miRNA-Mediated Mechanisms in the Generation of Effective and Safe Oncolytic Viruses
Abstract
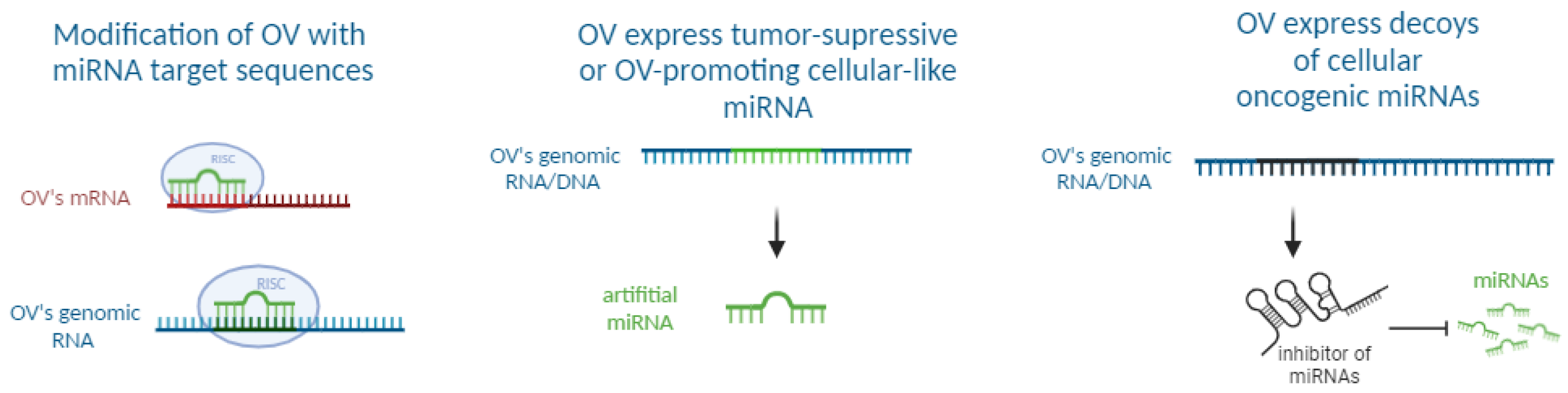
:1. Introduction
2. Control of Gene Expression via miRNAs
3. Diversity of miRNAs Used to Modify Oncolytic Viruses
4. Preclinical Studies of Oncolytic Viruses Modified with miRNA Recognition Sites
4.1. Application of miRTs for Oncolytic Viruses Detargeting
4.2. Issues Regarding miRT Usage and Their Solutions
4.3. Adenoviridae
4.4. Herpesviridae
4.5. Poxviridae
4.6. Picornaviridae
4.7. Togaviridae
4.8. Paramyxoviridae
5. Preclinical Studies of Oncolytic Viruses Expressing miRNAs
5.1. miRNA Precursors Delivered by Oncolytic Viruses
5.2. Some Other Findings on Oncolytic Viruses Encoding miRNAs
5.3. More Research Is Needed on OVs Expressing miRNAs
6. Preclinical Studies of Oncolytic Viruses Expressing miRNA Decoys
6.1. Long Non-Coding RNAs as miRNA Inhibitors Delivered by Oncolytic Viruses
6.2. A Protein as miRNA Decoy Delivered by Oncolytic Viruses
6.3. miRNA Decoys Can Enhance the Antitumor Effect of Oncolytic Viruses
6.4. A Combination of miRNA-Based Modification Approaches Is Possible
7. Clinical Studies of Oncolytic Viruses with miRNA Recognition Sites
8. Summary of the Current State of the Research
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today (Version 1.1). International Agency for Research on Cancer: Lyon, France. Available online: https://Gco.Iarc.Who.Int/Today (accessed on 25 June 2024).
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Debela, D.T.; Muzazu, S.G.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New approaches and procedures for cancer treatment: Current perspectives. SAGE Open Med. 2021, 9, 205031212110343. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.E.P.; Prasad, V. Targeted Cancer Therapies. Am. Fam. Physician 2021, 103, 155–163. [Google Scholar] [PubMed]
- Chiocca, E.A.; Rabkin, S.D. Oncolytic Viruses and Their Application to Cancer Immunotherapy. Cancer Immunol. Res. 2014, 2, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, P.F.; Pala, L.; Conforti, F.; Cocorocchio, E. Talimogene Laherparepvec (T-VEC): An Intralesional Cancer Immunotherapy for Advanced Melanoma. Cancers 2021, 13, 1383. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z. Current Status of Gendicine in China: Recombinant Human Ad-p53 Agent for Treatment of Cancers. Hum. Gene Ther. 2005, 16, 1016–1027. [Google Scholar] [CrossRef] [PubMed]
- Garber, K. China Approves World’s First Oncolytic Virus Therapy for Cancer Treatment. JNCI J. Natl. Cancer Inst. 2006, 98, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.M.; Lamm, D.L.; Meng, M.V.; Nemunaitis, J.J.; Stephenson, J.J.; Arseneau, J.C.; Aimi, J.; Lerner, S.; Yeung, A.W.; Kazarian, T.; et al. A First in Human Phase 1 Study of CG0070, a GM-CSF Expressing Oncolytic Adenovirus, for the Treatment of Nonmuscle Invasive Bladder Cancer. J. Urol. 2012, 188, 2391–2397. [Google Scholar] [CrossRef] [PubMed]
- Harrow, S.; Papanastassiou, V.; Harland, J.; Mabbs, R.; Petty, R.; Fraser, M.; Hadley, D.; Patterson, J.; Brown, S.M.; Rampling, R. HSV1716 injection into the brain adjacent to tumour following surgical resection of high-grade glioma: Safety data and long-term survival. Gene Ther. 2004, 11, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Carbonero, R.; Salazar, R.; Duran, I.; Osman-Garcia, I.; Paz-Ares, L.; Bozada, J.M.; Boni, V.; Blanc, C.; Seymour, L.; Beadle, J.; et al. Phase 1 study of intravenous administration of the chimeric adenovirus enadenotucirev in patients undergoing primary tumor resection. J. Immunother. Cancer 2017, 5, 71. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.A.; Morris, D.G.; Nicholas, G.; Tu, D.; Tehfe, M.; Goffin, J.R.; Shepherd, F.A.; Gregg, R.W.; Rothenstein, J.; Lee, C.; et al. Canadian Cancer Trials Group (CCTG) IND211: A randomized trial of pelareorep (Reolysin) in patients with previously treated advanced or metastatic non-small cell lung cancer receiving standard salvage therapy. Lung Cancer 2018, 120, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Malogolovkin, A.; Gasanov, N.; Egorov, A.; Weener, M.; Ivanov, R.; Karabelsky, A. Combinatorial Approaches for Cancer Treatment Using Oncolytic Viruses: Projecting the Perspectives through Clinical Trials Outcomes. Viruses 2021, 13, 1271. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.-O.; Hong, J.; Yoon, A.-R. Current clinical landscape of oncolytic viruses as novel cancer immunotherapeutic and recent preclinical advancements. Front. Immunol. 2022, 13, 953410. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.; Russell, S.J. History of Oncolytic Viruses: Genesis to Genetic Engineering. Mol. Ther. 2007, 15, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Sinkovics, J.; Horvath, J. New Developments in the Virus Therapy of Cancer: A Historical Review. Intervirology 1993, 36, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Dock, G. The influence of complicating diseases upon leukemia. Am. J. Med. Sci. 1904, 127, 563–592. [Google Scholar] [CrossRef]
- Pelner, L.; Fowler, G.A.; Nauts, H.C. Effects of concurrent infections and their toxins on the course of leukemia. Acta Med. Scand. Suppl. 1958, 162, 5–24. [Google Scholar]
- Smith, R.R.; Huebner, R.J.; Rowe, W.P.; Schatten, W.E.; Thomas, L.B. Studies on the Use of Viruses in the Treatment of Car-cinoma of the Cervix. Cancer 1956, 9, 1211–1218. [Google Scholar] [CrossRef]
- Tysome, J.; Lemoine, N.; Wang, Y. Update on oncolytic viral therapy—Targeting angiogenesis. OncoTargets Ther. 2013, 6, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Everts, A.; Bergeman, M.; McFadden, G.; Kemp, V. Simultaneous Tumor and Stroma Targeting by Oncolytic Viruses. Biomedicines 2020, 8, 474. [Google Scholar] [CrossRef] [PubMed]
- Hemminki, O.; dos Santos, J.M.; Hemminki, A. Oncolytic viruses for cancer immunotherapy. J. Hematol. Oncol. 2020, 13, 84. [Google Scholar] [CrossRef]
- Neault, S.; Bossow, S.; Achard, C.; Bell, J.; Diallo, J.; Leber, M.; Ungerechts, G. Robust envelope exchange platform for oncolytic measles virus. J. Virol. Methods 2022, 302, 114487. [Google Scholar] [CrossRef] [PubMed]
- Martikainen, M.; Ramachandran, M.; Lugano, R.; Ma, J.; Martikainen, M.-M.; Dimberg, A.; Yu, D.; Merits, A.; Essand, M. IFN-I-tolerant oncolytic Semliki Forest virus in combination with anti-PD1 enhances T cell response against mouse glioma. Mol. Ther. Oncolytics 2021, 21, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Hu, C.; Liu, Y.; Chen, X.; Song, D.; Shen, R.; Liu, Z.; Jia, X.; Zhang, Q.; Gao, Y.; et al. Directed natural evolution generates a next-generation oncolytic virus with a high potency and safety profile. Nat. Commun. 2023, 14, 3410. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ma, Z.; Xu, C.; Xie, Y.; Ouyang, D.; Song, S.; Zhao, X.; Liu, F. Progress in oncolytic viruses modified with nanomaterials for intravenous application. Cancer Biol. Med. 2023, 20, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Ajam-Hosseini, M.; Akhoondi, F.; Doroudian, M. Nano based-oncolytic viruses for cancer therapy. Crit. Rev. Oncol. Hematol. 2023, 185, 103980. [Google Scholar] [CrossRef] [PubMed]
- Chartouni, A.; Mouawad, A.; Boutros, M.; Attieh, F.; Medawar, N.; Kourie, H.R. Mesenchymal stem cells: A trojan horse to treat glioblastoma. Investig. New Drugs 2023, 41, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Fan, J.; Ding, Y.; Zhang, J.; Zhou, B.; Zhang, Y.; Huang, B. Oncolytic efficacy of thymidine kinase-deleted vaccinia virus strain Guang9. Oncotarget 2017, 8, 40533–40543. [Google Scholar] [CrossRef] [PubMed]
- Suryawanshi, Y.R.; Nace, R.A.; Russell, S.J.; Schulze, A.J. MicroRNA-detargeting proves more effective than leader gene deletion for improving safety of oncolytic Mengovirus in a nude mouse model. Mol. Ther. Oncolytics 2021, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Penza, V.; Russell, S.J.; Schulze, A.J. The long-lasting enigma of polycytidine (polyC) tract. PLoS Pathog. 2021, 17, e1009739. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhang, Q.; Li, Z.; Zhang, X.; Bao, S.; Fan, D.; Ru, Y.; Dong, S.; Zhang, Y.; Zhang, Y.; et al. Mesenchymal stem cells deliver and release conditionally replicative adenovirus depending on hepatic differentiation to eliminate hepatocellular carcinoma cells specifically. Cancer Lett. 2016, 381, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Delwar, Z.M.; Liu, G.; Kuo, Y.; Lee, C.; Bu, L.; Rennie, P.S.; Jia, W.W. Tumour-specific triple-regulated oncolytic herpes virus to target glioma. Oncotarget 2016, 7, 28658–28669. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, Y.; Liao, W.; Cao, Y.; Liu, Q.; Guo, Y.; Lu, Y.; Xie, Z. Oncolytic adenovirus programmed by synthetic gene circuit for cancer immunotherapy. Nat. Commun. 2019, 10, 4801. [Google Scholar] [CrossRef] [PubMed]
- Moaven, O.; Mangieri, C.W.; Stauffer, J.A.; Anastasiadis, P.Z.; Borad, M.J. Strategies to Develop Potent Oncolytic Viruses and Enhance Their Therapeutic Efficacy. JCO Precis. Oncol. 2021, 5, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, R.; Miest, T.; Shashkova, E.V.; Barry, M.A. Reprogrammed viruses as cancer therapeutics: Targeted, armed and shielded. Nat. Rev. Microbiol. 2008, 6, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Futami, M.; Sato, K.; Miyazaki, K.; Suzuki, K.; Nakamura, T.; Tojo, A. Efficacy and Safety of Doubly-Regulated Vaccinia Virus in a Mouse Xenograft Model of Multiple Myeloma. Mol. Ther. Oncolytics 2017, 6, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Wedge, M.-E.; Jennings, V.A.; Crupi, M.J.F.; Poutou, J.; Jamieson, T.; Pelin, A.; Pugliese, G.; de Souza, C.T.; Petryk, J.; Laight, B.J.; et al. Virally programmed extracellular vesicles sensitize cancer cells to oncolytic virus and small molecule therapy. Nat. Commun. 2022, 13, 1898. [Google Scholar] [CrossRef] [PubMed]
- Bommareddy, P.K.; Shettigar, M.; Kaufman, H.L. Integrating oncolytic viruses in combination cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Geoffroy, K.; Bourgeois-Daigneault, M.-C. The pros and cons of interferons for oncolytic virotherapy. Cytokine Growth Factor Rev. 2020, 56, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Jenks, N.; Myers, R.; Greiner, S.M.; Thompson, J.; Mader, E.K.; Greenslade, A.; Griesmann, G.E.; Federspiel, M.J.; Rakela, J.; Borad, M.J.; et al. Safety Studies on Intrahepatic or Intratumoral Injection of Oncolytic Vesicular Stomatitis Virus Expressing Interferon-β in Rodents and Nonhuman Primates. Hum. Gene Ther. 2010, 21, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Kurisetty, V.V.S.; Heiber, J.; Myers, R.; Pereira, G.S.; Goodwin, J.W.; Federspiel, M.J.; Russell, S.J.; Peng, K.W.; Barber, G.; Merchan, J.R. Preclinical safety and activity of recombinant VSV-IFN-β in an immunocompetent model of squamous cell carcinoma of the head and neck. Head Neck 2013, 36, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Nace, R.; Barber, G.N.; Russell, S.J. Potent systemic therapy of multiple myeloma utilizing oncolytic vesicular stomatitis virus coding for interferon-β. Cancer Gene Ther. 2012, 19, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Govindaraj, V.; Kar, S. Role of microRNAs in oncogenesis: Insights from computational and systems-level modeling approaches. Comput. Syst. Oncol. 2021, 1, e1028. [Google Scholar] [CrossRef]
- Jame-Chenarboo, F.; Ng, H.H.; Macdonald, D.; Mahal, L.K. High-Throughput Analysis Reveals miRNA Upregulating α-2,6-Sialic Acid through Direct miRNA–mRNA Interactions. ACS Central Sci. 2022, 8, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Long, D.; Lee, R.; Williams, P.; Chan, C.Y.; Ambros, V.; Ding, Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007, 14, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, G.G.R.; Mohammadi, Y.; Nazari, A.; Ataeinaeini, M.; Kazemi, P.; Yasamineh, S.; Al-Naqeeb, B.Z.T.; Zaidan, H.K.; Gholizadeh, O. A state-of-the-art review on the MicroRNAs roles in hematopoietic stem cell aging and longevity. Cell Commun. Signal. 2023, 21, 85. [Google Scholar] [CrossRef] [PubMed]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell Pharmacol. 2011, 3, 83–92. [Google Scholar] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Duchaine, T.F.; Fabian, M.R. Mechanistic Insights into MicroRNA-Mediated Gene Silencing. Cold Spring Harb. Perspect. Biol. 2018, 11, a032771. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 Chaperone Machinery Mediates ATP-Dependent RISC Loading of Small RNA Duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the Assembly of the RNAi Enzyme Complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Noland, C.L.; Doudna, J.A. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 2013, 19, 639–648. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Jungers, C.F.; Djuranovic, S. Modulation of miRISC-Mediated Gene Silencing in Eukaryotes. Front. Mol. Biosci. 2022, 9, 832916. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 2005, 11, 1640–1647. [Google Scholar] [CrossRef] [PubMed]
- Wightman, F.F.; Giono, L.E.; Fededa, J.P.; de la Mata, M. Target RNAs Strike Back on MicroRNAs. Front. Genet. 2018, 9, 435. [Google Scholar] [CrossRef] [PubMed]
- Ros, X.B.-D.; Rovira-Rigau, M.; Fillat, C. Implications of MicroRNAs in Oncolytic Virotherapy. Front. Oncol. 2017, 7, 142. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; Macrae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2018, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Croce, C.M. MicroRNA: Trends in clinical trials of cancer diagnosis and therapy strategies. Exp. Mol. Med. 2023, 55, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay Between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 2020, 10, 3079. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.S.; Grundhoff, A.; Tevethia, S.; Pipas, J.M.; Ganem, D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 2005, 435, 682–686. [Google Scholar] [CrossRef]
- Umbach, J.L.; Yen, H.-L.; Poon, L.L.M.; Cullen, B.R. Influenza A Virus Expresses High Levels of an Unusual Class of Small Viral Leader RNAs in Infected Cells. mBio 2010, 1, 00204-10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, M.; Geng, G.; Liu, B.; Huang, Z.; Luo, H.; Zhou, J.; Guo, X.; Cai, W.; Zhang, H. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology 2014, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Choy, E.Y.-W.; Siu, K.-L.; Kok, K.-H.; Lung, R.W.-M.; Tsang, C.M.; To, K.-F.; Kwong, D.L.-W.; Tsao, S.W.; Jin, D.-Y. An Epstein-Barr virus–encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 2008, 205, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Klase, Z.; Winograd, R.; Davis, J.; Carpio, L.; Hildreth, R.; Heydarian, M.; Fu, S.; McCaffrey, T.; Meiri, E.; Ayash-Rashkovsky, M.; et al. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 2009, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Bai, Z.; Ye, F.; Xie, J.; Kim, C.-G.; Huang, Y.; Gao, S.-J. Regulation of NF-κB inhibitor IκBα and viral replication by a KSHV microRNA. Nat. Cell Biol. 2010, 12, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Landais, I.; Pelton, C.; Streblow, D.; DeFilippis, V.; McWeeney, S.; Nelson, J.A. Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway. PLoS Pathog. 2015, 11, e1004881. [Google Scholar] [CrossRef] [PubMed]
- Hancock, M.H.; Hook, L.M.; Mitchell, J.; Nelson, J.A. Human Cytomegalovirus MicroRNAs miR-US5-1 and miR-UL112-3p Block Proinflammatory Cytokine Production in Response to NF-κB-Activating Factors through Direct Downregulation of IKKα and IKKβ. mBio 2017, 8, e00109-17. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.N.; Majumdar, N.; Williams, F.; Rajput, S.; Pokhrel, L.R.; Cook, P.P.; Akula, S.M. MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens. Biology 2023, 12, 1334. [Google Scholar] [CrossRef] [PubMed]
- Otani, Y.; Yoo, J.Y.; Chao, S.; Liu, J.; Jaime-Ramirez, A.C.; Lee, T.J.; Hurwitz, B.; Yan, Y.; Dai, H.; Glorioso, J.C.; et al. Oncolytic HSV-Infected Glioma Cells Activate NOTCH in Adjacent Tumor Cells Sensitizing Tumors to Gamma Secretase Inhibition. Clin. Cancer Res. 2020, 26, 2381–2392. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Cullen, B.R. Viral and Cellular MicroRNAs as Determinants of Viral Pathogenesis and Immunity. Cell Host Microbe 2008, 3, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Furuyama, W.; Lin, Z. RNA Virus-Encoded miRNAs: Current Insights and Future Challenges. Front. Microbiol. 2021, 12, 679210. [Google Scholar] [CrossRef] [PubMed]
- Varble, A.; Chua, M.A.; Perez, J.T.; Manicassamy, B.; García-Sastre, A.; Tenoever, B.R. Engineered RNA viral synthesis of microRNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 11519–11524. [Google Scholar] [CrossRef] [PubMed]
- Rouha, H.; Thurner, C.; Mandl, C.W. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010, 38, 8328–8337. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.S.; Langlois, R.A.; Pham, A.M.; Tenoever, B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA 2012, 18, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Anker, S.C.; Szczeponik, M.G.; Dessila, J.; Dittus, K.; Engeland, C.E.; Jäger, D.; Ungerechts, G.; Leber, M.F. Oncolytic Measles Virus Encoding MicroRNA for Targeted RNA Interference. Viruses 2023, 15, 308. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, M.; Jing, Q.; Georgel, P.; New, L.; Chen, J.; Mols, J.; Kang, Y.J.; Jiang, Z.; Du, X.; Cook, R.; et al. Hypersusceptibility to Vesicular Stomatitis Virus Infection in Dicer1-Deficient Mice Is Due to Impaired miR24 and miR93 Expression. Immunity 2007, 27, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Stern-Ginossar, N.; Saleh, N.; Goldberg, M.D.; Prichard, M.; Wolf, D.G.; Mandelboim, O. Analysis of Human Cytomegalovirus-Encoded MicroRNA Activity during Infection. J. Virol. 2009, 83, 10684–10693. [Google Scholar] [CrossRef] [PubMed]
- Alshahrani, S.H.; Alameri, A.A.; Kahar, F.; Ramírez-Coronel, A.A.; Obaid, R.F.; Alsaikhan, F.; Zabibah, R.S.; Qasim, Q.A.; Altalbawy, F.M.; Mustafa, Y.F.; et al. Overview of the role and action mechanism of microRNA-128 in viral infections. Microb. Pathog. 2023, 176, 106020. [Google Scholar] [CrossRef] [PubMed]
- Lagos, D.; Pollara, G.; Henderson, S.; Gratrix, F.; Fabani, M.; Milne, R.S.; Gotch, F.; Boshoff, C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 2010, 12, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Rovira-Rigau, M.; Raimondi, G.; Marín, M.; Gironella, M.; Alemany, R.; Fillat, C. Bioselection Reveals miR-99b and miR-485 as Enhancers of Adenoviral Oncolysis in Pancreatic Cancer. Mol. Ther. 2018, 27, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Hodzic, J.; Sie, D.; Vermeulen, A.; van Beusechem, V.W. Functional Screening Identifies Human miRNAs that Modulate Adenovirus Propagation in Prostate Cancer Cells. Hum. Gene Ther. 2017, 28, 766–780. [Google Scholar] [CrossRef]
- Brachtlova, T.; van Ginkel, J.-W.; Luinenburg, M.J.; de Menezes, R.X.; Koppers-Lalic, D.; Pegtel, D.M.; Dong, W.; de Gruijl, T.D.; van Beusechem, V.W. Expression of Oncolytic Adenovirus-Encoded RNAi Molecules Is Most Effective in a pri-miRNA Precursor Format. Mol. Ther. Oncolytics 2020, 19, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Geekiyanage, H.; Galanis, E. MiR-31 and miR-128 regulates poliovirus receptor-related 4 mediated measles virus infectivity in tumors. Mol. Oncol. 2016, 10, 1387–1403. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, S.; Pei, Y.; Hu, J.; Hu, Z.; Liu, X.; Wang, X.; Gu, M.; Hu, S.; Liu, X. Differential microRNA Expression in Newcastle Disease Virus-Infected HeLa Cells and Its Role in Regulating Virus Replication. Front. Oncol. 2021, 11, 616809. [Google Scholar] [CrossRef] [PubMed]
- Mishima, T.; Mizuguchi, Y.; Kawahigashi, Y.; Takizawa, T.; Takizawa, T. RT-PCR-based analysis of microRNA (miR-1 and -124) expression in mouse CNS. Brain Res. 2007, 1131, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, M.; Yu, D.; Dyczynski, M.; Baskaran, S.; Zhang, L.; Lulla, A.; Lulla, V.; Saul, S.; Nelander, S.; Dimberg, A.; et al. Safe and Effective Treatment of Experimental Neuroblastoma and Glioblastoma Using Systemically Delivered Triple MicroRNA-Detargeted Oncolytic Semliki Forest Virus. Clin. Cancer Res. 2017, 23, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Dong, X.; Zheng, D.; Nao, J. MiR-124 and the Underlying Therapeutic Promise of Neurodegenerative Disorders. Front. Pharmacol. 2020, 10, 1555. [Google Scholar] [CrossRef] [PubMed]
- Asghariazar, V.; Kadkhodayi, M.; Sarailoo, M.; Jolfayi, A.G.; Baradaran, B. MicroRNA-143 as a potential tumor suppressor in cancer: An insight into molecular targets and signaling pathways. Pathol. Res. Pract. 2023, 250, 154792. [Google Scholar] [CrossRef]
- Eniafe, J.; Jiang, S. MicroRNA-99 family in cancer and immunity. Wiley Interdiscip. Rev. RNA 2020, 12, e1635. [Google Scholar] [CrossRef] [PubMed]
- Ryu, I.S.; Kim, D.H.; Cho, H.-J.; Ryu, J.-H. The role of microRNA-485 in neurodegenerative diseases. Prog. Neurobiol. 2022, 34, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Gao, R.; Yan, B. Potential roles of microRNA-1 and microRNA-133 in cardiovascular disease. Rev. Cardiovasc. Med. 2020, 21, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Safa, A.; Bahroudi, Z.; Shoorei, H.; Majidpoor, J.; Abak, A.; Taheri, M.; Ghafouri-Fard, S. miR-1: A comprehensive review of its role in normal development and diverse disorders. Biomed. Pharmacother. 2020, 132, 110903. [Google Scholar] [CrossRef] [PubMed]
- Thakral, S.; Ghoshal, K. miR-122 is a Unique Molecule with Great Potential in Diagnosis, Prognosis of Liver Disease, and Therapy Both as miRNA Mimic and Antimir. Curr. Gene Ther. 2015, 15, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, M.; Kei, L. MIR-34C regulates the proliferation and apoptosis of lung cancer cells. Clin. Investig. Med. 2020, 43, E56–E62. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Banzhaf-Strathmann, J.; Edbauer, D. Good guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell Commun. Signal. 2014, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Misso, G.; Zarone, M.R.; Lombardi, A.; Grimaldi, A.; Cossu, A.M.; Ferri, C.; Russo, M.; Vuoso, D.C.; Luce, A.; Kawasaki, H.; et al. miR-125b Upregulates miR-34a and Sequentially Activates Stress Adaption and Cell Death Mechanisms in Multiple Myeloma. Mol. Ther. Nucleic Acids 2019, 16, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.; Feng, K.; Wang, S.; Chen, L.; Niu, S.; Lu, Q.; Fang, Y. Efficacy and safety of oncolytic virus combined with chemotherapy or immune checkpoint inhibitors in solid tumor patients: A meta-analysis. Front. Pharmacol. 2022, 13, 1023533. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, S.; Han, D.; Tang, B.; Ma, J. Delivery and Biosafety of Oncolytic Virotherapy. Front. Oncol. 2020, 10, 475. [Google Scholar] [CrossRef] [PubMed]
- Ros, X.B.-D.; Gironella, M.; Fillat, C. MiR-148a- and miR-216a-regulated Oncolytic Adenoviruses Targeting Pancreatic Tumors Attenuate Tissue Damage Without Perturbation of miRNA Activity. Mol. Ther. 2014, 22, 1665–1677. [Google Scholar] [CrossRef]
- Shayestehpour, M.; Moghim, S.; Salimi, V.; Jalilvand, S.; Yavarian, J.; Romani, B.; Mokhtari-Azad, T. Targeting human breast cancer cells by an oncolytic adenovirus using microRNA-targeting strategy. Virus Res. 2017, 240, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Leber, M.F.; Baertsch, M.-A.; Anker, S.C.; Henkel, L.; Singh, H.M.; Bossow, S.; Engeland, C.E.; Barkley, R.; Hoyler, B.; Albert, J.; et al. Enhanced Control of Oncolytic Measles Virus Using MicroRNA Target Sites. Mol. Ther. Oncolytics 2018, 9, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Farkaly, T.; Grzesik, P.; Lee, J.; Denslow, A.; Hewett, J.; Bryant, J.; Behara, P.; Goshert, C.; Wambua, D.; et al. Design of an Interferon-Resistant Oncolytic HSV-1 Incorporating Redundant Safety Modalities for Improved Tolerability. Mol. Ther. Oncolytics 2020, 18, 476–490. [Google Scholar] [CrossRef]
- Yao, W.; Guo, G.; Zhang, Q.; Fan, L.; Wu, N.; Bo, Y. The application of multiple miRNA response elements enables oncolytic adenoviruses to possess specificity to glioma cells. Virology 2014, 458–459, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-X.; Matsui, Y.; Lee, C.; Osamu, O.; Skinner, L.; Wang, J.; So, A.; Rennie, P.S.; Jia, W.W. Intravesical treatment of advanced urothelial bladder cancers with oncolytic HSV-1 co-regulated by differentially expressed microRNAs. Gene Ther. 2016, 23, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Baertsch, A.M.; Leber, M.F.; Bossow, S.; Singh, M.; Engeland, E.C.; Albert, J.; Grossardt, C.; Jäger, D.; von Kalle, C.; Ungerechts, G. MicroRNA-mediated multi-tissue detargeting of oncolytic measles virus. Cancer Gene Ther. 2014, 21, 373–380. [Google Scholar] [CrossRef]
- Ros, X.B.-D.; Villanueva, E.; Fillat, C. Late-phase miRNA-controlled oncolytic adenovirus for selective killing of cancer cells. Oncotarget 2015, 6, 6179–6190. [Google Scholar] [CrossRef]
- Sutto-Ortiz, P.; Eléouët, J.-F.; Ferron, F.; Decroly, E. Biochemistry of the Respiratory Syncytial Virus L Protein Embedding RNA Polymerase and Capping Activities. Viruses 2023, 15, 341. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.J.; Nace, R.; Barber, G.N.; Russell, S.J. Attenuation of Vesicular Stomatitis Virus Encephalitis through MicroRNA Targeting. J. Virol. 2010, 84, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Miyamoto, S.; Soda, Y.; Takishima, Y.; Sagara, M.; Liao, J.; Hirose, L.; Hijikata, Y.; Miura, Y.; Hara, K.; et al. Extremely Low Organ Toxicity and Strong Antitumor Activity of miR-34-Regulated Oncolytic Coxsackievirus B3. Mol. Ther. Oncolytics 2019, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Sagara, M.; Miyamoto, S.; Itoh, S.; Soda, Y.; Tani, K. Development of New Oncolytic Virotherapy Targeting Breast Cancer Using Coxsackievirus B3. Anticancer Res. 2021, 41, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Hazini, A.; Dieringer, B.; Klingel, K.; Pryshliak, M.; Geisler, A.; Kobelt, D.; Daberkow, O.; Kurreck, J.; van Linthout, S.; Fechner, H. Application Route and Immune Status of the Host Determine Safety and Oncolytic Activity of Oncolytic Coxsackievirus B3 Variant PD-H. Viruses 2021, 13, 1918. [Google Scholar] [CrossRef] [PubMed]
- Hazini, A.; Dieringer, B.; Pryshliak, M.; Knoch, K.-P.; Heimann, L.; Tolksdorf, B.; Pappritz, K.; El-Shafeey, M.; Solimena, M.; Beling, A.; et al. miR-375- and miR-1-Regulated Coxsackievirus B3 Has No Pancreas and Heart Toxicity but Strong Antitumor Efficiency in Colorectal Carcinomas. Hum. Gene Ther. 2021, 32, 216–230. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Yao, H.; Wang, J.; Xiao, Z.; Xin, L.; Liu, Z.; Ma, X.; Sun, J.; Jin, Q.; Liu, Z. Coxsackievirus B3 Engineered to Contain MicroRNA Targets for Muscle-Specific MicroRNAs Displays Attenuated Cardiotropic Virulence in Mice. J. Virol. 2015, 89, 908–916. [Google Scholar] [CrossRef]
- Liu, H.; Xue, Y.C.; Deng, H.; Mohamud, Y.; Ng, C.S.; Chu, A.; Lim, C.J.; Lockwood, W.W.; Jia, W.W.; Luo, H. MicroRNA Modification of Coxsackievirus B3 Decreases Its Toxicity, while Retaining Oncolytic Potency against Lung Cancer. Mol. Ther. Oncolytics 2020, 16, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Bahreyni, A.; Liu, H.; Mohamud, Y.; Xue, Y.C.; Zhang, J.; Luo, H. A new miRNA-Modified coxsackievirus B3 inhibits triple negative breast cancer growth with improved safety profile in immunocompetent mice. Cancer Lett. 2022, 548, 215849. [Google Scholar] [CrossRef] [PubMed]
- Bahreyni, A.; Liu, H.; Mohamud, Y.; Xue, Y.C.; Fan, Y.M.; Zhang, Y.L.; Luo, H. A combination of genetically engineered oncolytic virus and melittin-CpG for cancer viro-chemo-immunotherapy. BMC Med. 2023, 21, 193. [Google Scholar] [CrossRef] [PubMed]
- Elsedawy, N.B.; Nace, R.A.; Russell, S.J.; Schulze, A.J. Oncolytic Activity of Targeted Picornaviruses Formulated as Synthetic Infectious RNA. Mol. Ther. Oncolytics 2020, 17, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Sam, M.; Selman, M.; Zhao, W.; Jung, J.; Willingham, A.; Phan, U.; Starling, G.C.; Gao, Q. Engineering Oncolytic Coxsackievirus A21 with Small Transgenes and Enabling Cell-Mediated Virus Delivery by Integrating Viral cDNA into the Genome. J. Virol. 2023, 97, e0030923. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.J.; Hadac, E.M.; Nace, R.A.; Russell, S.J. MicroRNA-Detargeted Mengovirus for Oncolytic Virotherapy. J. Virol. 2016, 90, 4078–4092. [Google Scholar] [CrossRef]
- Bourhis, J.-M.; Canard, B.; Longhi, S. Structural disorder within the replicative complex of measles virus: Functional implications. Virology 2006, 344, 94–110. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.T.; Pham, A.M.; Lorini, M.H.; Chua, M.A.; Steel, J.; Tenoever, B.R. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat. Biotechnol. 2009, 27, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Penza, V.; Maroun, J.W.; Nace, R.A.; Schulze, A.J.; Russell, S.J. Polycytidine tract deletion from microRNA-detargeted oncolytic Mengovirus optimizes the therapeutic index in a murine multiple myeloma model. Mol. Ther. Oncolytics 2022, 28, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, Z.; Li, L.; Wu, J.; Zhang, H.; Zhang, H.; Lei, T.; Xu, B. Oncolytic Adenovirus: Prospects for Cancer Immunotherapy. Front. Microbiol. 2021, 12, 707290. [Google Scholar] [CrossRef] [PubMed]
- Elsner, L.; Heimann, L.; Geisler, A.; Dieringer, B.; Knoch, K.-P.; Hinze, L.; Klingel, K.; Solimena, M.; Kurreck, J.; Fechner, H. Fast Track Adaptation of Oncolytic Coxsackie B3 Virus to Resistant Colorectal Cancer Cells—A Method to Personalize Virotherapy. Biol. Proced. Online 2024, 26, 11. [Google Scholar] [CrossRef] [PubMed]
- Poutou, J.; Bunuales, M.; Gonzalezaparicio, M.; Garcia-Aragoncillo, E.; Quetglas, J.I.; Casado, R.; Bravo-Perez, C.; Alzuguren, P.; Hernandezalcoceba, R. Safety and antitumor effect of oncolytic and helper-dependent adenoviruses expressing interleukin-12 variants in a hamster pancreatic cancer model. Gene Ther. 2015, 22, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Chouljenko, D.V.; Murad, Y.M.; Lee, I.-F.; Delwar, Z.; Ding, J.; Liu, G.; Liu, X.; Bu, X.; Sun, Y.; Samudio, I.; et al. Targeting carcinoembryonic antigen-expressing tumors using a novel transcriptional and translational dual-regulated oncolytic herpes simplex virus type 1. Mol. Ther. Oncolytics 2023, 28, 334–348. [Google Scholar] [CrossRef] [PubMed]
- Marzulli, M.; Mazzacurati, L.; Zhang, M.; Goins, W.; Hatley, M.; Glorioso, J.; Cohen, J. A Novel Oncolytic Herpes Simplex Virus Design based on the Common Overexpression of microRNA-21 in Tumors. J. Gene Ther. 2018, 3, 1–8. [Google Scholar] [CrossRef]
- Mazzacurati, L.; Marzulli, M.; Reinhart, B.; Miyagawa, Y.; Uchida, H.; Goins, W.F.; Li, A.; Kaur, B.; Caligiuri, M.; Cripe, T.; et al. Use of miRNA Response Sequences to Block Off-target Replication and Increase the Safety of an Unattenuated, Glioblastoma-targeted Oncolytic HSV. Mol. Ther. 2015, 23, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Sarén, T.; Ramachandran, M.; Martikainen, M.; Yu, D. Insertion of the Type-I IFN Decoy Receptor B18R in a miRNA-Tagged Semliki Forest Virus Improves Oncolytic Capacity but Results in Neurotoxicity. Mol. Ther. Oncolytics 2017, 7, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Martikainen, M.; Niittykoski, M.; Fraunberg, M.v.U.z.; Immonen, A.; Koponen, S.; van Geenen, M.; Vähä-Koskela, M.; Ylösmäki, E.; Jääskeläinen, J.E.; Saksela, K.; et al. MicroRNA-Attenuated Clone of Virulent Semliki Forest Virus Overcomes Antiviral Type I Interferon in Resistant Mouse CT-2A Glioma. J. Virol. 2015, 89, 10637–10647. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.M.; Leber, M.F.; Bossow, S.; Engeland, C.E.; Dessila, J.; Grossardt, C.; Zaoui, K.; Bell, J.C.; Jäger, D.; von Kalle, C.; et al. MicroRNA-sensitive oncolytic measles virus for chemovirotherapy of pancreatic cancer. Mol. Ther. Oncolytics 2021, 21, 340–355. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Sun, Y.; Qi, M.; Li, W.; Zhang, Z.; Zhang, X.-E.; Cui, Z. Enterovirus A71 Oncolysis of Malignant Gliomas. Mol. Ther. 2020, 28, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Basnet, S.; Dai, Z.; Li, S.; Zhang, Z.; Ge, H. A Novel E1B55kDa-Deleted Oncolytic Adenovirus Carrying MicroRNA-143 Exerts Specific Antitumor Efficacy on Colorectal Cancer Cells. Am. J. Transl. Res. 2016, 8, 3822–3830. [Google Scholar] [PubMed]
- Lou, W.; Chen, Q.; Ma, L.; Liu, J.; Yang, Z.; Shen, J.; Cui, Y.; Bian, X.-W.; Qian, C. Oncolytic adenovirus co-expressing miRNA-34a and IL-24 induces superior antitumor activity in experimental tumor model. J. Mol. Med. 2013, 91, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Sakuda, T.; Kubo, T.; Johan, M.P.; Furuta, T.; Sakaguchi, T.; Adachi, N. Development of an Oncolytic Recombinant Vesicular Stomatitis Virus Encoding a Tumor-suppressor MicroRNA. Anticancer Res. 2020, 40, 6319–6325. [Google Scholar] [CrossRef] [PubMed]
- Sakuda, T.; Kubo, T.; Johan, M.P.; Furuta, T.; Sakaguchi, T.; Adachi, N. Oncolytic Effect of a Recombinant Vesicular Stomatitis Virus Encoding a Tumor-suppressor MicroRNA in an Osteosarcoma Mouse Model. Anticancer Res. 2023, 43, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Wang, S.; Yang, C.; Huang, X.; Chen, Z.; He, W.; Shen, J.; Liu, X.; Qian, W. Combined expression of miR-34a and Smac mediated by oncolytic vaccinia virus synergistically promote anti-tumor effects in Multiple Myeloma. Sci. Rep. 2016, 6, 32174. [Google Scholar] [CrossRef] [PubMed]
- Doerner, J.; Sallard, E.; Zhang, W.; Solanki, M.; Liu, J.; Ehrke-Schulz, E.; Zirngibl, H.; Lieber, A.; Ehrhardt, A. Novel Group C Oncolytic Adenoviruses Carrying a miRNA Inhibitor Demonstrate Enhanced Oncolytic Activity In Vitro and In Vivo. Mol. Cancer Ther. 2022, 21, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, G.; Gea-Sorlí, S.; Otero-Mateo, M.; Fillat, C. Inhibition of miR-222 by Oncolytic Adenovirus-Encoded miRNA Sponges Promotes Viral Oncolysis and Elicits Antitumor Effects in Pancreatic Cancer Models. Cancers 2021, 13, 3233. [Google Scholar] [CrossRef] [PubMed]
- Ang, L.; Guo, L.; Wang, J.; Huang, J.; Lou, X.; Zhao, M. Oncolytic virotherapy armed with an engineered interfering lncRNA exhibits antitumor activity by blocking the epithelial mesenchymal transition in triple-negative breast cancer. Cancer Lett. 2020, 479, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Su, Y.; Sun, B.; Ji, W.; Peng, Z.; Xu, Y.; Wu, M.; Su, C. An Artificially Designed Interfering lncRNA Expressed by Oncolytic Adenovirus Competitively Consumes OncomiRs to Exert Antitumor Efficacy in Hepatocellular Carcinoma. Mol. Cancer Ther. 2016, 15, 1436–1451. [Google Scholar] [CrossRef] [PubMed]
- Moshiri, F.; Callegari, E.; D’Abundo, L.; Corrà, F.; Lupini, L.; Sabbioni, S.; Negrini, M. Inhibiting the oncogenic mir-221 by microRNA sponge: Toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol. Hepatol. Bed. Bench. 2014, 7, 43–54. [Google Scholar] [PubMed]
- Ylosmaki, E.; Hakkarainen, T.; Hemminki, A.; Visakorpi, T.; Andino, R.; Saksela, K. Generation of a Conditionally Replicating Adenovirus Based on Targeted Destruction of E1A mRNA by a Cell Type-Specific MicroRNA. J. Virol. 2008, 82, 11009–11015. [Google Scholar] [CrossRef] [PubMed]
- Girard, M.; Jacquemin, E.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. miR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 2008, 48, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Qi, M.; Kim, A.S.; Lizhar, E.M.; Sun, O.W.; Al-Abdullah, I.H.; Riggs, A.D. Reassessing the Abundance of miRNAs in the Human Pancreas and Rodent Cell Lines and Its Implication. Non-Coding RNA 2023, 9, 20. [Google Scholar] [CrossRef]
- Aldrak, N.; Alsaab, S.; Algethami, A.; Bhere, D.; Wakimoto, H.; Shah, K.; Alomary, M.N.; Zaidan, N. Oncolytic Herpes Simplex Virus-Based Therapies for Cancer. Cells 2021, 10, 1541. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.; Rabkin, S.D. Designing herpes viruses as oncolytics. Mol. Ther. Oncolytics 2015, 2, 15010. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves First Oncolytic Virus Therapy. Oncol. Times UK 2015, 37, 36. [CrossRef]
- Xu, L.; Sun, H.; Lemoine, N.R.; Xuan, Y.; Wang, P. Oncolytic vaccinia virus and cancer immunotherapy. Front. Immunol. 2024, 14, 1324744. [Google Scholar] [CrossRef] [PubMed]
- Kaynarcalidan, O.; Mascaraque, S.M.; Drexler, I. Vaccinia Virus: From Crude Smallpox Vaccines to Elaborate Viral Vector Vaccine Design. Biomedicines 2021, 9, 1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Luo, H. Development of Group B Coxsackievirus as an Oncolytic Virus: Opportunities and Challenges. Viruses 2021, 13, 1082. [Google Scholar] [CrossRef] [PubMed]
- Au, G.G.; Beagley, L.G.; Haley, E.S.; Barry, R.D.; Shafren, D.R. Oncolysis of malignant human melanoma tumors by Coxsackieviruses A13, A15 and A18. Virol. J. 2011, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Bradley, S.; Jakes, A.D.; Harrington, K.; Pandha, H.; Melcher, A.; Errington-Mais, F. Applications of coxsackievirus A21 in oncology. Oncolytic Virother. 2014, 3, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Geisler, A.; Hazini, A.; Heimann, L.; Kurreck, J.; Fechner, H. Coxsackievirus B3—Its Potential as an Oncolytic Virus. Viruses 2021, 13, 718. [Google Scholar] [CrossRef] [PubMed]
- Sin, J.; Mangale, V.; Thienphrapa, W.; Gottlieb, R.A.; Feuer, R. Recent progress in understanding coxsackievirus replication, dissemination, and pathogenesis. Virology 2015, 484, 288–304. [Google Scholar] [CrossRef]
- Ruiz, A.J.; Russell, S.J. MicroRNAs and oncolytic viruses. Curr. Opin. Virol. 2015, 13, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Sutton, P. Treatment of cancer by infectious nucleic acid. Lancet 1991, 337, 1553. [Google Scholar] [CrossRef] [PubMed]
- Hadac, E.M.; Kelly, E.J.; Russell, S.J. Myeloma Xenograft Destruction by a Nonviral Vector Delivering Oncolytic Infectious Nucleic Acid. Mol. Ther. 2011, 19, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Lewthwaite, P.; Perera, D.; Cardosa, M.J.; McMinn, P.; Ooi, M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010, 10, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Carocci, M.; Bakkali-Kassimi, L. The encephalomyocarditis virus. Virulence 2012, 3, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Fazakerley, J.K.; Cotterill, C.L.; Lee, G.; Graham, A. Virus tropism, distribution, persistence and pathology in the corpus callosum of the Semliki Forest virus-infected mouse brain: A novel system to study virus–oligodendrocyte interactions. Neuropathol. Appl. Neurobiol. 2006, 32, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Bailey, K.; Fielding, A. Measles to the Rescue: A Review of Oncolytic Measles Virus. Viruses 2016, 8, 294. [Google Scholar] [CrossRef]
- Di Leva, G.; Croce, C.M. miRNA profiling of cancer. Curr. Opin. Genet. Dev. 2013, 23, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Chen, Y.; Leong, K.W. MicroRNA delivery for regenerative medicine. Adv. Drug Deliv. Rev. 2015, 88, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Curtin, C.M.; Castaño, I.M.; O’Brien, F.J. Scaffold-Based microRNA Therapies in Regenerative Medicine and Cancer. Adv. Healthc. Mater. 2017, 7, 695. [Google Scholar] [CrossRef] [PubMed]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Chen, H.-Y.; Hao, N.-B.; Tang, B.; Guo, H.; Yong, X.; Dong, H.; Yang, S.-M. microRNA inhibitors: Natural and artificial sequestration of microRNA. Cancer Lett. 2017, 407, 139–147. [Google Scholar] [CrossRef] [PubMed]
- St-Cyr, G.; Penarroya, D.; Daniel, L.; Giguère, H.; Alkayyal, A.A.; Tai, L.-H. Remodeling the tumor immune microenvironment with oncolytic viruses expressing miRNAs. Front. Immunol. 2023, 13, 1071223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, L.; Xu, T.; Zhang, W.; Li, J.; Rao, N.; Le, T.D. Time to infer miRNA sponge modules. Wiley Interdiscip. Rev. RNA 2021, 13, e1686. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Rauschhuber, C.; Mueck-Haeusl, M.; Zhang, W.; Nettelbeck, D.M.; Ehrhardt, A. RNAi suppressor P19 can be broadly exploited for enhanced adenovirus replication and microRNA knockdown experiments. Sci. Rep. 2013, 3, 1363. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.Gov. Available online: https://Clinicaltrials.Gov (accessed on 25 June 2024).
- Elicera Therapeutics Continues Phase I/IIa Study with Oncolytic Virus as Planned, Following Safety Review in Cohort 3. Available online: https://www.elicera.com/Press-Releases/Elicera-Therapeutics-Continues-Phase-i-Iia-Study-with-Oncolytic-Virus-as-Planned-Following-Safety-Review-in-Cohort-3 (accessed on 25 June 2024).
- Yu, D.; Leja-Jarblad, J.; Loskog, A.; Hellman, P.; Giandomenico, V.; Oberg, K.; Essand, M. Preclinical Evaluation of AdVince, an Oncolytic Adenovirus Adapted for Treatment of Liver Metastases from Neuroendocrine Cancer. Neuroendocrinology 2017, 105, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Nettelbeck, D.M.; Leber, M.F.; Altomonte, J.; Angelova, A.; Beil, J.; Berchtold, S.; Delic, M.; Eberle, J.; Ehrhardt, A.; Engeland, C.E.; et al. Virotherapy in Germany—Recent Activities in Virus Engineering, Preclinical Development, and Clinical Studies. Viruses 2021, 13, 1420. [Google Scholar] [CrossRef] [PubMed]
- The Victory NET Foundation. Available online: https://www.victorynet.org/de/ (accessed on 25 June 2024).
- Haines, B.B.; Denslow, A.; Grzesik, P.; Lee, J.S.; Farkaly, T.; Hewett, J.; Wambua, D.; Kong, L.; Behera, P.; Jacques, J.; et al. ONCR-177, an Oncolytic HSV-1 Designed to Potently Activate Systemic Antitumor Immunity. Cancer Immunol. Res. 2021, 9, 291–308. [Google Scholar] [CrossRef] [PubMed]
- Park, J.C.; Soliman, H.; Falchook, G.; Owonikoko, T.; Spreafico, A.; Massarelli, E.; McKean, M.; Chow, L.; Ott, P.; Wesolowski, R.; et al. 511 Initial Results of a Phase 1 Study of Intratumoral ONCR-177, an Oncolytic Herpes-Simplex Virus-1 Expressing Five Immunomodulatory Transgenes, in Subjects with Advanced Injectable Tumors. In Proceedings of the Regular and Young Investigator Award Abstracts; BMJ Publishing Group Ltd.: London, UK, 2021; p. A542. [Google Scholar]
- Oncorus Announces Portfolio Reprioritization to Focus on IV-Administered, Self-Amplifying RNA Medicines for Patients with Cancer. Available online: https://investors.oncorus.com/Node/7976/Pdf (accessed on 25 June 2024).
- Shen, Y.; Rahimian, S.; Qin, A.; Qiu, Y.; Fang, T.; Liang, X.; Li, Y.; Tan, Q.; Zhao, R.; Bai, X.; et al. 83P The updated report of phase I trial of VG2025, a non-attenuated HSV-1 oncolytic virus expressing IL-12 and IL-15/RA payloads, in patients with advanced solid tumors. Ann. Oncol. 2023, 34, S1499. [Google Scholar] [CrossRef]
- Fu, S.; Kundranda, M.N.; Rahimian, S.; Shen, Y.; Tan, Q.; Zhao, R.; Esmaeili, N.; Singh, M.; Ding, J.; Murad, Y.; et al. The initial report of phase I trial of VG2025, a non-attenuated HSV-1 oncolytic virus expressing IL-12 and IL-15/RA payloads, in patients with advanced solid tumors. J. Clin. Oncol. 2023, 41, 2580. [Google Scholar] [CrossRef]
- Zhang, W.-H.; Jiang, L.; Li, M.; Liu, J. MicroRNA-124: An emerging therapeutic target in central nervous system disorders. Exp. Brain Res. 2023, 241, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Ching, A.-S.; Ahmad-Annuar, A. A Perspective on the Role of microRNA-128 Regulation in Mental and Behavioral Disorders. Front. Cell. Neurosci. 2015, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Budi, H.S.; Younus, L.A.; Lafta, M.H.; Parveen, S.; Mohammad, H.J.; Al-Qaim, Z.H.; Jawad, M.A.; Parra, R.M.R.; Mustafa, Y.F.; Alhachami, F.R.; et al. The role of miR-128 in cancer development, prevention, drug resistance, and immunotherapy. Front. Oncol. 2023, 12, 1067974. [Google Scholar] [CrossRef] [PubMed]
- Lanza, M.; Cuzzocrea, S.; Oddo, S.; Esposito, E.; Casili, G. The Role of miR-128 in Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 6024. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, X.; Zeng, X.; Peng, X. The Role of Mir-148a in Cancer. J. Cancer 2016, 7, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ko, Y.; Park, H.; Zhang, H.; Jeong, Y.; Kim, Y.; Noh, M.; Park, S.; Kim, Y.-M.; Kwon, Y.-G. MicroRNA-148a/b-3p regulates angiogenesis by targeting neuropilin-1 in endothelial cells. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, Y.; Dang, H.; Wu, X. MicroRNA-148a-3p inhibits the proliferation of cervical cancer cells by regulating the expression levels of DNMT1 and UTF1. Oncol. Lett. 2021, 22, 617. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Ovcharenko, D.; Grossmann, R.; Lauková, M.; Mlynček, M. Identification of MicroRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J. Cell. Physiol. 2009, 219, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, G.; Jiang, Y. The Emerging Roles of miR-125b in Cancers. Cancer Manag. Res. 2020, 12, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Latreille, M.; Hausser, J.; Stützer, I.; Zhang, Q.; Hastoy, B.; Gargani, S.; Kerr-Conte, J.; Pattou, F.; Zavolan, M.; Esguerra, J.L.; et al. MicroRNA-7a regulates pancreatic β cell function. J. Clin. Investig. 2014, 124, 2722–2735. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhou, Y.; Guo, M.; Yue, D.; Chen, C.; Liang, G.; Xu, L. MicroRNA-7: Expression and function in brain physiological and pathological processes. Cell Biosci. 2020, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhang, T.; Kusumanchi, P.; Huda, N.; Jiang, Y.; Liangpunsakul, S.; Yang, Z. Role of microRNA-7 in liver diseases: A comprehensive review of the mechanisms and therapeutic applications. J. Investig. Med. 2020, 68, 1208–1216. [Google Scholar] [CrossRef]
- Jopling, C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Elia, L.; Quintavalle, M.; Zhang, J.; Contu, R.; Cossu, L.; Latronico, M.V.G.; Peterson, K.L.; Indolfi, C.; Catalucci, D.; Chen, J.; et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ. 2009, 16, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Vacante, F.; Denby, L.; Sluimer, J.C.; Baker, A.H. The function of miR-143, miR-145 and the MiR-143 host gene in cardiovascular development and disease. Vascul. Pharmacol. 2019, 112, 24–30. [Google Scholar] [CrossRef]
- Guo, H.; Chen, Y.; Hu, X.; Qian, G.; Ge, S.; Zhang, J. The regulation of toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol. Cancer 2013, 12, 77–100. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lu, Y.; Li, Z.; Wang, Q. microRNA-133: Expression, Function and Therapeutic Potential in Muscle Diseases and Cancer. Curr. Drug Targets 2014, 15, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Erener, S.; Ellis, C.E.; Ramzy, A.; Glavas, M.M.; O’dwyer, S.; Pereira, S.; Wang, T.; Pang, J.; Bruin, J.E.; Riedel, M.J.; et al. Deletion of pancreas-specific miR-216a reduces beta-cell mass and inhibits pancreatic cancer progression in mice. Cell Rep. Med. 2021, 2, 100434. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, A.A.; Taghehchian, N.; Zangouei, A.S.; Akhlaghipour, I.; Maharati, A.; Basirat, Z.; Moghbeli, M. Molecular mechanisms of microRNA-216a during tumor progression. Cancer Cell Int. 2023, 23, 19. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Hanazawa, T.; Nohata, N.; Okamoto, Y.; Seki, N. The functional significance of microRNA-375 in human squamous cell carcinoma: Aberrant expression and effects on cancer pathways. J. Hum. Genet. 2012, 57, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Bhinge, A.; Namboori, S.C.; Bithell, A.; Soldati, C.; Buckley, N.J.; Stanton, L.W. MiR-375 is Essential for Human Spinal Motor Neuron Development and May Be Involved in Motor Neuron Degeneration. Stem Cells 2015, 34, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Lu, Y.; Wang, R.; Xu, X.; Liu, Q.; He, S.; Pan, H.; Liu, X.; Yuan, B.; Ding, Y.; et al. MicroRNA-375: Potential cancer suppressor and therapeutic drug. Biosci. Rep. 2021, 41, 1494. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y.; Zhang, B.; Xiong, L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci. Rep. 2014, 4, 5150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-G.; Zheng, J.-N.; Pei, D.-S. P53/microRNA-34-induced metabolic regulation: New opportunities in anticancer therapy. Mol. Cancer 2014, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, X.; He, J.; Cao, Q.; Du, D.; Zhan, X.; Zeng, Y.; Yuan, S.; Sun, L. The comprehensive landscape of miR-34a in cancer research. Cancer Metastasis Rev. 2021, 40, 925–948. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Wang, Y.; Li, Y.; Cui, L.; Zhao, Y.; Zhao, B.; Li, K. MiR-206, a Key Modulator of Skeletal Muscle Development and Disease. Int. J. Biol. Sci. 2015, 11, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Khalilian, S.; Imani, S.Z.H.; Ghafouri-Fard, S. Emerging roles and mechanisms of miR-206 in human disorders: A comprehensive review. Cancer Cell Int. 2022, 22, 412. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Y.; Sun, X. The functions of microRNA-208 in the heart. Diabetes Res. Clin. Pract. 2020, 160, 108004. [Google Scholar] [CrossRef] [PubMed]
- Chapnik, E.; Rivkin, N.; Mildner, A.; Beck, G.; Pasvolsky, R.; Metzl-Raz, E.; Birger, Y.; Amir, G.; Tirosh, I.; Porat, Z.; et al. miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. eLife 2014, 3, e01964. [Google Scholar] [CrossRef] [PubMed]
- Zareifar, P.; Ahmed, H.M.; Ghaderi, P.; Farahmand, Y.; Rahnama, N.; Esbati, R.; Moradi, A.; Yazdani, O.; Sadeghipour, Y. miR-142-3p/5p role in cancer: From epigenetic regulation to immunomodulation. Cell Biochem. Funct. 2024, 42, e3931. [Google Scholar] [CrossRef] [PubMed]
- Paterson, M.R.; Kriegel, A.J. MiR-146a/b: A family with shared seeds and different roots. Physiol. Genom. 2017, 49, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Rusca, N.; Monticelli, S. MiR-146a in Immunity and Disease. Mol. Biol. Int. 2011, 2011, 437301. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-F.; Zhang, L.; Waye, M.M.Y.; Fu, W.-M.; Zhang, J.-F. MiR-218 Mediates tumorigenesis and metastasis: Perspectives and implications. Exp. Cell Res. 2015, 334, 173–182. [Google Scholar] [CrossRef]
- Schell, G.; Roy, B.; Prall, K.; Dwivedi, Y. miR-218: A Stress-Responsive Epigenetic Modifier. Non-Coding RNA 2022, 8, 55. [Google Scholar] [CrossRef]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, B.; Sun, H.; Zhao, X.; Wang, X.; Zhao, N.; Zhang, Y.; Li, Y.; Gu, Q.; Liu, F.; et al. Regulation of proliferation, angiogenesis and apoptosis in hepatocellular carcinoma by miR-26b-5p. Tumor Biol. 2016, 37, 10965–10979. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-K.; Lin, H.-Y.; Dou, X.-W.; Chen, M.; Wei, X.-L.; Zhang, Y.-Q.; Wu, Y.; Chen, C.-F.; Bai, J.-W.; Xiao, Y.-S.; et al. MiR-221/222 promote epithelial-mesenchymal transition by targeting Notch3 in breast cancer cell lines. npj Breast Cancer 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.T.; Arbitrio, M.; Caracciolo, D.; Cordua, A.; Cuomo, O.; Grillone, K.; Riillo, C.; Caridà, G.; Scionti, F.; Labanca, C.; et al. miR-221/222 as biomarkers and targets for therapeutic intervention on cancer and other diseases: A systematic review. Mol. Ther. Nucleic Acids 2022, 27, 1191–1224. [Google Scholar] [CrossRef] [PubMed]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Tao, X.; Gao, F.; Fan, C.; Wu, D. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol. Lett. 2012, 4, 483–488. [Google Scholar] [CrossRef]
- Chen, W.; Fan, X.-M.; Mao, L.; Zhang, J.-Y.; Li, J.; Wu, J.-Z.; Tang, J.-H. MicroRNA-224: As a potential target for miR-based therapy of cancer. Tumor Biol. 2015, 36, 6645–6652. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wu, X.; Xiang, S.; Qiao, M.; Cen, X.; Pan, X.; Huang, X.; Zhao, Z. Regulatory mechanism of miR-20a-5p expression in Cancer. Cell Death Discov. 2022, 8, 262. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.K. miR-106b as an emerging therapeutic target in cancer. Genes Dis. 2021, 9, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Li, J.; Li, Y.; Yang, M.; Nie, S.; Zhou, M.; Zhou, Z.; Yang, X.; Liu, Y.; Hou, F.F. MicroRNA-10 negatively regulates inflammation in diabetic kidney via targeting activation of the NLRP3 inflammasome. Mol. Ther. 2021, 29, 2308–2320. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Shi, Y.; Liu, Z.; Li, Z.; Xu, W. The emerging role of miR-10 family in gastric cancer. Cell Cycle 2021, 20, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, A.; Derakhshan, S.M.; Ghavi, D.; Foruzandeh, Z.; Hashemi, S. The role of mir-151a-5p in tumorigenesis; A systematic review. Pathol. Res. Pract. 2023, 249, 154576. [Google Scholar] [CrossRef] [PubMed]
- Mashima, R. Physiological roles of miR-155. Immunology 2015, 145, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, R.; Van Roosbroeck, K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2017, 37, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Indrieri, A.; Carrella, S.; Carotenuto, P.; Banfi, S.; Franco, B. The Pervasive Role of the miR-181 Family in Development, Neurodegeneration, and Cancer. Int. J. Mol. Sci. 2020, 21, 2092. [Google Scholar] [CrossRef]
- Rezaei, T.; Amini, M.; Hashemi, Z.S.; Mansoori, B.; Rezaei, S.; Karami, H.; Mosafer, J.; Mokhtarzadeh, A.; Baradaran, B. microRNA-181 serves as a dual-role regulator in the development of human cancers. Free Radic. Biol. Med. 2019, 152, 432–454. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, M.; Rezaee, D.; Fakhari, F.; Najafi, S.; Aghaei-Zarch, S.M.; Beyranvand, P.; Rashidi, M.A.; Bagheri-Mohammadi, S.; Zamani-Rarani, F.; Bakhtiari, M.; et al. microRNA-184 in the landscape of human malignancies: A review to roles and clinical significance. Cell Death Discov. 2023, 9, 423. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Deng, W.; Zhao, Q.; Zhuang, H.; Zhang, C.; Jian, Z. miR-501 acts as an independent prognostic factor that promotes the epithelial-mesenchymal transition through targeting JDP2 in hepatocellular carcinoma. Hum. Cell 2019, 32, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Xing, T.; Gao, X.; Liu, F. miR-501-3p promotes colorectal cancer progression via activation of Wnt/β-catenin signaling. Int. J. Oncol. 2019, 55, 671–683. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chen, S.; Ying, Y.; Xie, H.; Li, J.; Ma, X.; Wang, W.; Shen, H.; Wang, X.; Zheng, X.; et al. MicroRNA-501-3p inhibits the proliferation of kidney cancer cells by targeting WTAP. Cancer Med. 2021, 10, 7222–7232. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.; Katz, S.; Bally-Cuif, L. miR-9: A versatile regulator of neurogenesis. Front. Cell. Neurosci. 2013, 7, 220. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Tan, H.-Y.; Feng, Y.-G.; Zhang, C.; Chen, F.; Feng, Y. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers 2018, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Yao, X.; Lin, Y.; Zhang, D.; Cui, R.; Zhang, X. Interactive functions of microRNAs in the miR-23a-27a-24-2 cluster and the potential for targeted therapy in cancer. J. Cell. Physiol. 2020, 235, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-Y.; Zhang, Q.; Wang, D.-D.; Yan, W.; Sha, H.-H.; Zhao, J.-H.; Yang, S.-J.; Zhang, H.-D.; Hou, J.-C.; Xu, H.-Z.; et al. MiR-29a: A potential therapeutic target and promising biomarker in tumors. Biosci. Rep. 2018, 38, 1265. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of miR-29 in Cancer. Mol. Ther. Oncolytics 2018, 12, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Yang, Y.-L.; Wang, F.-S. The Role of miR-29a in the Regulation, Function, and Signaling of Liver Fibrosis. Int. J. Mol. Sci. 2018, 19, 1889. [Google Scholar] [CrossRef] [PubMed]
- Shad, A.N.; Fanoodi, A.; Maharati, A.; Akhlaghipour, I.; Moghbeli, M. Molecular mechanisms of microRNA-301a during tumor progression and metastasis. Pathol. Res. Pract. 2023, 247, 154538. [Google Scholar] [CrossRef]
- Wei, F.; Cao, C.; Xu, X.; Wang, J. Diverse functions of miR-373 in cancer. J. Transl. Med. 2015, 13, 162. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ye, B.; Wang, P.; Yao, F.; Zhang, C.; Yu, G. Overview of microRNA-199a Regulation in Cancer. Cancer Manag. Res. 2019, 11, 10327–10335. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-Y.M.; Obrietan, K. Revealing a Role of MicroRNAs in the Regulation of the Biological Clock. Cell Cycle 2007, 6, 3034–3038. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Li, H.; Huang, H.; Qiu, M. MicroRNAs and Glial Cell Development. Neurosci. 2011, 18, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Hong, L.; Chen, Z.; Nie, Y.; Fan, D. Epigenetic Regulation of microRNAs in Gastric Cancer. Dig. Dis. Sci. 2014, 59, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhu, C.; Pang, Q.; Liu, H.C. MicroRNA-217: A regulator of human cancer. Biomed. Pharmacother. 2021, 133, 110943. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, F.; Gopalan, V.; Smith, R.A.; Lam, A.K.-Y. miR-126 in human cancers: Clinical roles and current perspectives. Exp. Mol. Pathol. 2014, 96, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Soofiyani, S.R.; Hosseini, K.; Ebrahimi, T.; Forouhandeh, H.; Sadeghi, M.; Beirami, S.M.; Ghasemnejad, T.; Tarhriz, V.; Montazersaheb, S. Prognostic Value and Biological Role of miR-126 in Breast Cancer. MicroRNA 2022, 11, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, E.; Cairns, M.J. MiR-137: An important player in neural development and neoplastic transformation. Mol. Psychiatry 2016, 22, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Pan, H.; Li, R. The dual regulatory role of miR-204 in cancer. Tumor Biol. 2016, 37, 11667–11677. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Bian, Z.; Xu, P.; Sun, S.; Huang, Z. MicroRNA-204-5p: A pivotal tumor suppressor. Cancer Med. 2023, 12, 3185–3200. [Google Scholar] [CrossRef] [PubMed]

| Virus | Viral Family | Oncolytic Virus | Genome | Target Cancer Type | Type of miR-Associated Modification | miR | Features | Combo Agent 1 | Combo Agent 2 | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Coxsackievirus B3 | Picornaviridae | PD-SK | Linear ss(+)RNA 1 | Colorectal cancer | Target for miR | miRT-375 in the 3′ UTR of the genome | - | - | - | [132] |
| Coxsackievirus B3 | Picornaviridae | miR-CVB3 | Linear ss(+)RNA | Breast cancer and melanoma | Target for miR | Four copies of miRT-145, two copies of miRT-143, two copies of miR-1, and four copies of miR-216 in the 5′UTR of the CVB3 genome | - | Pore-forming lytic peptide melittin | Synthetic Toll-like receptor 9 ligand-CpG oligodeoxynucleotides | [123] |
| Coxsackievirus B3 | Picornaviridae | miR-CVB3-1.1 | Linear ss(+)RNA | Breast cancer and small-cell lung cancer | Target for miR | Four copies of miRT-145, two copies of miRT-143, two copies of miR-1, and four copies of miR-216 in the 5′UTR of the CVB3 genome | - | - | - | [124] |
| Coxsackievirus B3 | Picornaviridae | PD-H-375TS | Linear ss(+)RNA | Colorectal cancer | Target for miR | Two copies of miRT-375 in the 3′UTR of the OV’s genome | - | - | - | [119] |
| Coxsackievirus B3 | Picornaviridae | H3N-375TS with miRT-375 and H3N-375/1TS with miRT-375 and miRT-1 | Linear ss(+)RNA | Colorectal carcinoma | Target for miR | miRT-375 and miRT-1 | - | - | - | [120] |
| Coxsackievirus B3 | Picornaviridae | CVB3-HP | Linear ss(+)RNA | Triple-negative breast cancer (TNBC) | Target for miR | miRT-1 and miRT-217 in the 3′UTR of the viral genome | - | - | - | [118] |
| Coxsackievirus B3 | Picornaviridae | Sveveral viruses | Linear ss(+)RNA | Lung adenocarcinoma | Target for miR | Four copies of miRT-145 and two copies of miRT-143 in either the 5′ UTR or 3′ UTR of the CVB3 genome | - | - | - | [122] |
| Coxsackievirus A21 | Picornaviridae | Several viruses | Linear ss(+)RNA | Melanoma | Target for miR | Two copies each of sequences completely complementary to miR-133 and miR-206, each separated by 4–6 nt long linkers in different positions of the viral genome | - | - | - | [125] |
| Coxsackievirus B3 | Picornaviridae | Several viruses | Linear ss(+)RNA | Lung cancer | Target for miR | Four tandem miRT-34a and/or miRT-34c in the 5′ UTR or 3′ UTR of the CVB3 genome | - | - | - | [117] |
| Coxsackievirus B3 | Picornaviridae | Several viruses | Linear ss(+)RNA | Non-specific | Target for miR | miRT-133 and miRT-206 in different positions of the viral genome | - | - | - | [121] |
| Adenovirus | Adenoviridae | Several viruses | Linear dsDNA 2 | Hepatocellular carcinoma | Target for miR | miRT-21, miRT-199-3p, and miRT-142 | GM-CSF, IL-2, and single-chain variable fragments (scFvs) against either programmed death-1 (PD-1) or programmed death-ligand 1 (PD-L1) | - | - | [34] |
| Adenovirus | Adenoviridae | Ad5-5miR145T and Ad5-10miR145T | Linear dsDNA | Breast cancer | Target for miR | Five or ten copies of miRT-145-5p downstream of E1A gene | GFP | - | - | [108] |
| Adenovirus | Adenoviridae | Conditionally replicative adenovirus (CRAd) l | Linear dsDNA | Hepatocellular carcinoma | Target for miR | miRNA-122 in 3′UTR of the E1A gene | Adenovirus loaded on human umbilical cord-derived mesenchymal stem cells, with E1A gene dual regulated by α-fetoprotein promoter and microRNA-122 target sequence. | - | - | [32] |
| Adenovirus | Adenoviridae | OAV-scIL12-miR, OAV-scIL12-GPI, and OAV-scIL12-TM | Linear dsDNA | Pancreatic cancer metastatic to the liver | Target for miR | miRT-122 in the 3′ UTR of the scIL-12 encoding sequence | scIL-12 encoding gene; OAV-scIL12-GPI contained GPI anchor signal from the folate receptor; OAV-scIL12-TM contained TM (transmembrane) domain from human CD4 | - | - | [133] |
| Adenovirus | Adenoviridae | Ad-L5-8miR148aT | Linear dsDNA | Pancreatic cancer | Target for miR | Eight miRT-148a of perfect complementarity in the 3′UTR of the L5 coding sequence | - | - | - | [114] |
| Adenovirus | Adenoviridae | Several viruses | Linear dsDNA | Pancreatic ductal adenocarcinoma | Target for miR | miRT-148a and miRT-216a in the 3′UTR of the E1A gene | - | - | - | [107] |
| Adenovirus | Adenoviridae | A-4MREs | Linear dsDNA | Glioma | Target for miR | Two copies each of miRT-124, miRT-128, miRT-146b, and miRT-218 in the 3′UTR of the E1A gene | - | - | - | [111] |
| Herpes simplex virus | Herpesviridae | VG2025 | Linear dsDNA | Non-specific | Target for miR | Tandem repeats of binding sites with perfect complementarity to miR-124 and miR-143 in 3′UTR of the ICP34.5 gene. | Truncated gB to enhance fusogenicity, control of ICP27 by CEA promoter; IL-12, IL-15, and the IL-15Rα subunit, CXCR4 promoter driving cytokine expression | - | - | [134] |
| Herpes simplex virus | Herpesviridae | ONCR-159 | Linear dsDNA | Non-specific | Target for miR | miRT-128, miRT-204, and miRT-219a in 3′ UTR of the ICP34.5 gene; miRT-217, miRT-137, and miRT-126 in 3′ UTR of the UL8 gene; miRT-128, miRT-219a, and miRT-122 in 3′UTR of the ICP27 gene; and miRT-124, miRT-1, and miRT-143 in 3′UTR of the ICP4 gene | Deletion of the joint region, the introduction of a gateway recombination cassette in the UL3/UL4 intergenic region, a null mutation in US12, the mutation of amino acids D285N and A549T in the fusogenic glycoprotein B (gB), null mutations in ICP47, and mutations in UL37 | - | - | [110] |
| Herpes simplex virus | Herpesviridae | 3Ldn9T21 | Linear dsDNA | Non-specific | Targets for miR: miR induces viral replication | miRT-21in UL9-C535C 3′UTR | - | - | - | [135] |
| Herpes simplex virus | Herpesviridae | SU4-124 HSV-1 | Linear dsDNA | Glioma | Target for miR | Five copies of miRT-124 in 3′UTR of the ICP4 gene | ICP4 was placed under a tumor-specific survivin promoter and a 5′UTR of rat fibroblast growth factor 2 was added in front of the viral ICP4 gene | - | - | [33] |
| Herpes simplex virus | Herpesviridae | Several HSV-1 viruses | Linear dsDNA | Urothelial bladder cancer | Target for miR | Five copies each miRT-143 and/or miR-124 in 3′UTR of the ICP4 gene | - | - | - | [112] |
| Herpes simplex virus | Herpesviridae | KG4:T124, KGE-4:T124 | Linear dsDNA | Glioblastoma multiforme | Target for miR | Four copies of miRT-124 in the 3′ UTR of ICP4 gene | Fusion of the complete gC ORF to GFP via a 2A peptide sequence; for KGE-4:T124, the introduction of scFv antibodies specific to EGFR | - | - | [136] |
| Semliki Forest virus | Togaviridae | SFV-AM6-124T | Linear ss(+)RNA | Glioblastoma | Target for miR | Six copies of miRT-124 between the viral nonstructural protein 3 and 4 (NSP3 and NSP4) genes | 4 aa substitutions as compared to the parental SFV4; that is, nsP3A70G, nsP3Y369F, nsP4R611K, and E2K162E | Anti-PD1 therapy | - | [24] |
| Semliki Forest virus | Togaviridae | SFV4B18RmiRT | Linear ss(+)RNA | Glioblastoma | Target for miR | Two copies of each miRT-124, miRT-125, and miRT-134 in the beginning of 3′UTR of the SFV genome | B18R gene coding for a decoy receptor, which binds to type-I IFN | - | - | [137] |
| Semliki Forest virus | Togaviridae | SFV4miRT | Linear ss(+)RNA | Glioblastoma multiforme and high-risk neuroblastoma | Target for miR | Two copies of each miRT-124, miRT-125, and miRT-134 in the beginning of 3′UTR of the SFV genome | - | - | - | [91] |
| Semliki Forest virus | Togaviridae | SFV4 | Linear ss(+)RNA | Gliomas | Target for miR | miRT-124 | - | - | - | [138] |
| Measles virus | Paramyxoviridae | MeV-CD-FmiRTS148a | Linear ss(-)RNA | Pancreatic ductal adenocarcinoma | Target for miR | miRT-148a in 3′UTR of the fusion (F) gene | Cytosine deaminase–uracil phosphoribosyl transferase | 5-fluorocytosine (5-FC) | - | [139] |
| Measles virus | Paramyxoviridae | Several viruses | Linear ss(-)RNA | Non-specific | Target for miR | Viruses with target sites for miR-124-3p, miR-125b-5p, and miR-7-5p in the 3′UTRs of either the N, F, H, or L genes | EGFP | - | - | [109] |
| Measles virus | Paramyxoviridae | MV-EGFPmtd (multi-tissue-detargeted) | Linear ss(-)RNA | Pancreatic carcinoma | Target for miR | Three copies of fully complementary miRT-148a in 3′UTR of the F gene; three copies each of fully complementary miRT-122 and miRT-7 in 3′UTR of the hemagglutinin (H) | EGFP | - | - | [113] |
| Mengovirus | Picornaviridae | Several viruses | Linear ss(+)RNA | Myeloma, plasmacytoma | Target for miR | Two copies each of miRT-133b, miRT-208a, and miRT-124 | Varying polyC tract lengths | - | - | [130] |
| Mengovirus | Picornaviridae | vMC24NC, vMC24ΔL | Linear ss(+)RNA | Glioblastoma | Target for miR | Two copies of miRT-124 in the 5′ UTR and two copies each of miRT-133 and miRT-208 in the 3′ UTR of the viral genome | vMC24ΔL-deletion of the leader gene, truncated polyC | - | - | [30] |
| Mengovirus | Picornaviridae | Several viruses | Linear ss(+)RNA | Non-specific | Target for miR | Several combinations of miRT-124, miRT-125, miRT-133, miRT-208, and miRT-142 in 3′UTR and/or 5′UTR | Truncated polyC | - | - | [127] |
| Enterovirus A71 | Picornaviridae | EV-A71-miR124T | Linear ss(+)RNA | Malignant gliomas | Target for miR | Three copies of miR124T in the location between the 5ʹ-UTR and the coding sequences | - | - | - | [140] |
| Vaccinia virus | Poxviridae | ΔTK let-7a-targeted VV | Linear dsDNA | Multiple myeloma | Target for miR | miRT-let-7a in the 3′ UTR of B5R gene | Deleted thymidine-kinase | - | - | [37] |
| Adenovirus | Adenoviridae | Several viruses | Linear dsDNA | Non-specific | miR precursor | miR-1-3p in short hairpin RNA (shRNA), precursor microRNA (pre-miRNA), and primary miRNA (pri-miRNA) format; miR-26b-5p in pri-miRNA format | A 24 bp deletion in the early gene E1A | - | - | [87] |
| Adenovirus | Adenoviridae | ICOVIR15 miR-99b, ICOVIR15 miR-485 | Linear dsDNA | Pancreatic cancer | miR precursor | pri-miR-99b and pri-miR-485—miRNA genomic sequences under the control of the cytomegalovirus promoter | A 24 bp deletion and E2F-responsive elements in the E1A region; the fiber contains an RGD motif inserted in the HI-loop region. | - | - | [85] |
| Adenovirus | Adenoviridae | Ad-ZD55-miR-143 | Linear dsDNA | Colorectal cancer | miR precursor | miR-143 in pri-miRNA format | E1B55kDa encoding gene was deleted | - | - | [141] |
| Adenovirus | Adenoviridae | AdCN205-IL-24-miR-34a | Linear dsDNA | Hepatocellular carcinoma | miR precursor | miR-34a | IL24 gene controlled by the adenovirus endogenous E3 promoter | - | - | [142] |
| VSV | Rhabdoviridae | rVSV-miR143 | Linear ss(-)RNA | Osteosarcoma | miR precursor | miR-143 | - | - | - | [143,144] |
| VSV | Rhabdoviridae | VSV∆51-amiR-4, VSVΔ51-shPD-L1 | Linear ss(-)RNA | Non-specific | miR precursor | Artificial “amiR-4” or hairpin RNA targeting the immune response suppressor PD-L1 | - | GSK126 | - | [38] |
| Measles | Paramyxoviridae | Several viruses | Linear ss(-)RNA | Non-specific | miR precursor | Precursor miR-122 sequence flanked by 50 and 49 nucleotides of pri-miR-122 at its 5′ and 3′ ends, respectively. | - | - | - | [80] |
| Vaccinia virus | Poxviridae | VV-miR-34a | Linear dsDNA | Multiple myeloma | miR precursor | miR-34a in pri-miRNA format inserted into the thymidine kinase (TK) gene | Disrupted sequence of TK | VV encoding SMAC | - | [145] |
| Adenovirus | Adenoviridae | Several viruses | Linear dsDNA | Non-specific | miR inhibitors | Plant-derived RNAi inhibitor P19 under the control of the major late promoter and connected to the fiber coding sequence | A 24 bp deletion in the pRB binding region in the early gene E1A | - | - | [146] |
| Adenovirus | Adenoviridae | AdNuPAR-E-miR222-S | Linear dsDNA | Pancreatic cancer | miR inhibitors | miR-222 sponges | - | - | - | [147] |
| Adenovirus | Adenoviridae | AdSVP-lncRNAi9 | Linear dsDNA | Triple negative breast cancer | miR inhibitors | Artificial lncRNA, which contains 10 copies of the complementary sequences of nine OncomiRs: mir-9-5p, miR10b-5p, miR-21–5p, miR-23a-3p miR-29a-3p, miR-155-5p, miR-222–3p, miR-301a-3p, and miR-373-3p | A 24 bp deletion in the early gene E1A | - | - | [148] |
| Adenovirus | Adenoviridae | AdSVPE1a-lncR | Linear dsDNA | Hepatocellular carcinoma | miR inhibitors | An artificially designed lncRNAi with a tandem of complementary binding sequences to the seed sequences of the 12 OncomiRs: miR21, miR221/222, miR224, miR17-5p/20a, miR10b, miR106b, miR151-5p, miR155, miR181a/181b, miR184, miR1, and miR501-5p. The sequences were repeated six times. | E1A gene under tumor-specific survivin promoter | - | - | [149] |
| Adenovirus and AAV 3 | Adenovirus–Adenoviridae; AAV–Parvoviridae | rAd-199T-miR-221 sponge, rAAV.miR-221 sponge | Adenovirus–linear dsDNA; AAV–linear ssDNA | Hepatocellular carcinoma | miR inhibitors, target for miR | mir-221 sponge with four copies of the miR-221 binding sites and four-nucleotide-long spacers; the adenivirus contained miRT-199 | - | - | - | [150] |
| Virus | Oncolytic Virus Name | miRTs | Features | Clinical Trial (ID) | Monotherapy or Combination | Combo Agent | Cancer Type | Phase | Recruitment Status |
|---|---|---|---|---|---|---|---|---|---|
| Adenovirus | AdVince | miRT-122 in 3′UTR of the E1A gene | Gene promoter from human chromogranin A, a cell-penetrating peptide in the capsid (the protein transduction domain (PTD) from the trans-activator of transcription (Tat) protein of human immunodeficiency virus (HIV)-1) | NCT02749331 | Mono | No | Metastatic neuroendocrine tumors (NETs) | 1/2 | Recruiting |
| HSV | ONCR-177 | miRT-124-3p, miR-T-1-3p, and miR-T-143-3p in 3′UTR of the ICP4 gene; miR-T-128-3p, miR-T-219a-5p, and miR-T-122-5p in 3′UTR of the ICP27 gene; miR-T-217-5p, miR-T-137-3p, and miR-T-126-3p in 3′UTR of the UL8 gene; and miR-T-128-3p, miR-T-204-5p, and miR-T-219a-5p in 3′UTR of the ICP34.5 gene | Genes coding for IL12, FLT3LG, CCL4, PD-1, CTLA-4, and UL37 mutation | NCT04348916 | Combo | Pembrolizumab | Melanoma, solid tumors, head and neck squmous cell carcinoma, and breast cancer | 1 | Terminated |
| HSV | VG2025 | miRT-124 and miRT-143 in 3′UTR of the ICO34.5 gene | CXCR4 promoter driving cytokine expression, truncated gB to enhance fusogenicity, control of ICP27 gene by CEA promoter; IL-12, IL-15, and the IL-15Rα subunit coding sequences | NCT05477849 | Mono | No | Advanced malignant solid tumor | 1 | Recruiting |
| HSV | VG2025 | miRT-124 and miRT-143 in 3′UTR of the ICO34.5 gene | CXCR4 promoter driving cytokine expression, truncated gB to enhance fusogenicity, control of ICP27 gene by CEA promoter; IL-12, IL-15, and the IL-15Rα subunit coding sequences | NCT05266612 | Mono, combo | Nivolumab | Advanced malignant solid tumor | 1 | Recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toropko, M.; Chuvpilo, S.; Karabelsky, A. miRNA-Mediated Mechanisms in the Generation of Effective and Safe Oncolytic Viruses. Pharmaceutics 2024, 16, 986. https://doi.org/10.3390/pharmaceutics16080986
Toropko M, Chuvpilo S, Karabelsky A. miRNA-Mediated Mechanisms in the Generation of Effective and Safe Oncolytic Viruses. Pharmaceutics. 2024; 16(8):986. https://doi.org/10.3390/pharmaceutics16080986
Chicago/Turabian StyleToropko, Mariia, Sergey Chuvpilo, and Alexander Karabelsky. 2024. "miRNA-Mediated Mechanisms in the Generation of Effective and Safe Oncolytic Viruses" Pharmaceutics 16, no. 8: 986. https://doi.org/10.3390/pharmaceutics16080986





