenkiron, C.; Chamley, L.W. Biodistribution of extracellular vesicles following administration into animals: A systematic review. J. Extracell. Vesicles 2021, 10, e12085. [Google Scholar] [CrossRef]
- Kanchanapally, R.; Deshmukh, S.K.; Chavva, S.R.; Tyagi, N.; Srivastava, S.K.; Patel, G.K.; Singh, A.P.; Singh, S. Drug-loaded exosomal preparations from different cell types exhibit distinctive loading capability, yield, and antitumor efficacies: A comparative analysis. Int. J. Nanomed. 2019, 14, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, S.; Andrzejewska, A.; Janowski, M.; Lukomska, B. Immunomodulatory and Regenerative Effects of Mesenchymal Stem Cells and Extracellular Vesicles: Therapeutic Outlook for Inflammatory and Degenerative Diseases. Front. Immunol. 2020, 11, 591065. [Google Scholar] [CrossRef]
- Kou, M.; Huang, L.; Yang, J.; Chiang, Z.; Chen, S.; Liu, J.; Guo, L.; Zhang, X.; Zhou, X.; Xu, X.; et al. Mesenchymal stem cell-derived extracellular vesicles for immunomodulation and regeneration: A next generation therapeutic tool? Cell Death Dis. 2022, 13, 580. [Google Scholar] [CrossRef]
- Xu, R.; Feng, Z.; Wang, F.S. Mesenchymal stem cell treatment for COVID-19. eBioMedicine 2022, 77, 103920. [Google Scholar] [CrossRef]
- Kalimuthu, S.; Gangadaran, P.; Li, X.J.; Oh, J.M.; Lee, H.W.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. In Vivo therapeutic potential of mesenchymal stem cell-derived extracellular vesicles with optical imaging reporter in tumor mice model. Sci. Rep. 2016, 6, 30418. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Kolluri, K.K.; Gowers, K.H.; Janes, S.M. TRAIL delivery by MSC-derived extracellular vesicles is an effective anticancer therapy. J. Extracell. Vesicles 2017, 6, 1265291. [Google Scholar] [CrossRef]
- Wei, H.; Chen, J.; Wang, S.; Fu, F.; Zhu, X.; Wu, C.; Liu, Z.; Zhong, G.; Lin, J. A Nanodrug Consisting Of Doxorubicin And Exosome Derived From Mesenchymal Stem Cells For Osteosarcoma Treatment In Vitro. Int. J. Nanomed. 2019, 14, 8603–8610. [Google Scholar] [CrossRef]
- Cocce, V.; Farronato, D.; Brini, A.T.; Masia, C.; Gianni, A.B.; Piovani, G.; Sisto, F.; Alessandri, G.; Angiero, F.; Pessina, A. Drug Loaded Gingival Mesenchymal Stromal Cells (GinPa-MSCs) Inhibit In Vitro Proliferation of Oral Squamous Cell Carcinoma. Sci. Rep. 2017, 7, 9376. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, A.; Steimberg, N.; Benetti, A.; Berenzi, A.; Alessandri, G.; Pascucci, L.; Boniotti, J.; Cocce, V.; Sordi, V.; Pessina, A.; et al. Paclitaxel-releasing mesenchymal stromal cells inhibit the growth of multiple myeloma cells in a dynamic 3D culture system. Hematol. Oncol. 2017, 35, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, A.; Ghezzi, E.; Pascucci, L.; Aralla, M.; Ceserani, V.; Pettinari, L.; Cocce, V.; Guercio, A.; Alessandri, G.; Parati, E.; et al. Effect of canine mesenchymal stromal cells loaded with paclitaxel on growth of canine glioma and human glioblastoma cell lines. Vet. J. 2017, 223, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Seeger, R.C.; Fabbri, M.; Wang, L.; Wayne, A.S.; Jong, A.Y. Biological roles and potential applications of immune cell-derived extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1400370. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef]
- Silva, A.K.; Luciani, N.; Gazeau, F.; Aubertin, K.; Bonneau, S.; Chauvierre, C.; Letourneur, D.; Wilhelm, C. Combining magnetic nanoparticles with cell derived microvesicles for drug loading and targeting. Nanomedicine 2015, 11, 645–655. [Google Scholar] [CrossRef]
- Haney, M.J.; Zhao, Y.; Jin, Y.S.; Li, S.M.; Bago, J.R.; Klyachko, N.L.; Kabanov, A.V.; Batrakova, E.V. Macrophage-Derived Extracellular Vesicles as Drug Delivery Systems for Triple Negative Breast Cancer (TNBC) Therapy. J. Neuroimmune Pharmacol. 2020, 15, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Wu, J.Y.; Wang, J.M.; Hu, X.B.; Cai, J.X.; Xiang, D.X. Gemcitabine loaded autologous exosomes for effective and safe chemotherapy of pancreatic cancer. Acta Biomater. 2020, 101, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Chulpanova, D.S.; Kitaeva, K.V.; James, V.; Rizvanov, A.A.; Solovyeva, V.V. Therapeutic Prospects of Extracellular Vesicles in Cancer Treatment. Front. Immunol. 2018, 9, 1534. [Google Scholar] [CrossRef]
- Zhu, L.; Kalimuthu, S.; Oh, J.M.; Gangadaran, P.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. Enhancement of antitumor potency of extracellular vesicles derived from natural killer cells by IL-15 priming. Biomaterials 2019, 190–191, 38–50. [Google Scholar] [CrossRef]
- Campanella, C.; Caruso Bavisotto, C.; Logozzi, M.; Marino Gammazza, A.; Mizzoni, D.; Cappello, F.; Fais, S. On the Choice of the Extracellular Vesicles for Therapeutic Purposes. Int. J. Mol. Sci. 2019, 20, 236. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, Y.; Tang, K.; Zhang, H.; Yin, X.; Li, Y.; Xu, P.; Sun, Y.; Ma, R.; Ji, T.; et al. Reversing drug resistance of soft tumor-repopulating cells by tumor cell-derived chemotherapeutic microparticles. Cell Res. 2016, 26, 713–727. [Google Scholar] [CrossRef]
- Liang, Q.; Bie, N.; Yong, T.; Tang, K.; Shi, X.; Wei, Z.; Jia, H.; Zhang, X.; Zhao, H.; Huang, W.; et al. The softness of tumour-cell-derived microparticles regulates their drug-delivery efficiency. Nat. Biomed. Eng. 2019, 3, 729–740. [Google Scholar] [CrossRef]
- Liu, J.; Ma, J.; Tang, K.; Huang, B. Therapeutic Use of Tumor Cell-Derived Extracellular Vesicles. Methods Mol. Biol. 2017, 1660, 433–440. [Google Scholar]
- Yang, Y.; Chen, Y.; Zhang, F.; Zhao, Q.; Zhong, H. Increased anti-tumour activity by exosomes derived from doxorubicin-treated tumour cells via heat stress. Int. J. Hyperth. 2015, 31, 498–506. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, C.; Pang, D.W.; Zhang, Z.L. Controlled Release of Therapeutic Agents with Near-Infrared Laser for Synergistic Photochemotherapy toward Cervical Cancer. Anal. Chem. 2019, 91, 6555–6560. [Google Scholar] [CrossRef]
- Liu, H.; Shen, M.; Zhao, D.; Ru, D.; Duan, Y.; Ding, C.; Li, H. The Effect of Triptolide-Loaded Exosomes on the Proliferation and Apoptosis of Human Ovarian Cancer SKOV3 Cells. BioMed Res. Int. 2019, 2019, 2595801. [Google Scholar] [CrossRef]
- Saari, H.; Lazaro-Ibanez, E.; Viitala, T.; Vuorimaa-Laukkanen, E.; Siljander, P.; Yliperttula, M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J. Control. Release 2015, 220 Pt B, 727–737. [Google Scholar] [CrossRef]
- Salarpour, S.; Forootanfar, H.; Pournamdari, M.; Ahmadi-Zeidabadi, M.; Esmaeeli, M.; Pardakhty, A. Paclitaxel incorporated exosomes derived from glioblastoma cells: Comparative study of two loading techniques. Daru 2019, 27, 533–539. [Google Scholar] [CrossRef]
- Chitti, S.V.; Nedeva, C.; Manickam, R.; Fonseka, P.; Mathivanan, S. Extracellular vesicles as drug targets and delivery vehicles for cancer therapy. Pharmaceutics 2022, 14, 2822. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef]
- Lener, T.; Gimona, M.; Aigner, L.; Börger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; Del Portillo, H.A.; et al. Applying extracellular vesicles based therapeutics in clinical trials—An ISEV position paper. J. Extracell. Vesicles 2015, 4, 30087. [Google Scholar] [CrossRef]
- Wan, Y.; Wang, L.; Zhu, C.; Zheng, Q.; Wang, G.; Tong, J.; Fang, Y.; Xia, Y.; Cheng, G.; He, X.; et al. Aptamer-Conjugated Extracellular Nanovesicles for Targeted Drug Delivery. Cancer Res. 2018, 78, 798–808. [Google Scholar] [CrossRef]
- Morishita, M.; Takahashi, Y.; Matsumoto, A.; Nishikawa, M.; Takakura, Y. Exosome-based tumor antigens-adjuvant co-delivery utilizing genetically engineered tumor cell-derived exosomes with immunostimulatory CpG DNA. Biomaterials 2016, 111, 55–65. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Li, S.; Li, J.; Deng, T.; Ying, G.; Ba, Y. Cell-derived Exosomes as Promising Carriers for Drug Delivery and Targeted Therapy. Curr. Cancer Drug Targets 2018, 18, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mo, H.; He, Z.; Chen, A.; Cheng, P. Extracellular vesicles as an emerging drug delivery system for cancer treatment: Current strategies and recent advances. Biomed. Pharmacother. 2022, 153, 113480. [Google Scholar] [CrossRef]
- Li, J.; Chen, X.; Yi, J.; Liu, Y.; Li, D.; Wang, J.; Hou, D.; Jiang, X.; Zhang, J.; Wang, J.; et al. Identification and Characterization of 293T Cell-Derived Exosomes by Profiling the Protein, mRNA and MicroRNA Components. PLoS ONE 2016, 11, e0163043. [Google Scholar] [CrossRef]
- Wang, C.; Li, N.; Li, Y.; Hou, S.; Zhang, W.; Meng, Z.; Wang, S.; Jia, Q.; Tan, J.; Wang, R.; et al. Engineering a HEK-293T exosome-based delivery platform for efficient tumor-targeting chemotherapy/internal irradiation combination therapy. J. Nanobiotechnol. 2022, 20, 247. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, X.; Xiu, H.; Sun, Y.; Chen, J.; Cheng, G.; Song, Z.; Peng, Y.; Shen, Y.; Wang, J.; et al. Extracellular vesicles: Natural liver-accumulating drug delivery vehicles for the treatment of liver diseases. J. Extracell. Vesicles 2020, 10, e12030. [Google Scholar] [CrossRef]
- García-Martínez, J.; Pérez-Castillo, Í.M.; Salto, R.; López-Pedrosa, J.M.; Rueda, R.; Girón, M.D. Beneficial Effects of Bovine Milk Exosomes in Metabolic Interorgan Cross-Talk. Nutrients 2022, 14, 1442. [Google Scholar] [CrossRef]
- Tian, M.Y.; Hao, D.X.; Liu, Y.; He, J.; Zhao, Z.H.; Guo, T.Y.; Li, X.; Zhang, Y. Milk exosomes: An oral drug delivery system with great application potential. Food Funct. 2023, 14, 1320–1337. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef]
- Somiya, M.; Yoshioka, Y.; Ochiya, T. Biocompatibility of highly purified bovine milk-derived extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1440132. [Google Scholar] [CrossRef]
- Betker, J.L.; Angle, B.M.; Graner, M.W.; Anchordoquy, T.J. The Potential of Exosomes From Cow Milk for Oral Delivery. J. Pharm. Sci. 2019, 108, 1496–1505. [Google Scholar] [CrossRef]
- Zhang, M.; Xiao, B.; Wang, H.; Han, M.K.; Zhang, Z.; Viennois, E.; Xu, C.; Merlin, D. Edible ginger-derived nano-lipids loaded with doxorubicin as a novel drug-delivery approach for colon cancer therapy. Mol. Ther. 2016, 24, 1783–1796. [Google Scholar] [CrossRef]
- Wang, Q.; Ren, Y.; Mu, J.; Egilmez, N.K.; Zhuang, X.; Deng, Z.; Zhang, L.; Yan, J.; Miller, D.; Zhang, H.G. Grapefruit-Derived Nanovectors Use an Activated Leukocyte Trafficking Pathway to Deliver Therapeutic Agents to Inflammatory Tumor Sites. Cancer Res. 2015, 75, 2520–2529. [Google Scholar] [CrossRef]
- Kuerban, K.; Gao, X.; Zhang, H.; Liu, J.; Dong, M.; Wu, L.; Ye, R.; Feng, M.; Ye, L. Doxorubicin-loaded bacterial outer-membrane vesicles exert enhanced anti-tumor efficacy in non-small-cell lung cancer. Acta Pharm. Sinica. B 2020, 10, 1534–1548. [Google Scholar] [CrossRef]
- Kim, O.Y.; Dinh, N.T.; Park, H.T.; Choi, S.J.; Hong, K.; Gho, Y.S. Bacterial protoplast-derived nanovesicles for tumor targeted delivery of chemotherapeutics. Biomaterials 2017, 113, 68–79. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.; Erdbrügger, U. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Nieuwland, R.; Falcón-Pérez, J.M.; Théry, C.; Witwer, K.W. Rigor and standardization of extracellular vesicle research: Paving the road towards robustness. J. Extracell. Vesicles 2020, 10, e12037. [Google Scholar] [CrossRef]
- De Sousa, K.P.; Rossi, I.; Abdullahi, M.; Ramirez, M.I.; Stratton, D.; Inal, J.M. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1835. [Google Scholar] [CrossRef]
- Du, Y.; Qiu, R.; Chen, L.; Chen, Y.; Zhong, Z.; Li, P.; Fan, F.; Cheng, Y. Identification of serum exosomal metabolomic and proteomic profiles for remote ischemic preconditioning. J. Transl. Med. 2023, 21, 241. [Google Scholar] [CrossRef]
- Brezgin, S.; Parodi, A.; Kostyusheva, A.; Ponomareva, N.; Lukashev, A.; Sokolova, D.; Pokrovsky, V.S.; Slatinskaya, O.; Maksimov, G.; Zamyatnin, A.A., Jr.; et al. Technological aspects of manufacturing and analytical control of biological nanoparticles. Biotechnol. Adv. 2023, 64, 108122. [Google Scholar] [CrossRef]
- Duong, P.; Chung, A.; Bouchareychas, L.; Raffai, R.L. Cushioned-Density Gradient Ultracentrifugation (C-DGUC) improves the isolation efficiency of extracellular vesicles. PLoS ONE 2019, 14, e0215324. [Google Scholar] [CrossRef]
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.G.; Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017, 13, 2061–2065. [Google Scholar] [CrossRef]
- Linares, R.; Tan, S.; Gounou, C.; Arraud, N.; Brisson, A.R. High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell. Vesicles 2015, 4, 29509. [Google Scholar] [CrossRef]
- Benedikter, B.J.; Bouwman, F.G.; Vajen, T.; Heinzmann, A.C.A.; Grauls, G.; Mariman, E.C.; Wouters, E.F.M.; Savelkoul, P.H.; Lopez-Iglesias, C.; Koenen, R.R.; et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci. Rep. 2017, 7, 15297. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Ramos, I.; Bancu, I.; Oliveira-Tercero, A.; Armengol, M.P.; Menezes-Neto, A.; Del Portillo, H.A.; Lauzurica-Valdemoros, R.; Borras, F.E. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles 2015, 4, 27369. [Google Scholar] [CrossRef] [PubMed]
- Onodi, Z.; Pelyhe, C.; Terezia Nagy, C.; Brenner, G.B.; Almasi, L.; Kittel, A.; Mancek-Keber, M.; Ferdinandy, P.; Buzas, E.I.; Giricz, Z. Isolation of High-Purity Extracellular Vesicles by the Combination of Iodixanol Density Gradient Ultracentrifugation and Bind-Elute Chromatography From Blood Plasma. Front. Physiol. 2018, 9, 1479. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Mazzega, E.; Toffoletto, B.; de Marco, A. Isolation of anti-extra-cellular vesicle single-domain antibodies by direct panning on vesicle-enriched fractions. Microb. Cell Factories 2018, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef] [PubMed]
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 2015, 199, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Zhang, S.; Huang, R.; Yi, H.; Wang, J.-W. Extracellular vesicles as a novel photosensitive drug delivery system for enhanced photodynamic therapy. Front. Bioeng. Biotechnol. 2022, 10, 1032318. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Zhang, X.; Shi, M.; Li, F.; Wang, S.; Wang, Y.; Wang, Y.; Wei, W.; Ma, G. Tumor Exosomes Reprogrammed by Low pH Are Efficient Targeting Vehicles for Smart Drug Delivery and Personalized Therapy against their Homologous Tumor. Adv. Sci. 2021, 8, 2002787. [Google Scholar] [CrossRef]
- Cao, H.; Gao, H.; Wang, L.; Cheng, Y.; Wu, X.; Shen, X.; Wang, H.; Wang, Z.; Zhan, P.; Liu, J.; et al. Biosynthetic Dendritic Cell-Exocytosed Aggregation-Induced Emission Nanoparticles for Synergistic Photodynamic Immunotherapy. ACS Nano 2022, 16, 13992–14006. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, T.; He, W.; Jin, H.; Liu, C.; Yang, Z.; Ren, J. Methotrexate-Loaded Extracellular Vesicles Functionalized with Therapeutic and Targeted Peptides for the Treatment of Glioblastoma Multiforme. ACS Appl. Mater. Interfaces 2018, 10, 12341–12350. [Google Scholar] [CrossRef]
- Kim, S.; Kang, J.H.; Nguyen Cao, T.G.; Kang, S.J.; Jeong, K.; Kang, H.C.; Kwon, Y.J.; Rhee, W.J.; Ko, Y.T.; Shim, M.S. Extracellular vesicles with high dual drug loading for safe and efficient combination chemo-phototherapy. Biomater. Sci. 2022, 10, 2817–2830. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wu, F.; Hu, G.; Chen, L.; Xu, J.; Xu, P.; Wang, X.; Li, Y.; Liu, S.; Zhang, S.; et al. Autologous tumor cell-derived microparticle-based targeted chemotherapy in lung cancer patients with malignant pleural effusion. Sci. Transl. Med. 2019, 11, eaat5690. [Google Scholar] [CrossRef] [PubMed]
- Fukuta, T.; Nishikawa, A.; Kogure, K. Low level electricity increases the secretion of extracellular vesicles from cultured cells. Biochem. Biophys. Rep. 2020, 21, 100713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Bai, M.; Wang, J.; Zhu, K.; Liu, R.; Ge, S.; Li, J.; Ning, T.; Deng, T. Exosomes serve as nanoparticles to suppress tumor growth and angiogenesis in gastric cancer by delivering hepatocyte growth factor si RNA. Cancer Sci. 2018, 109, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Limoni, S.K.; Moghadam, M.F.; Moazzeni, S.M.; Gomari, H.; Salimi, F. Engineered Exosomes for Targeted Transfer of siRNA to HER2 Positive Breast Cancer Cells. Appl. Biochem. Biotechnol. 2019, 187, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.D.; Sia, C.L.; Kirwin, K.; Haupt, S.; Mahimkar, G.; Zi, T.; Xu, K.; Dooley, K.; Jang, S.C.; Choi, B.; et al. Exosome Surface Display of IL12 Results in Tumor-Retained Pharmacology with Superior Potency and Limited Systemic Exposure Compared with Recombinant IL12. Mol. Cancer Ther. 2021, 20, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.H.; Liang, J.; Czarny, B.; Wacker, M.G.; Yu, V.; Wang, J.W.; Pastorin, G. Extracellular Vesicle (EV) biohybrid systems for cancer therapy: Recent advances and future perspectives. Semin. Cancer Biol. 2021, 74, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Aqil, F.; Munagala, R.; Jeyabalan, J.; Agrawal, A.K.; Gupta, R. Exosomes for the Enhanced Tissue Bioavailability and Efficacy of Curcumin. AAPS J. 2017, 19, 1691–1702. [Google Scholar] [CrossRef]
- Zhang, M.; Shao, W.; Yang, T.; Liu, H.; Guo, S.; Zhao, D.; Weng, Y.; Liang, X.J.; Huang, Y. Conscription of immune cells by light-activatable silencing NK-derived exosome (LASNEO) for synergetic tumor eradication. Adv. Sci. 2022, 9, 2201135. [Google Scholar] [CrossRef]
- Lara, P.; Huis in ‘t Veld, R.V.; Jorquera-Cordero, C.; Chan, A.B.; Ossendorp, F.; Cruz, L.J. Zinc-phthalocyanine-loaded extracellular vesicles increase efficacy and selectivity of photodynamic therapy in co-culture and preclinical models of colon cancer. Pharmaceutics 2021, 13, 1547. [Google Scholar] [CrossRef]
- Huis In ‘t Veld, R.V.; Lara, P.; Jager, M.J.; Koning, R.I.; Ossendorp, F.; Cruz, L.J. M1-derived extracellular vesicles enhance photodynamic therapy and promote immunological memory in preclinical models of colon cancer. J. Nanobiotechnol. 2022, 20, 252. [Google Scholar] [CrossRef]
- Gong, C.; Tian, J.; Wang, Z.; Gao, Y.; Wu, X.; Ding, X.; Qiang, L.; Li, G.; Han, Z.; Yuan, Y. Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy. J. Nanobiotechnol. 2019, 17, 93. [Google Scholar] [CrossRef]
- Jorquera-Cordero, C.; Lara, P.; Cruz, L.J.; Schomann, T.; van Hofslot, A.; de Carvalho, T.G.; Guedes, P.M.D.M.; Creemers, L.; Koning, R.I.; Chan, A.B. Extracellular vesicles from M1-polarized macrophages combined with hyaluronic acid and a β-blocker potentiate doxorubicin’s antitumor activity by downregulating tumor-associated macrophages in breast cancer. Pharmaceutics 2022, 14, 1068. [Google Scholar] [CrossRef]
- Li, H.; Xu, W.; Li, F.; Zeng, R.; Zhang, X.; Wang, X.; Zhao, S.; Weng, J.; Li, Z.; Sun, L. Amplification of anticancer efficacy by co-delivery of doxorubicin and lonidamine with extracellular vesicles. Drug Deliv. 2022, 29, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Isazadeh, H.; Oruji, F.; Shabani, S.; Behroozi, J.; Nasiri, H.; Isazadeh, A.; Akbari, M. Advances in siRNA delivery approaches in cancer therapy: Challenges and opportunities. Mol. Biol. Rep. 2023, 50, 9529–9543. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, L.; Piontek, K.; Sakaguchi, M.; Selaru, F.M. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology 2018, 67, 940–954. [Google Scholar] [CrossRef]
- Yang, T.; Fogarty, B.; LaForge, B.; Aziz, S.; Pham, T.; Lai, L.; Bai, S. Delivery of small interfering RNA to inhibit vascular endothelial growth factor in zebrafish using natural brain endothelia cell-secreted exosome nanovesicles for the treatment of brain cancer. AAPS J. 2017, 19, 475–486. [Google Scholar] [CrossRef]
- Zhang, D.; Lee, H.; Zhu, Z.; Minhas, J.K.; Jin, Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L110–L121. [Google Scholar] [CrossRef]
- O’Loughlin, A.J.; Mager, I.; de Jong, O.G.; Varela, M.A.; Schiffelers, R.M.; El Andaloussi, S.; Wood, M.J.A.; Vader, P. Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles. Mol. Ther. 2017, 25, 1580–1587. [Google Scholar] [CrossRef]
- Didiot, M.C.; Hall, L.M.; Coles, A.H.; Haraszti, R.A.; Godinho, B.M.; Chase, K.; Sapp, E.; Ly, S.; Alterman, J.F.; Hassler, M.R.; et al. Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing. Mol. Ther. 2016, 24, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Meng, Y.; Wang, B.; Wang, C.-X.; Hou, C.-X.; Zhu, Q.-H.; Tang, Y.-T.; Ye, J.-H. In vitro experimental study on the formation of microRNA-34a loaded exosomes and their inhibitory effect in oral squamous cell carcinoma. Cell Cycle 2022, 21, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Oshchepkova, A.; Zenkova, M.; Vlassov, V. Extracellular Vesicles for Therapeutic Nucleic Acid Delivery: Loading Strategies and Challenges. Int. J. Mol. Sci. 2023, 24, 7287. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control. Release 2015, 205, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Albero, M.; Encabo-Berzosa, M.D.M.; Beltran-Visiedo, M.; Fernandez-Messina, L.; Sebastian, V.; Sanchez-Madrid, F.; Arruebo, M.; Santamaria, J.; Martin-Duque, P. Efficient encapsulation of theranostic nanoparticles in cell-derived exosomes: Leveraging the exosomal biogenesis pathway to obtain hollow gold nanoparticle-hybrids. Nanoscale 2019, 11, 18825–18836. [Google Scholar] [CrossRef] [PubMed]
- Podolak, I.; Galanty, A.; Sobolewska, D. Saponins as cytotoxic agents: A review. Phytochem. Rev. 2010, 9, 425–474. [Google Scholar] [CrossRef] [PubMed]
- Nazimek, K.; Bryniarski, K. Perspectives in Manipulating EVs for Therapeutic Applications: Focus on Cancer Treatment. Int. J. Mol. Sci. 2020, 21, 4623. [Google Scholar] [CrossRef]
- Lennaard, A.J.; Mamand, D.R.; Wiklander, R.J.; El Andaloussi, S.; Wiklander, O.P.B. Optimised Electroporation for Loading of Extracellular Vesicles with Doxorubicin. Pharmaceutics 2021, 14, 38. [Google Scholar] [CrossRef]
- Krishn, S.R.; Garcia, V.; Naranjo, N.M.; Quaglia, F.; Shields, C.D.; Harris, M.A.; Kossenkov, A.V.; Liu, Q.; Corey, E.; Altieri, D.C. Small extracellular vesicle-mediated ITGB6 siRNA delivery downregulates the αVβ6 integrin and inhibits adhesion and migration of recipient prostate cancer cells. Cancer Biol. Ther. 2022, 23, 173–185. [Google Scholar] [CrossRef]
- Draguet, F.; Dubois, N.; Bouland, C.; Pieters, K.; Bron, D.; Meuleman, N.; Stamatopoulos, B.; Lagneaux, L. Extracellular vesicles derived from human umbilical cord mesenchymal stromal cells as an efficient nanocarrier to deliver siRNA or drug to pancreatic cancer cells. Cancers 2023, 15, 2901. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Huang, H.; Liu, D.; Wen, S.; Shen, L.; Lin, Q. Augmented cellular uptake and homologous targeting of exosome-based drug loaded IOL for posterior capsular opacification prevention and biosafety improvement. Bioact. Mater. 2022, 15, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Christiansen, G.; Gurevich, L.; Moos, T.; Duroux, M. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology 2016, 68, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Jeyaram, A.; Patel, D.B.; Parajuli, B.; Livingston, N.K.; Arumugasaamy, N.; Schardt, J.S.; Jay, S.M. Oncogene knockdown via active loading of small RNAs into extracellular vesicles by sonication. Cell. Mol. Bioeng. 2016, 9, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wan, Z.; Wang, C.; Lu, F.; Wei, M.; Wang, D.; Hao, Q. Designer exosomes for targeted and efficient ferroptosis induction in cancer via chemo-photodynamic therapy. Theranostics 2021, 11, 8185–8196. [Google Scholar] [CrossRef] [PubMed]
- Yerneni, S.S.; Yalcintas, E.P.; Smith, J.D.; Averick, S.; Campbell, P.G.; Ozdoganlar, O.B. Skin-targeted delivery of extracellular vesicle-encapsulated curcumin using dissolvable microneedle arrays. Acta Biomater. 2022, 149, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.J.; Klyachko, N.L.; Harrison, E.B.; Zhao, Y.; Kabanov, A.V.; Batrakova, E.V. TPP1 Delivery to Lysosomes with Extracellular Vesicles and their Enhanced Brain Distribution in the Animal Model of Batten Disease. Adv. Healthc. Mater. 2019, 8, e1801271. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, W.; Li, Y.; Chang, J.; Tian, F.; Zhao, F.; Ma, Y.; Sun, J. Microfluidic Sonication To Assemble Exosome Membrane-Coated Nanoparticles for Immune Evasion-Mediated Targeting. Nano Lett. 2019, 19, 7836–7844. [Google Scholar] [CrossRef]
- Cho, N.J.; Hwang, L.Y.; Solandt, J.J.R.; Frank, C.W. Comparison of Extruded and Sonicated Vesicles for Planar Bilayer Self-Assembly. Materials 2013, 6, 3294–3308. [Google Scholar] [CrossRef]
- Narayanan, E. Exosomes as drug delivery vehicles for cancer treatment. Curr. Nanosci. 2020, 16, 15–26. [Google Scholar] [CrossRef]
- Kalimuthu, S.; Gangadaran, P.; Rajendran, R.L.; Zhu, L.; Oh, J.M.; Lee, H.W.; Gopal, A.; Baek, S.H.; Jeong, S.Y.; Lee, S.-W. A new approach for loading anticancer drugs into mesenchymal stem cell-derived exosome mimetics for cancer therapy. Front. Pharmacol. 2018, 9, 1116. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, G.; Mao, Y.; Luo, J.; Cao, Y.; Tan, W.; Li, W.; Yu, H.; Jia, X.; Li, H. Engineering extracellular vesicles mimetics for targeted chemotherapy of drug-resistant ovary cancer. Nanomedicine 2024, 19, 25–41. [Google Scholar] [CrossRef]
- Qiao, L.; Hu, S.; Huang, K.; Su, T.; Li, Z.; Vandergriff, A.; Cores, J.; Dinh, P.-U.; Allen, T.; Shen, D. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics 2020, 10, 3474. [Google Scholar] [CrossRef]
- Tran, P.H.; Xiang, D.; Tran, T.T.; Yin, W.; Zhang, Y.; Kong, L.; Chen, K.; Sun, M.; Li, Y.; Hou, Y. Exosomes and nanoengineering: A match made for precision therapeutics. Adv. Mater. 2020, 32, 1904040. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef]
- Costa, A.P.; Xu, X.; Burgess, D.J. Freeze-anneal-thaw cycling of unilamellar liposomes: Effect on encapsulation efficiency. Pharm. Res. 2014, 31, 97–103. [Google Scholar] [CrossRef]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.-a.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016, 6, 21933. [Google Scholar] [CrossRef]
- Goh, W.J.; Lee, C.K.; Zou, S.; Woon, E.C.; Czarny, B.; Pastorin, G. Doxorubicin-loaded cell-derived nanovesicles: An alternative targeted approach for anti-tumor therapy. Int. J. Nanomed. 2017, 12, 2759–2767. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Lee, D.-Y.; Lu, Y.-L.; Delila, L.; Nebie, O.; Barro, L.; Changou, C.A.; Lu, L.-S.; Goubran, H.; Burnouf, T. Platelet extracellular vesicles are efficient delivery vehicles of doxorubicin, an anti-cancer drug: Preparation and in vitro characterization. Platelets 2023, 34, 2237134. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, D.; Li, G.; Chen, J.; Yang, Y.; Bian, L.; Zhou, J.; Wu, Y.; Chen, Y. Enhanced Therapeutic Potential of Hybrid Exosomes Loaded with Paclitaxel for Cancer Therapy. Int. J. Mol. Sci. 2024, 25, 3645. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, H.; Huang, Q.; Peng, C.; Yao, L.; Chen, H.; Qiu, Z.; Wu, Y.; Wang, L.; Chen, W. Exosomes from M1-Polarized Macrophages Enhance Paclitaxel Antitumor Activity by Activating Macrophages-Mediated Inflammation. Theranostics 2019, 9, 1714–1727. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zeng, Q.; Han, Q.; Xia, W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell 2019, 10, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Rhim, W.K.; Kim, J.Y.; Lee, S.Y.; Cha, S.G.; Park, J.M.; Park, H.J.; Park, C.G.; Han, D.K. Recent advances in extracellular vesicle engineering and its applications to regenerative medicine. Biomater. Res. 2023, 27, 130. [Google Scholar] [CrossRef] [PubMed]
- Kanuma, T.; Yamamoto, T.; Kobiyama, K.; Moriishi, E.; Masuta, Y.; Kusakabe, T.; Ozasa, K.; Kuroda, E.; Jounai, N.; Ishii, K.J. CD63-mediated antigen delivery into extracellular vesicles via DNA vaccination results in robust CD8+ T cell responses. J. Immunol. 2017, 198, 4707–4715. [Google Scholar] [CrossRef] [PubMed]
- Yim, N.; Ryu, S.W.; Choi, K.; Lee, K.R.; Lee, S.; Choi, H.; Kim, J.; Shaker, M.R.; Sun, W.; Park, J.H.; et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 2016, 7, 12277. [Google Scholar] [CrossRef] [PubMed]
- Bui, S.; Dancourt, J.; Lavieu, G. Virus-free method to control and enhance extracellular vesicle cargo loading and delivery. ACS Appl. Bio Mater. 2023, 6, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-K.; Youn, Y.-J.; Lee, Y.-B.; Kim, S.-H.; Song, D.-K.; Shin, M.; Jin, H.K.; Bae, J.-s.; Shrestha, S.; Hong, C.-W. Extracellular vesicles from dHL-60 cells as delivery vehicles for diverse therapeutics. Sci. Rep. 2021, 11, 8289. [Google Scholar] [CrossRef]
- Qiu, Y.; Sun, J.; Qiu, J.; Chen, G.; Wang, X.; Mu, Y.; Li, K.; Wang, W. Antitumor activity of cabazitaxel and MSC-TRAIL derived extracellular vesicles in drug-resistant oral squamous cell carcinoma. Cancer Manag. Res. 2020, 12, 10809–10820. [Google Scholar] [CrossRef]
- O’brien, K.; Khan, S.; Gilligan, K.; Zafar, H.; Lalor, P.; Glynn, C.; O’flatharta, C.; Ingoldsby, H.; Dockery, P.; De Bhulbh, A. Employing mesenchymal stem cells to support tumor-targeted delivery of extracellular vesicle (EV)-encapsulated microRNA-379. Oncogene 2018, 37, 2137–2149. [Google Scholar] [CrossRef]
- Guo, Y.; Hu, G.; Xia, Y.; Li, H.; Yuan, J.; Zhang, J.; Chen, Y.; Guo, H.; Yang, Y.; Wang, Y. Eliminating the original cargos of glioblastoma cell-derived small extracellular vesicles for efficient drug delivery to glioblastoma with improved biosafety. Bioact. Mater. 2022, 16, 204–217. [Google Scholar] [CrossRef]
- Simhadri, V.R.; Reiners, K.S.; Hansen, H.P.; Topolar, D.; Simhadri, V.L.; Nohroudi, K.; Kufer, T.A.; Engert, A.; Pogge von Strandmann, E. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS ONE 2008, 3, e3377. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/macrosialin: Not just a histochemical marker. Lab. Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Greenwald, D.R.; Ruoslahti, E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science 2010, 328, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef]
- Zhao, Z.; Shuang, T.; Gao, Y.; Lu, F.; Zhang, J.; He, W.; Qu, L.; Chen, B.; Hao, Q. Targeted delivery of exosomal miR-484 reprograms tumor vasculature for chemotherapy sensitization. Cancer Lett. 2022, 530, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Gecys, D.; Kazlauskas, A.; Gecyte, E.; Pauziene, N.; Kulakauskiene, D.; Lukminaite, I.; Jekabsone, A. Internalisation of RGD-Engineered Extracellular Vesicles by Glioblastoma Cells. Biology 2022, 11, 1483. [Google Scholar] [CrossRef]
- Zhou, Y.; Yuan, Y.; Liu, M.; Hu, X.; Quan, Y.; Chen, X. Tumor-specific delivery of KRAS siRNA with iRGD-exosomes efficiently inhibits tumor growth. ExRNA 2019, 1, 28. [Google Scholar] [CrossRef]
- Lin, D.; Zhang, H.; Liu, R.; Deng, T.; Ning, T.; Bai, M.; Yang, Y.; Zhu, K.; Wang, J.; Duan, J.; et al. iRGD-modified exosomes effectively deliver CPT1A siRNA to colon cancer cells, reversing oxaliplatin resistance by regulating fatty acid oxidation. Mol. Oncol. 2021, 15, 3430–3446. [Google Scholar] [CrossRef]
- Liu, Q.; Dai, G.; Wu, Y.; Zhang, M.; Yang, M.; Wang, X.; Song, M.; Li, X.; Xia, R.; Wu, Z. iRGD-modified exosomes-delivered BCL6 siRNA inhibit the progression of diffuse large B-cell lymphoma. Front. Oncol. 2022, 12, 822805. [Google Scholar] [CrossRef] [PubMed]
- Roth, L.; Agemy, L.; Kotamraju, V.R.; Braun, G.; Teesalu, T.; Sugahara, K.N.; Hamzah, J.; Ruoslahti, E. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene 2012, 31, 3754–3763. [Google Scholar] [CrossRef] [PubMed]
- Jubb, A.M.; Strickland, L.A.; Liu, S.D.; Mak, J.; Schmidt, M.; Koeppen, H. Neuropilin-1 expression in cancer and development. J. Pathol. 2012, 226, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Duan, J.; Liu, R.; Du, Y.; Luo, Q.; Cui, Y.; Su, Z.; Xu, J.; Xie, Y.; Lu, W. Engineered targeting tLyp-1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J. Pharm. Sci. 2020, 15, 461–471. [Google Scholar] [PubMed]
- Nievergall, E.; Ramshaw, H.S.; Yong, A.S.; Biondo, M.; Busfield, S.J.; Vairo, G.; Lopez, A.F.; Hughes, T.P.; White, D.L.; Hiwase, D.K. Monoclonal antibody targeting of IL-3 receptor alpha with CSL362 effectively depletes CML progenitor and stem cells. Blood 2014, 123, 1218–1228. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Riccioni, R.; Militi, S.; Coccia, E.; Stellacci, E.; Samoggia, P.; Latagliata, R.; Mariani, G.; Rossini, A.; Battistini, A. Elevated expression of IL-3Rα in acute myelogenous leukemia is associated with enhanced blast proliferation, increased cellularity, and poor prognosis. Blood J. Am. Soc. Hematol. 2002, 100, 2980–2988. [Google Scholar]
- Bellavia, D.; Raimondo, S.; Calabrese, G.; Forte, S.; Cristaldi, M.; Patinella, A.; Memeo, L.; Manno, M.; Raccosta, S.; Diana, P.; et al. Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 2017, 7, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Nord, K.; Gunneriusson, E.; Ringdahl, J.; Ståhl, S.; Uhlén, M.; Nygren, P.A. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat. Biotechnol. 1997, 15, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Wikman, M.; Steffen, A.C.; Gunneriusson, E.; Tolmachev, V.; Adams, G.P.; Carlsson, J.; Stahl, S. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des. Sel. 2004, 17, 455–462. [Google Scholar]
- Manri, C.; Yokoi, T.; Nishida, H. Size-Selective Harvesting of Extracellular Vesicles for Strategic Analyses Towards Tumor Diagnoses. Appl. Biochem. Biotechnol. 2017, 182, 609–623. [Google Scholar] [CrossRef]
- Zahnd, C.; Wyler, E.; Schwenk, J.M.; Steiner, D.; Lawrence, M.C.; McKern, N.M.; Pecorari, F.; Ward, C.W.; Joos, T.O.; Pluckthun, A. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J. Mol. Biol. 2007, 369, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Gomari, H.; Forouzandeh Moghadam, M.; Soleimani, M.; Ghavami, M.; Khodashenas, S. Targeted delivery of doxorubicin to HER2 positive tumor models. Int. J. Nanomed. 2019, 14, 5679–5690. [Google Scholar] [CrossRef] [PubMed]
- Molavipordanjani, S.; Khodashenas, S.; Abedi, S.M.; Moghadam, M.F.; Mardanshahi, A.; Hosseinimehr, S.J. (99m)Tc-radiolabeled HER2 targeted exosome for tumor imaging. Eur. J. Pharm. Sci. 2020, 148, 105312. [Google Scholar] [CrossRef] [PubMed]
- Beerli, R.R.; Bauer, M.; Buser, R.B.; Gwerder, M.; Muntwiler, S.; Maurer, P.; Saudan, P.; Bachmann, M.F. Isolation of human monoclonal antibodies by mammalian cell display. Proc. Natl. Acad. Sci. USA 2008, 105, 14336–14341. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.; Nagata, S.; Pastan, I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc. Natl. Acad. Sci. USA 2006, 103, 9637–9642. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, R.; Wu, X.; Sun, Y.; Yao, M.; Li, J.; Xu, Y.; Gu, J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005, 19, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Takanashi, M.; Sudo, K.; Ueda, S.; Ishikawa, A.; Matsuyama, N.; Fujita, K.; Mizutani, T.; Ohgi, T.; Ochiya, T.; et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013, 21, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Lopez, P.; Ribas-Aparicio, R.M.; Becerra-Baez, E.I.; Fraga-Perez, K.; Flores-Martinez, L.F.; Mateos-Chavez, A.A.; Luria-Perez, R. Single-Chain Fragment Variable: Recent Progress in Cancer Diagnosis and Therapy. Cancers 2022, 14, 4206. [Google Scholar] [CrossRef] [PubMed]
- Longatti, A.; Schindler, C.; Collinson, A.; Jenkinson, L.; Matthews, C.; Fitzpatrick, L.; Blundy, M.; Minter, R.; Vaughan, T.; Shaw, M.; et al. High affinity single-chain variable fragments are specific and versatile targeting motifs for extracellular vesicles. Nanoscale 2018, 10, 14230–14244. [Google Scholar] [CrossRef]
- Cheng, Q.; Shi, X.; Han, M.; Smbatyan, G.; Lenz, H.J.; Zhang, Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J. Am. Chem. Soc. 2018, 140, 16413–16417. [Google Scholar] [CrossRef]
- Delcayre, A.; Estelles, A.; Sperinde, J.; Roulon, T.; Paz, P.; Aguilar, B.; Villanueva, J.; Khine, S.; Le Pecq, J.B. Exosome Display technology: Applications to the development of new diagnostics and therapeutics. Blood Cells Mol. Dis. 2005, 35, 158–168. [Google Scholar] [CrossRef]
- Thorne, S.H.; Barak, Y.; Liang, W.; Bachmann, M.H.; Rao, J.; Contag, C.H.; Matin, A. CNOB/ChrR6, a new prodrug enzyme cancer chemotherapy. Mol. Cancer Ther. 2009, 8, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Forterre, A.V.; Zhao, J.; Frimannsson, D.O.; Delcayre, A.; Antes, T.J.; Efron, B.; Jeffrey, S.S.; Pegram, M.D.; Matin, A.C. Anti-HER2 scFv-Directed Extracellular Vesicle-Mediated mRNA-Based Gene Delivery Inhibits Growth of HER2-Positive Human Breast Tumor Xenografts by Prodrug Activation. Mol. Cancer Ther. 2018, 17, 1133–1142. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Zou, W.; Wu, Y.; Zhao, J.; Chen, X. Exosomes containing miRNAs targeting HER2 synthesis and engineered to adhere to HER2 on tumor cells surface exhibit enhanced antitumor activity. J. Nanobiotechnol. 2020, 18, 153. [Google Scholar] [CrossRef]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Sung, B.H.; von Lersner, A.; Guerrero, J.; Krystofiak, E.S.; Inman, D.; Pelletier, R.; Zijlstra, A.; Ponik, S.M.; Weaver, A.M. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun. 2020, 11, 2092. [Google Scholar] [CrossRef] [PubMed]
- Urban, S.; Zieseniss, S.; Werder, M.; Hauser, H.; Budzinski, R.; Engelmann, B. Scavenger receptor BI transfers major lipoprotein-associated phospholipids into the cells. J. Biol. Chem. 2000, 275, 33409–33415. [Google Scholar] [CrossRef]
- Yamamoto, S.; Fukuhara, T.; Ono, C.; Uemura, K.; Kawachi, Y.; Shiokawa, M.; Mori, H.; Wada, M.; Shima, R.; Okamoto, T.; et al. Lipoprotein Receptors Redundantly Participate in Entry of Hepatitis C Virus. PLoS Pathog. 2016, 12, e1005610. [Google Scholar] [CrossRef]
- Kharaziha, P.; Chioureas, D.; Rutishauser, D.; Baltatzis, G.; Lennartsson, L.; Fonseca, P.; Azimi, A.; Hultenby, K.; Zubarev, R.; Ullen, A.; et al. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget 2015, 6, 21740–21754. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Ducoux-Petit, M.; Denat, L.; Soldan, V.; Dauvillier, S.; Balor, S.; Burlet-Schiltz, O.; Larue, L.; Muller, C.; et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment. Cell Melanoma Res. 2015, 28, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Kan, S.; Zhu, Y.; Feng, S.; Feng, W.; Gao, S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomed. 2018, 13, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Shi, J.; Xie, J.; Wang, Y.; Sun, J.; Liu, T.; Zhao, Y.; Zhao, X.; Wang, X.; Ma, Y.; et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020, 4, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M. Exosomes and GPI-anchored proteins: Judicious pairs for investigating biomarkers from body fluids. Adv. Drug Deliv. Rev. 2020, 161–162, 110–123. [Google Scholar] [CrossRef] [PubMed]
- De Gassart, A.; Géminard, C.; Février, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef]
- Kooijmans, S.A.; Aleza, C.G.; Roffler, S.R.; van Solinge, W.W.; Vader, P.; Schiffelers, R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L. Post isolation modification of exosomes for nanomedicine applications. Nanomedicine 2016, 11, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Han, Y.; An, Y.; Ding, Y.; He, C.; Wang, X.; Tang, Q. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials 2018, 178, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Shi, M.; Liu, X.; Jin, C.; Xing, X.; Qiu, L.; Tan, W. Aptamer-Functionalized Exosomes: Elucidating the Cellular Uptake Mechanism and the Potential for Cancer-Targeted Chemotherapy. Anal. Chem. 2019, 91, 2425–2430. [Google Scholar] [CrossRef]
- Lee, E.S.; Sul, J.H.; Shin, J.M.; Shin, S.; Lee, J.A.; Kim, H.K.; Cho, Y.; Ko, H.; Son, S.; Lee, J.; et al. Reactive oxygen species-responsive dendritic cell-derived exosomes for rheumatoid arthritis. Acta Biomater. 2021, 128, 462–473. [Google Scholar] [CrossRef]
- Cao, Y.; Wu, T.; Zhang, K.; Meng, X.; Dai, W.; Wang, D.; Dong, H.; Zhang, X. Engineered Exosome-Mediated Near-Infrared-II Region V(2)C Quantum Dot Delivery for Nucleus-Target Low-Temperature Photothermal Therapy. ACS Nano 2019, 13, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Xiong, Z.; Wang, C.; Xiao, W.; Xiao, H.; Xie, K.; Chen, K.; Liang, H.; Zhang, X.; Yang, H. Folic acid-modified Exosome-PH20 enhances the efficiency of therapy via modulation of the tumor microenvironment and directly inhibits tumor cell metastasis. Bioact. Mater. 2021, 6, 963–974. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Yuan, D.; Deygen, I.; Klyachko, N.L.; Kabanov, A.V.; Batrakova, E.V. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomedicine 2018, 14, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Mongelard, F.; Bouvet, P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. Ther. 2010, 12, 107–114. [Google Scholar] [PubMed]
- Wang, Y.; Chen, X.; Tian, B.; Liu, J.; Yang, L.; Zeng, L.; Chen, T.; Hong, A.; Wang, X. Nucleolin-targeted Extracellular Vesicles as a Versatile Platform for Biologics Delivery to Breast Cancer. Theranostics 2017, 7, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Pi, F.; Binzel, D.W.; Lee, T.J.; Li, Z.; Sun, M.; Rychahou, P.; Li, H.; Haque, F.; Wang, S.; Croce, C.M.; et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018, 13, 82–89. [Google Scholar] [CrossRef]
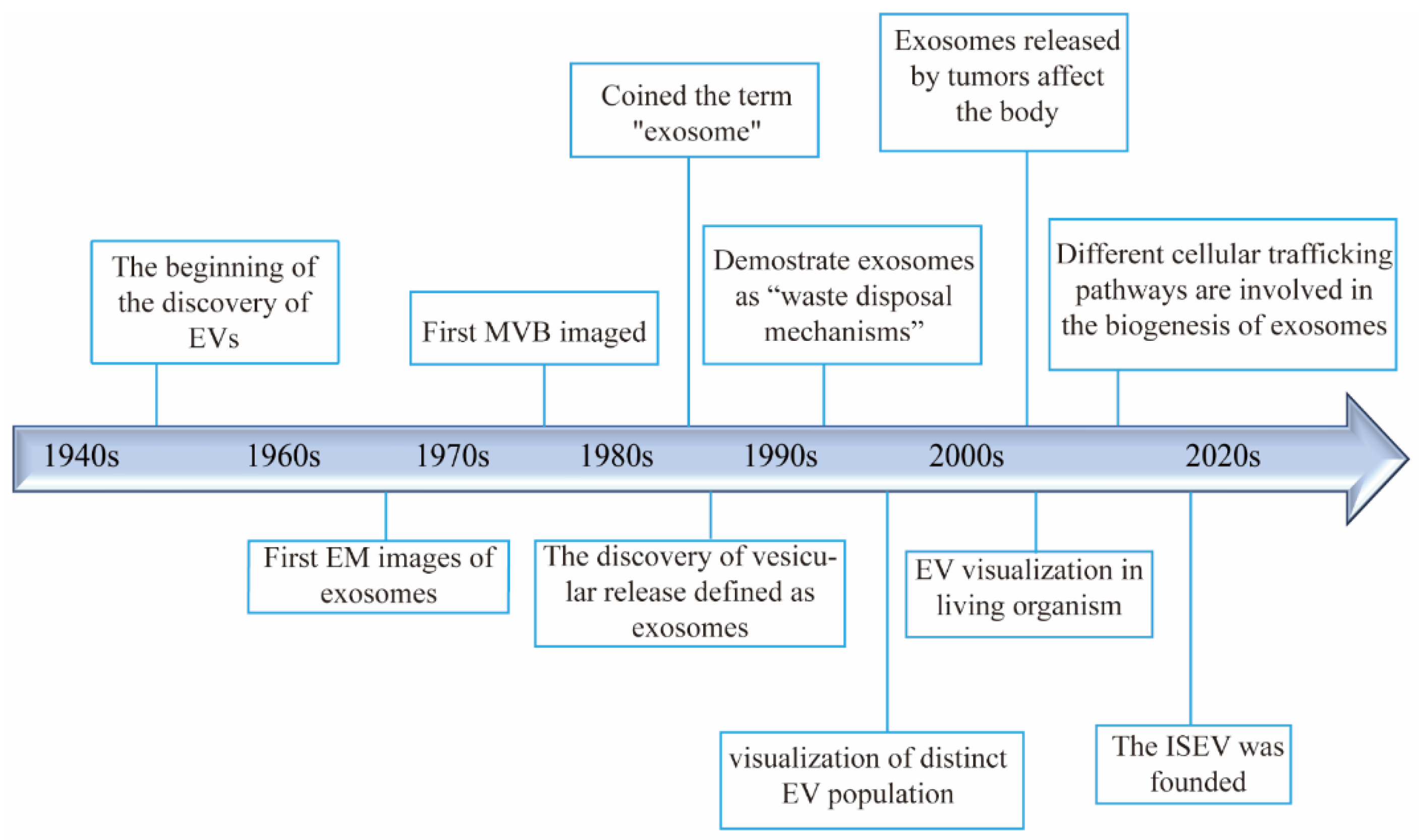
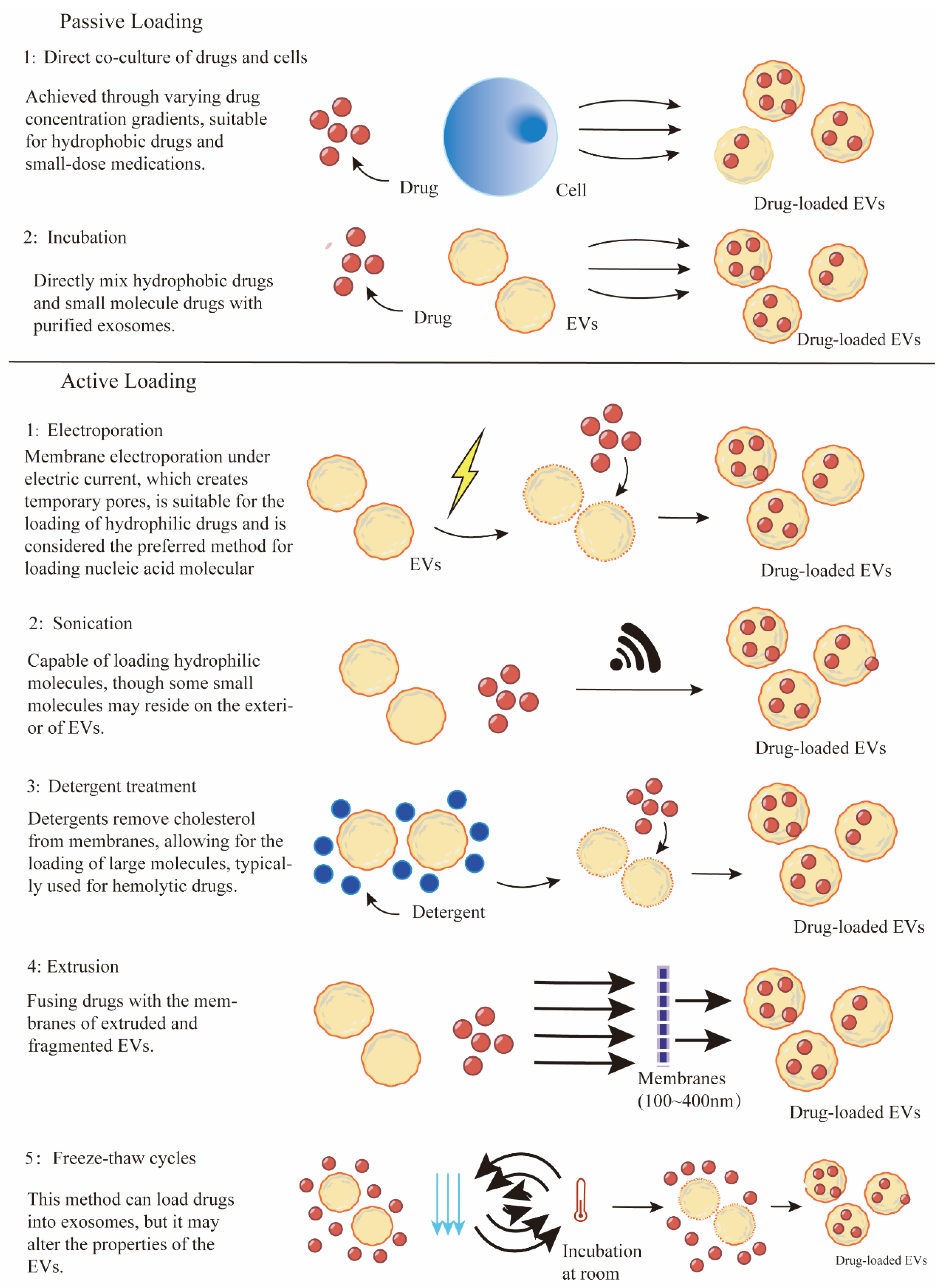
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Yin, B.; Lian, J.; Wang, X. Extracellular Vesicles as Drug Delivery System for Cancer Therapy. Pharmaceutics 2024, 16, 1029. https://doi.org/10.3390/pharmaceutics16081029
Wang J, Yin B, Lian J, Wang X. Extracellular Vesicles as Drug Delivery System for Cancer Therapy. Pharmaceutics. 2024; 16(8):1029. https://doi.org/10.3390/pharmaceutics16081029
Chicago/Turabian StyleWang, Jin, Bohang Yin, Jiabing Lian, and Xia Wang. 2024. "Extracellular Vesicles as Drug Delivery System for Cancer Therapy" Pharmaceutics 16, no. 8: 1029. https://doi.org/10.3390/pharmaceutics16081029







