HER-2 Receptor and αvβ3 Integrin Dual-Ligand Surface-Functionalized Liposome for Metastatic Breast Cancer Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
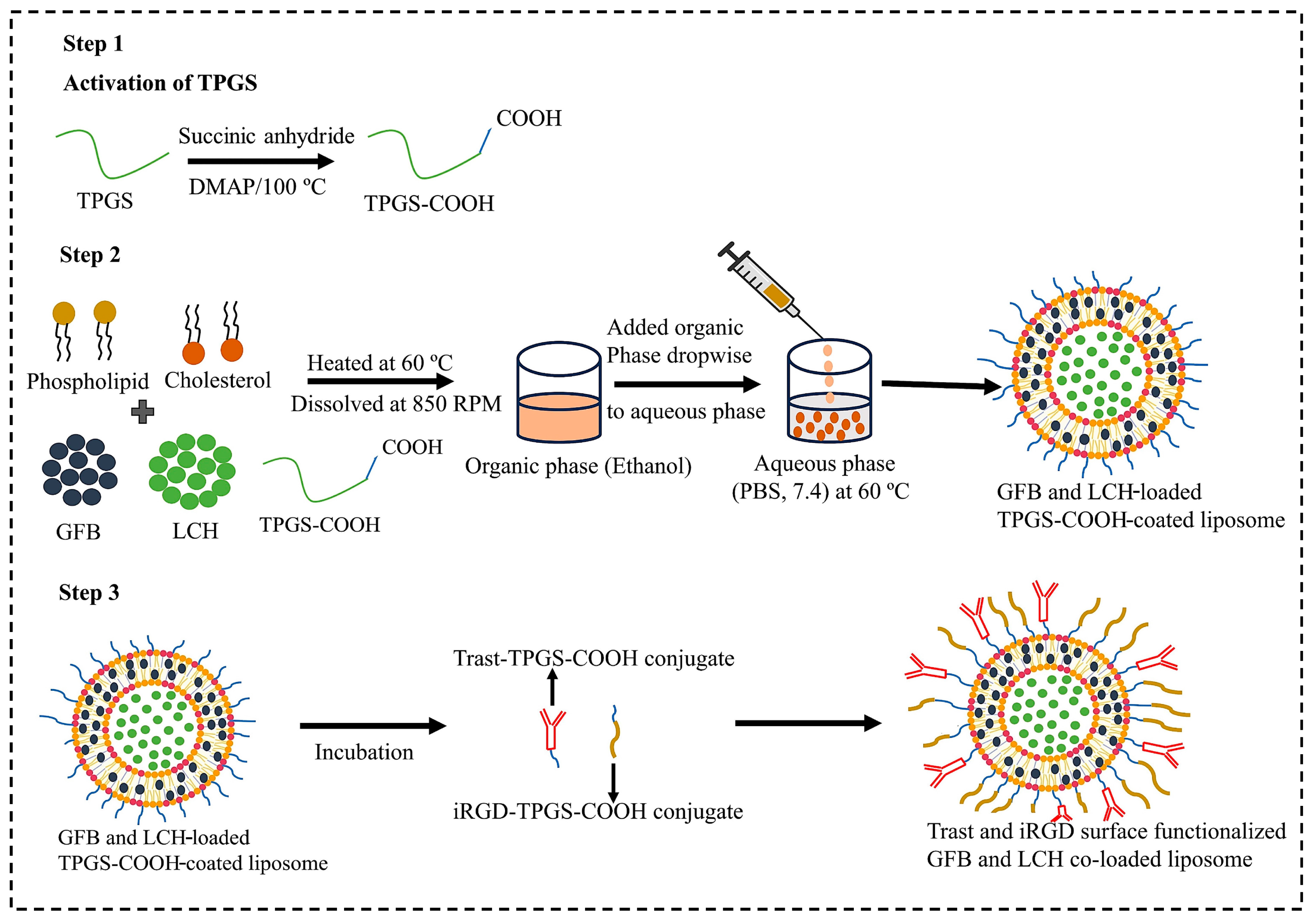
2.2.1. Synthesis of TPGS-COOH
2.2.2. Synthesis of Trast-TPGS-COOH Hybrid Conjugate
2.2.3. iRGD-TPGS-COOH Hybrid Conjugate
2.2.4. Experimental Design and Optimization of GFB and LCH Dual Drug-Loaded Liposome
2.2.5. Surface Functionalization of Optimized Liposome by iRGD and Trast
3. Characterization of DSDLs
3.1. Zeta Analysis
3.2. Scanning Electron Microscopy (SEM) Analysis
3.3. Transmission Electron Microscopy (TEM) Study
3.4. X-ray Diffraction (XRD) Study
3.5. Fourier Transform Infrared Spectrophotometer (FTIR) Analysis
3.6. Determination of Encapsulation Efficiency (EE%) and Drug Loading (DL%)
3.7. Stability Studies
3.8. In-Vitro Drug Release Study
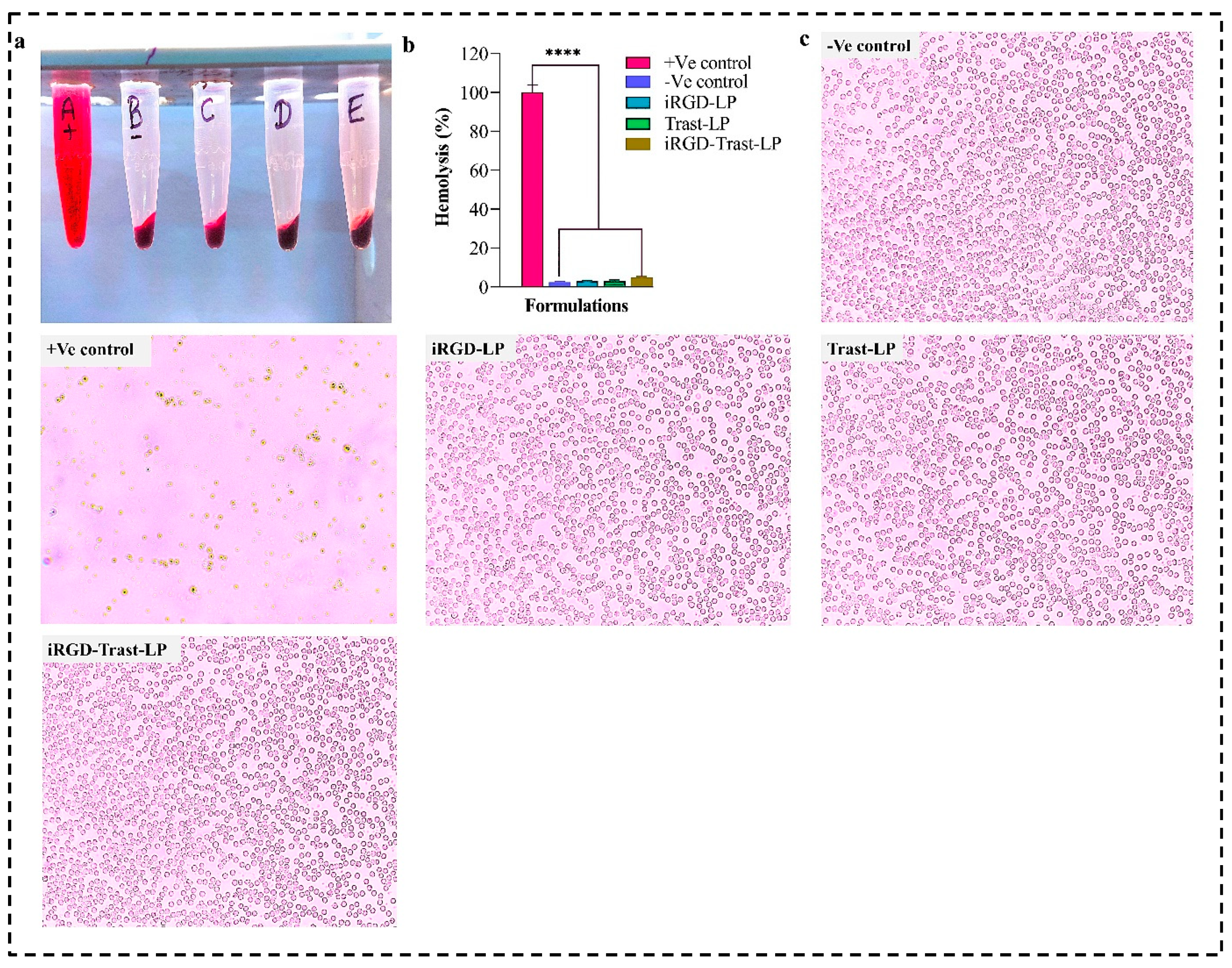
3.9. Blood Compatibility Study
3.10. In-Vitro Cell Study
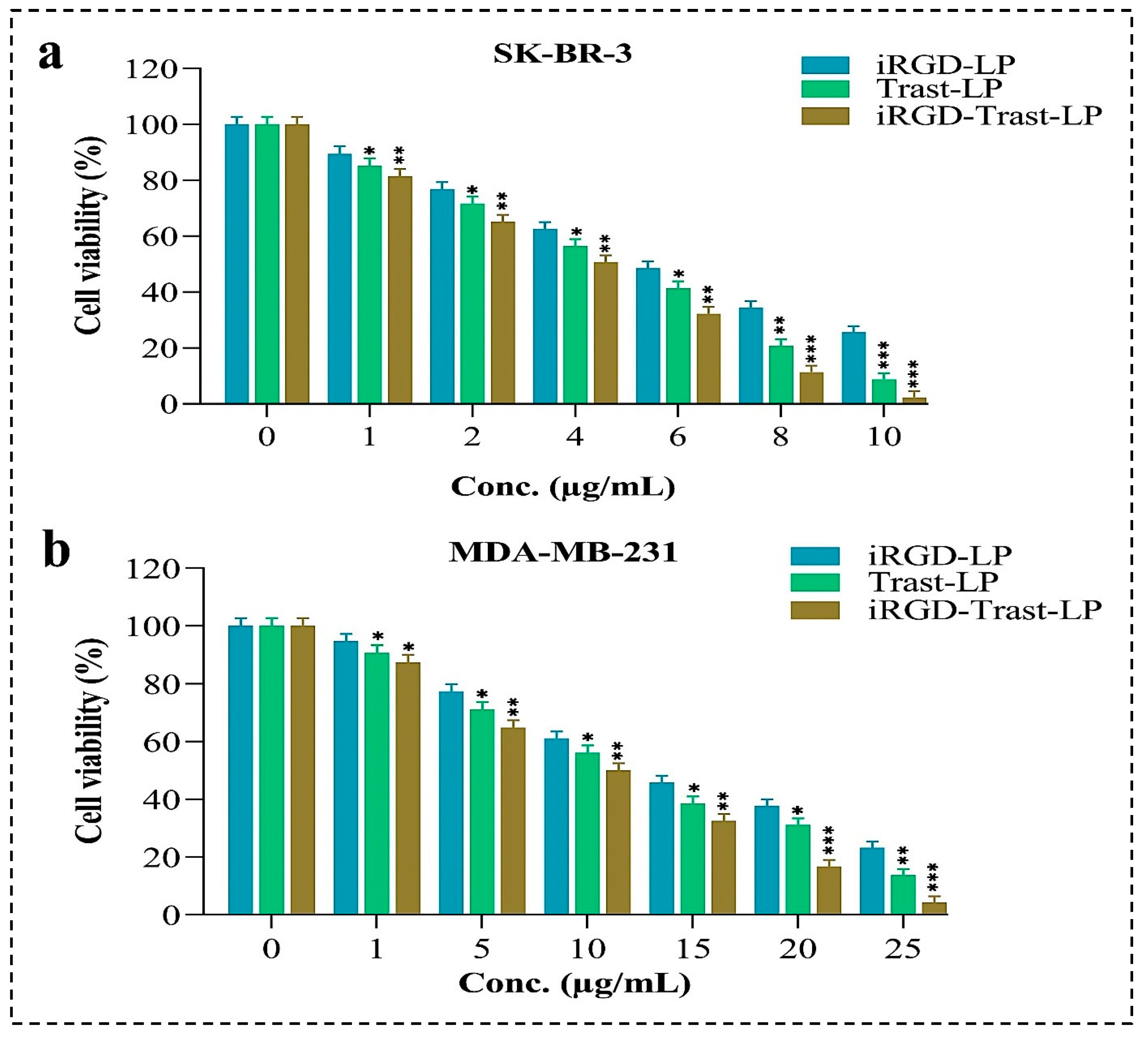
3.10.1. In-Vitro Cytotoxicity (MTT Assay)
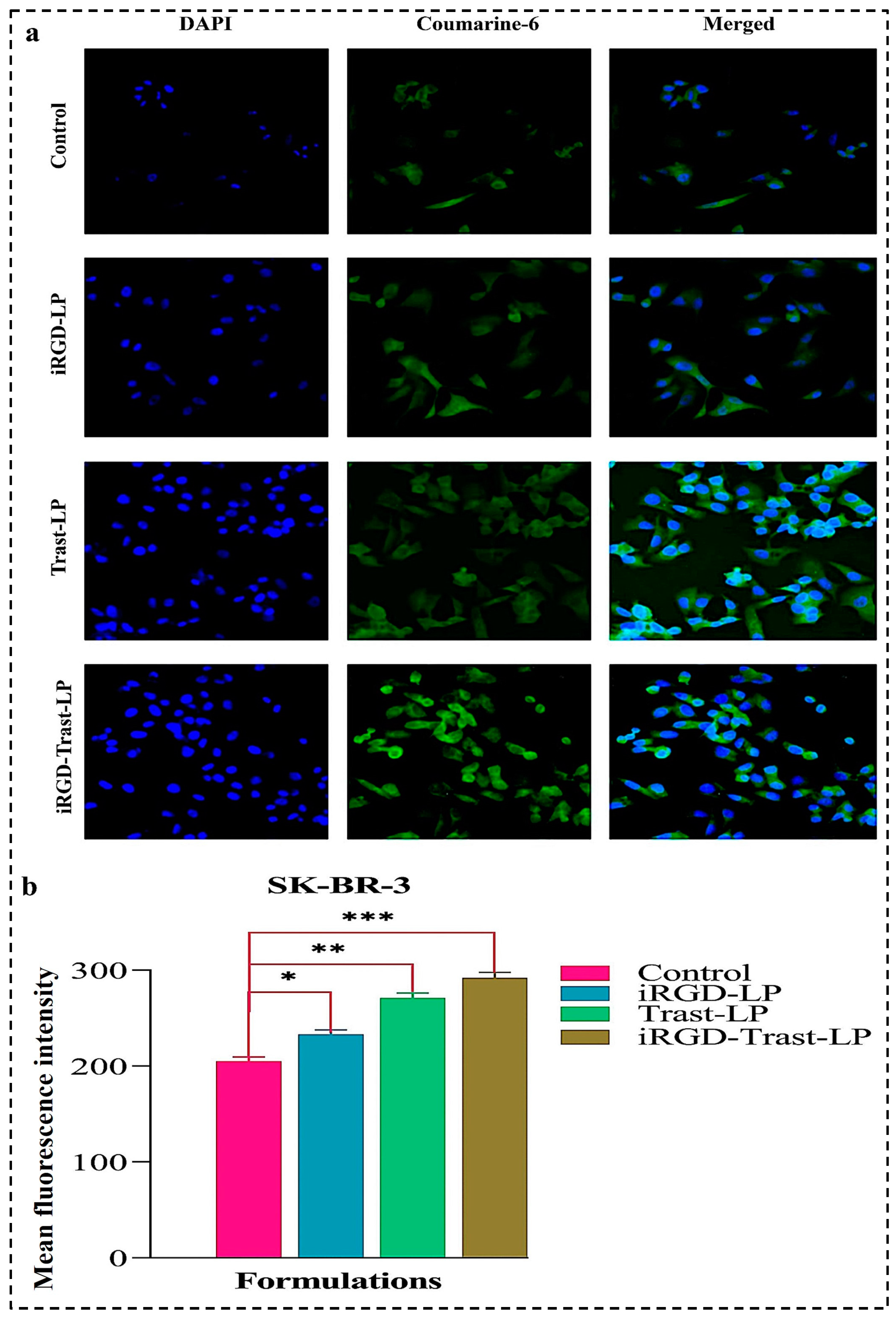
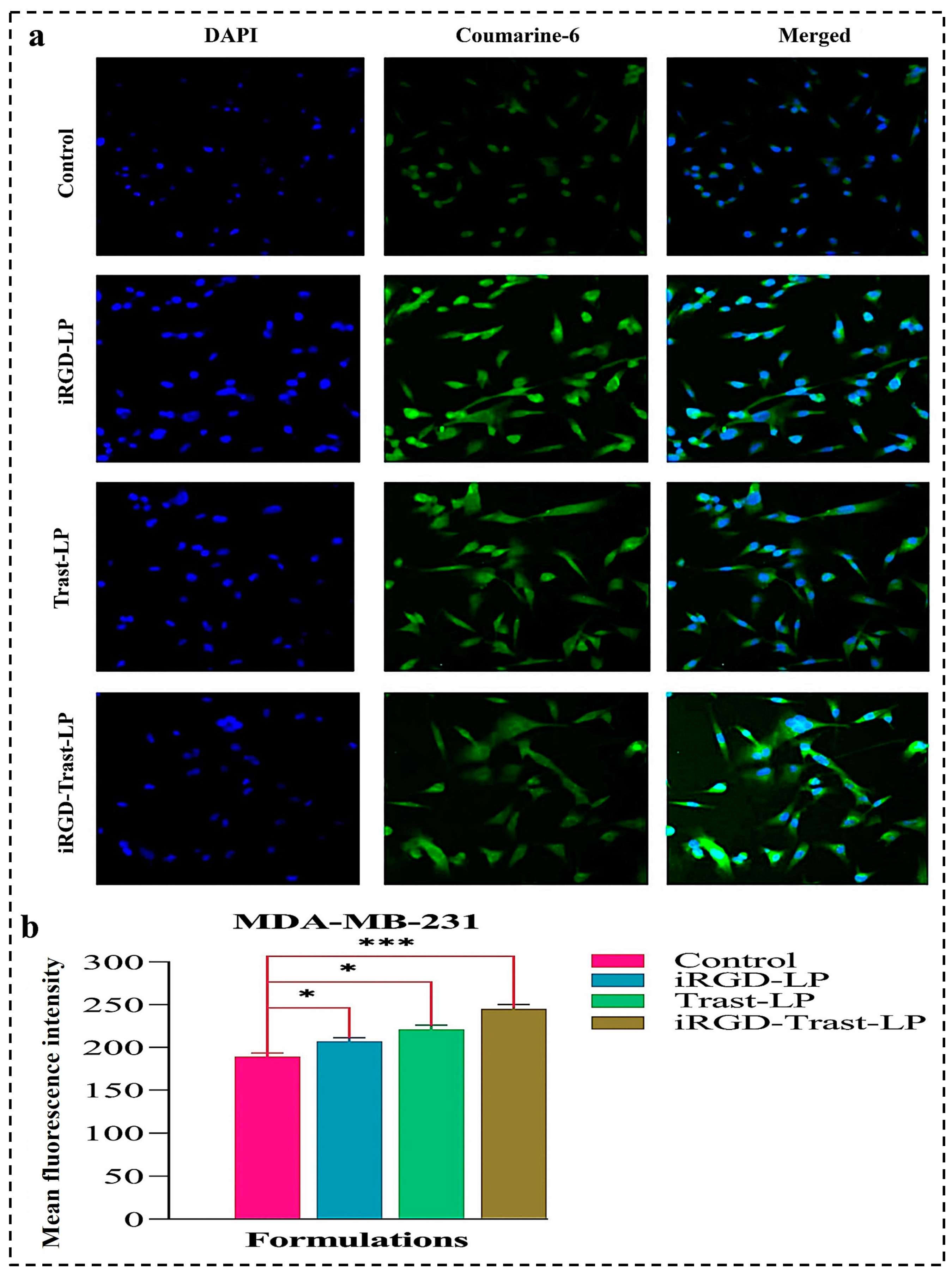
3.10.2. Cellular Uptake by Confocal Laser Scanning Microscopy (CLSM)
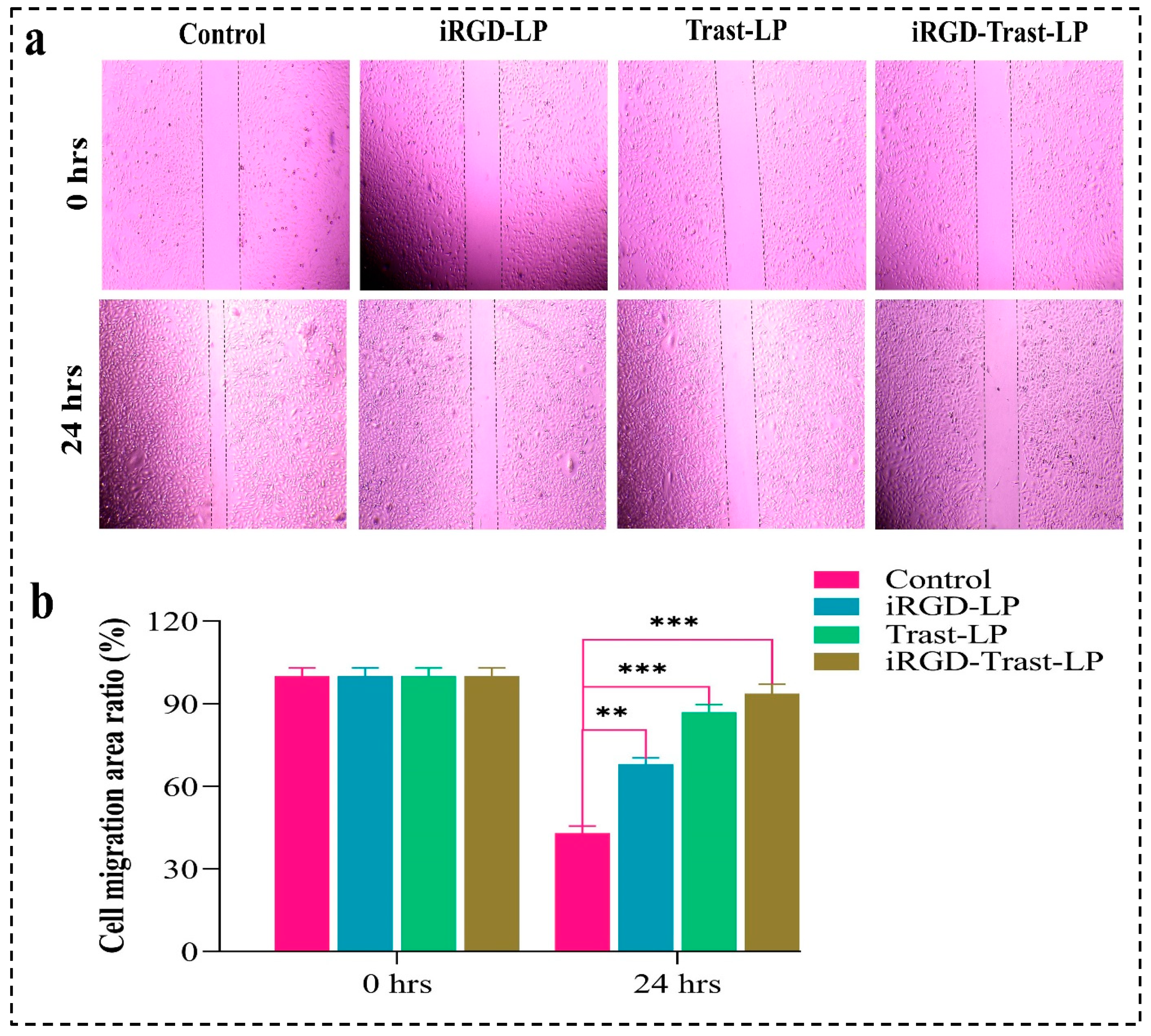
3.10.3. Wound-Healing Assay
3.11. Statistical Analysis
4. Results
4.1. Experimental Design and Optimization of Liposomes
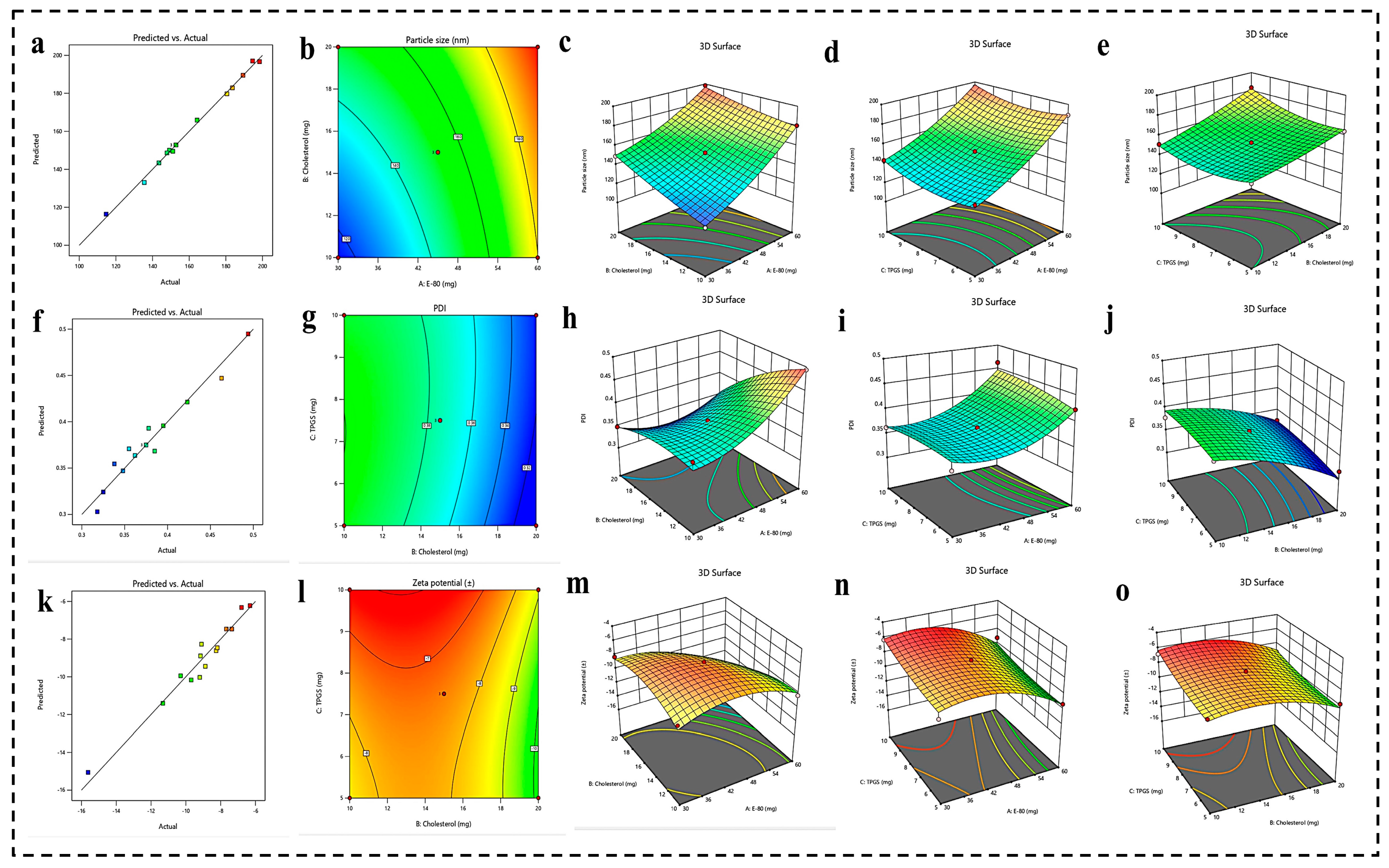
4.1.1. BBD 3 Factor-3 Level Approach
Effect of Formulation Parameters (E-80, Cholesterol, and TPGS) on PS
Effect of Formulation Parameters (E-80, Cholesterol, and TPGS) on PDI
Effect of Formulation Parameters (E-80, Cholesterol, and TPGS) on ZP
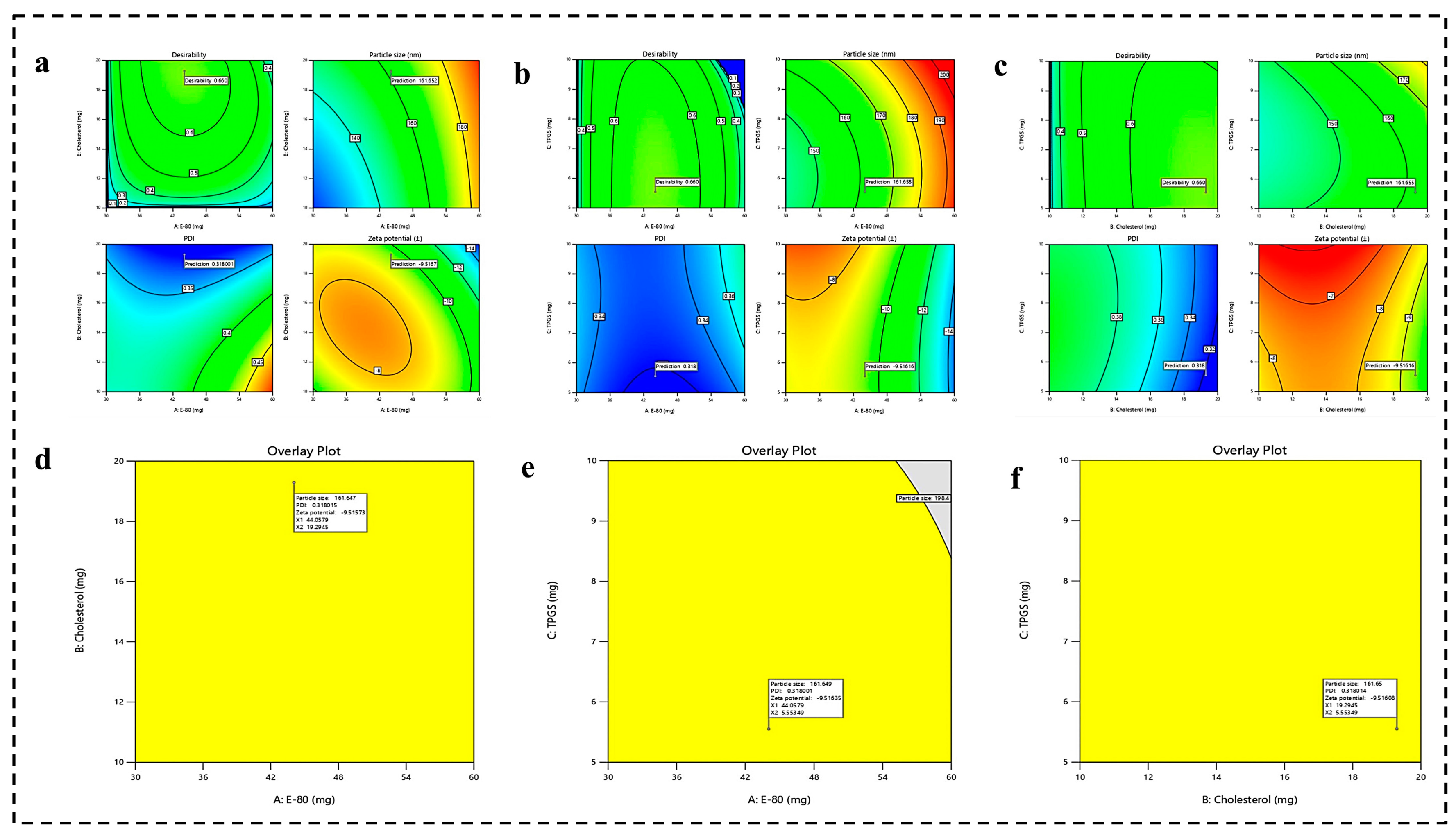
4.1.2. Optimization of Formulation by Numerical Analysis and Point Prediction
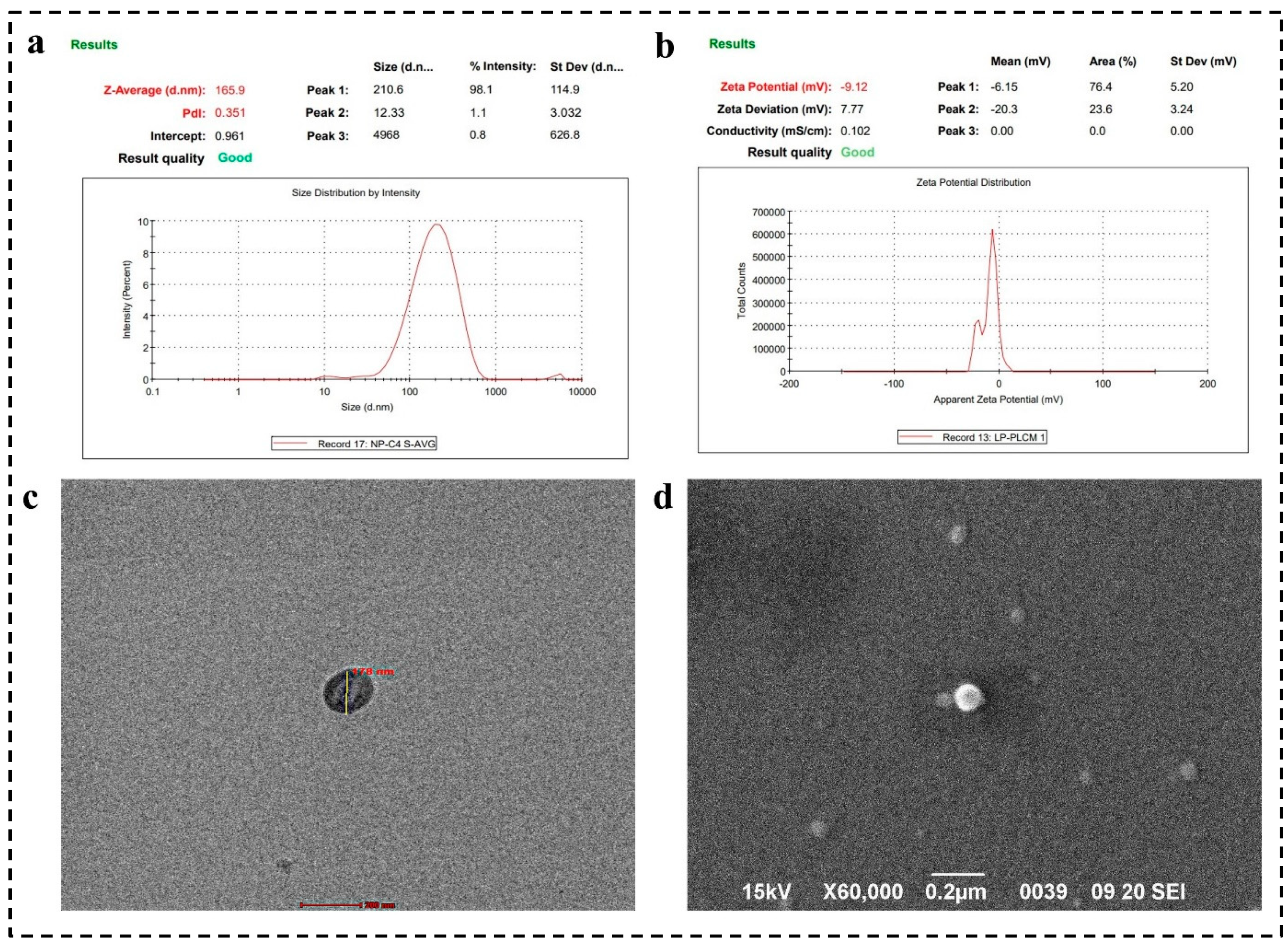
4.2. Zeta Analysis
4.3. Surface Morphology by SEM and TEM Analysis
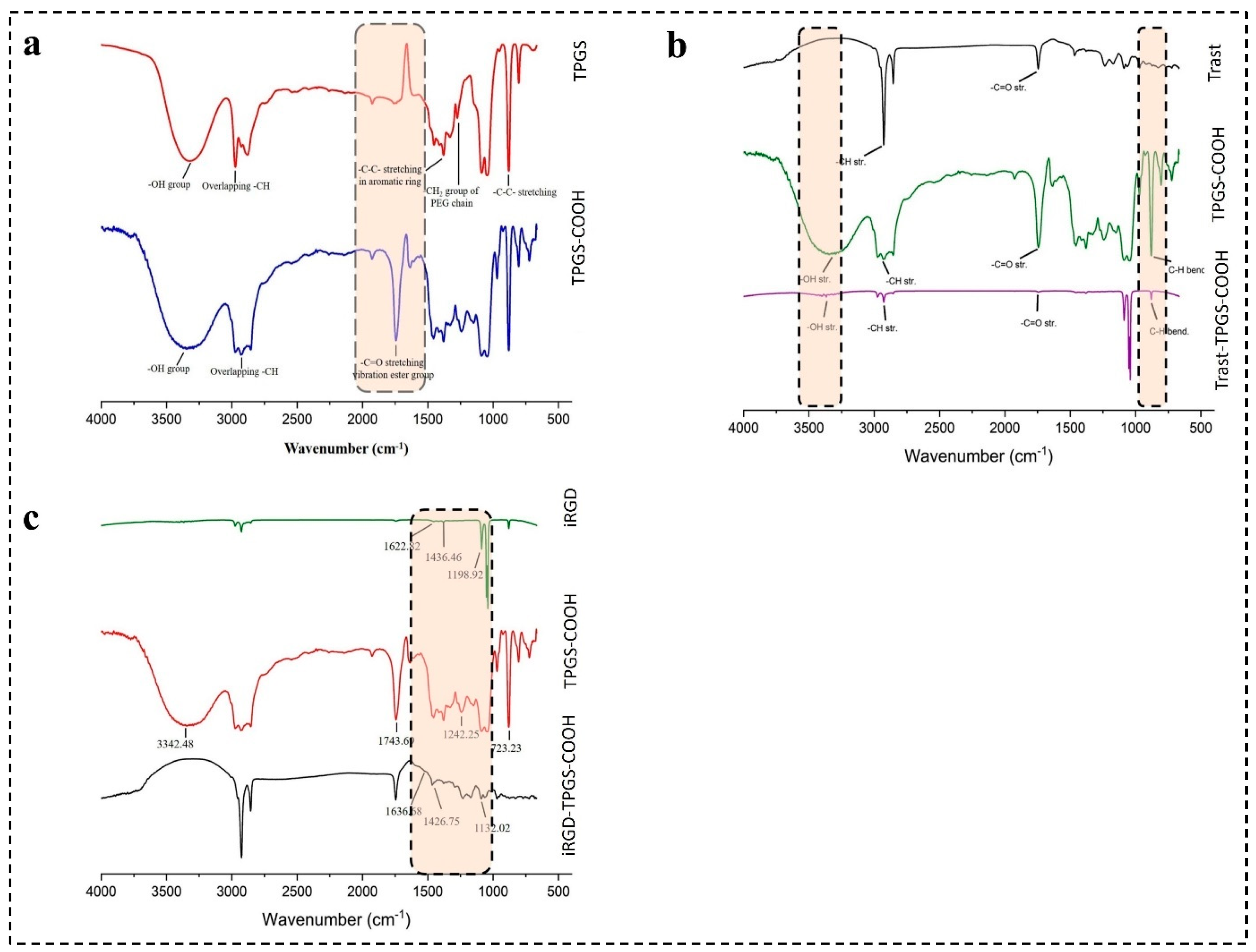
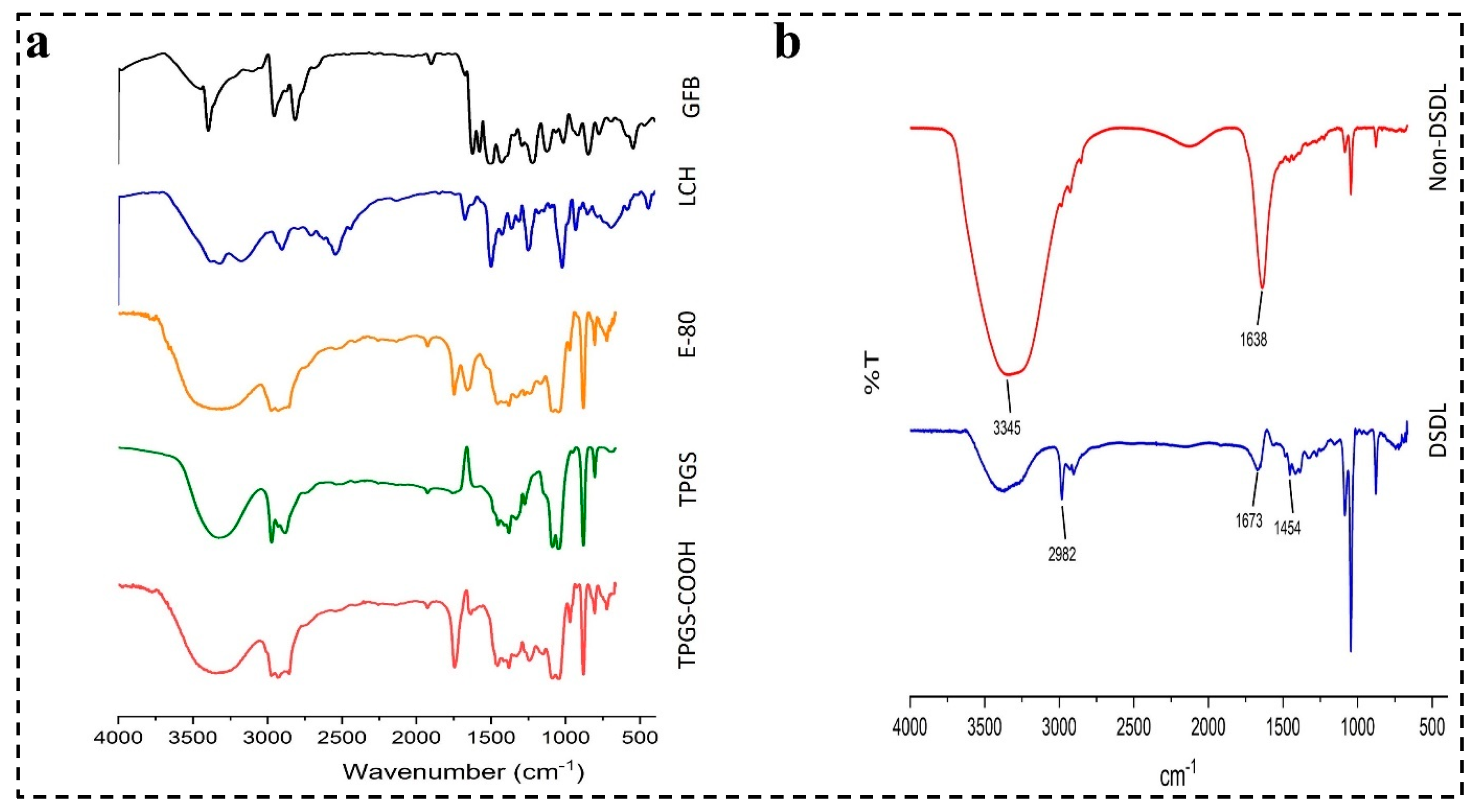
4.4. Characterization of Hybrid Conjugates FTIR Analysis
4.4.1. Characterization of TPGS-COOH by FTIR
4.4.2. Characterization of iRGD-TPGS-COOH by FTIR
4.4.3. Characterization of Trast-TPGS-COOH by FTIR
4.4.4. Characterization of DSDL Formulation
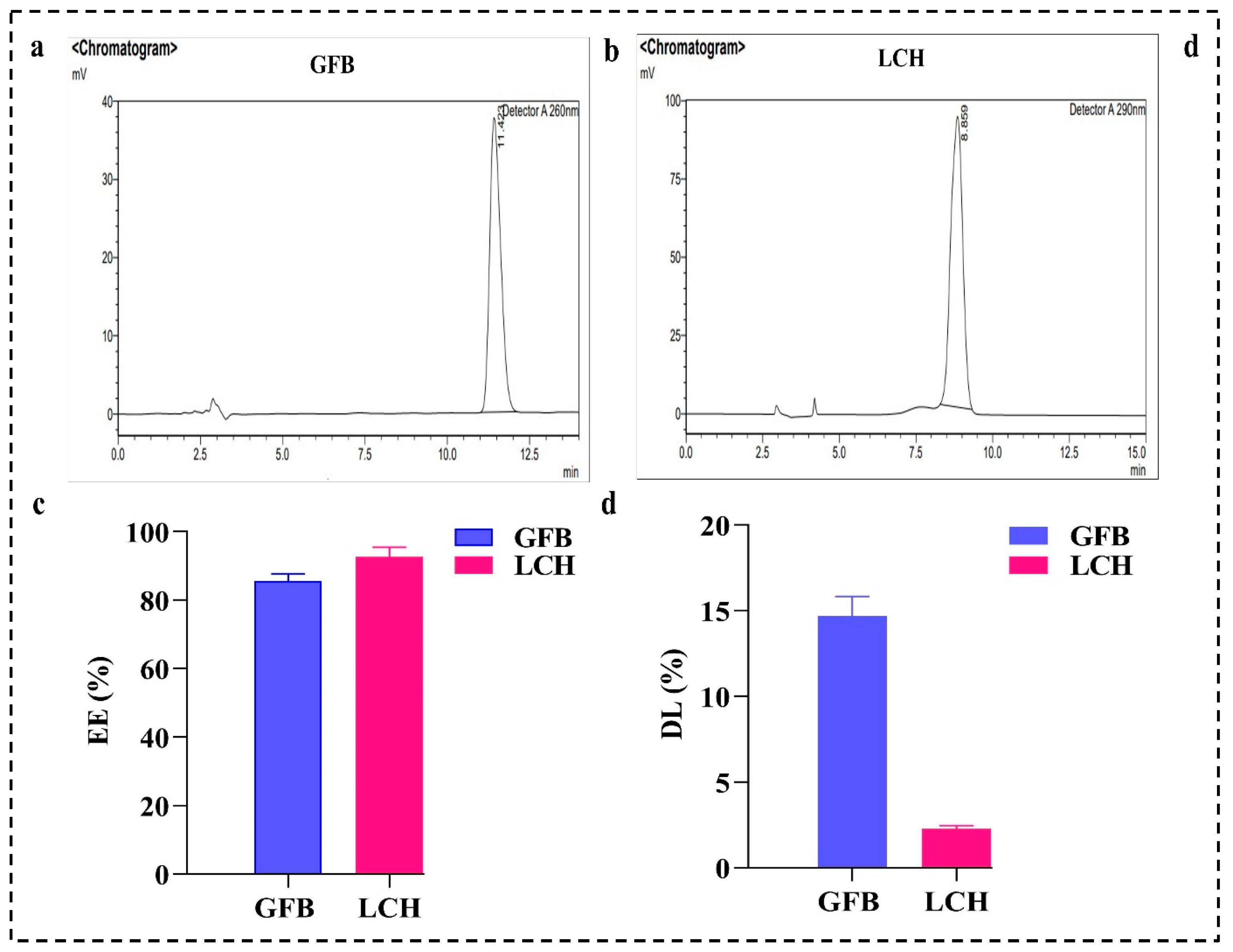
4.5. EE (%) and DL (%)
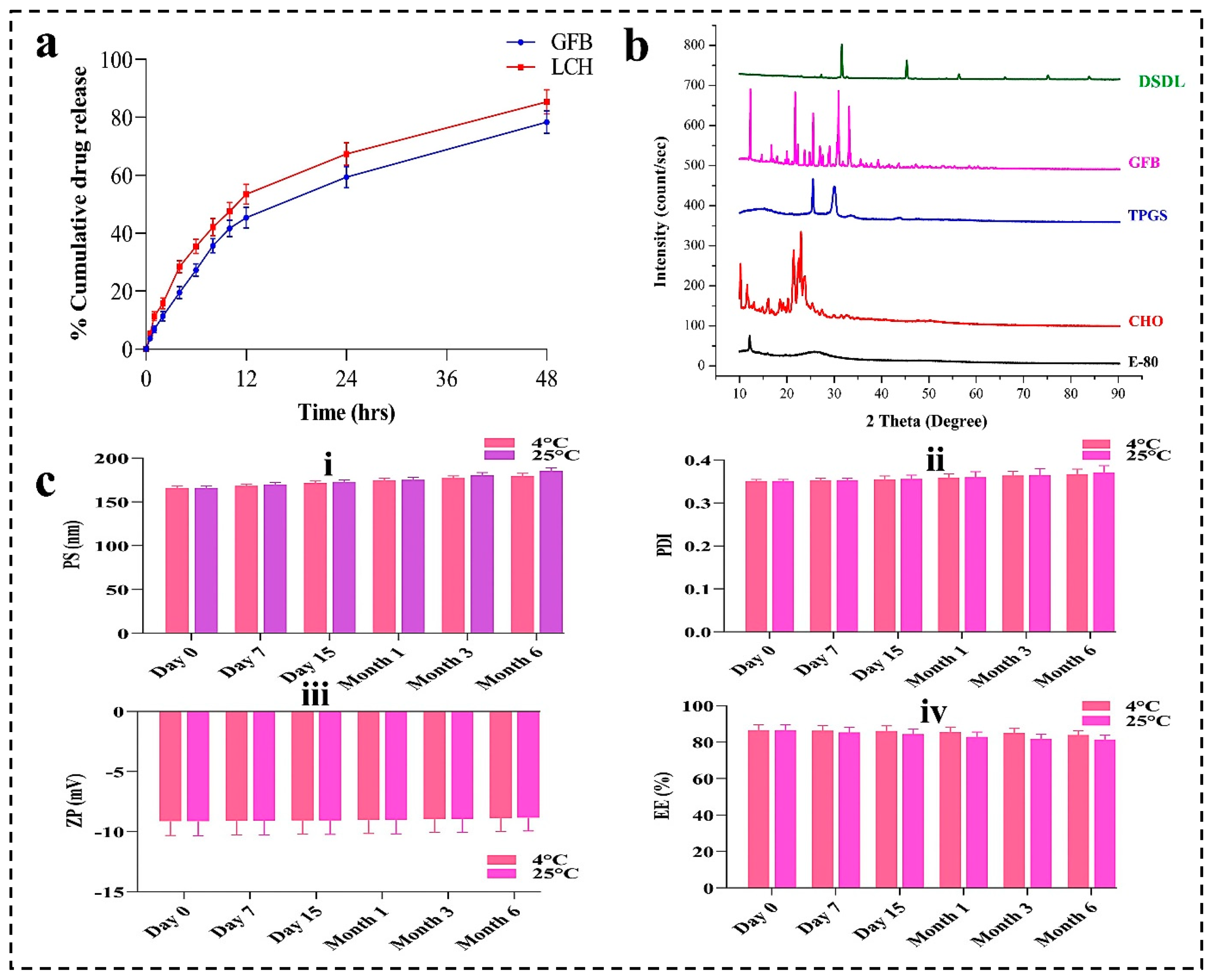
4.6. In-Vitro Drug Release Profile
4.7. XRD Analysis
4.8. Stability Studies
4.9. Biocompatibility Study
4.10. In-Vitro Cell Line
4.10.1. Cytotoxicity Studies
4.10.2. Cellular Uptake of SDLs and DSDLs on SK-BR-3 and MDA-MB-231
4.10.3. Wound-Healing Assay
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HER-2 | Human epidermal growth factor receptor-2 |
| Trast | Trastuzumab |
| GFB | Gefitinib |
| LCH | Lycorine hydrochloride |
| TPGS-COOH | Activated D-Alpha-Tocopheryl polyethylene glycol 1000 succinate |
| DSDL | Dual surface-decorated liposomes |
| PS | Particle size |
| PDI | Polydispersity index |
| ZP | Zeta potential |
| IC-50 | Half-maximum inhibitory concentration |
| SDL | Single decorated liposomes |
| CLSM | Confocal laser scanning microscopy |
| ErbB-2 | Erythroblastic leukemia viral oncogene homolog-2 |
| RGDK | Cryptic CendR motif |
| NRP-1 | Neuropilin-1 |
| TCRP1 | Tongue cancer resistance-associated protein |
| Akt | Protein kinase B |
| mTOR | Mammalian target of rapamycin |
| BCS | Biopharmaceutical classification system |
| MAPK/PI3K | Mitogen-activated protein kinase/phosphatidylinositol 3-kinase |
| TME | Tumor microenvironment |
| TPGS | D-Alpha-Tocopheryl polyethylene glycol 1000 succinate |
| HLB | Hydrophilic and lipophilic balance |
| FDA | Food and drug administration |
| DSPE-PEG2000 | 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] |
| CAFs | Cancer-associated fibroblasts |
| ECM | Extracellular matrix |
| TAMs | Tumor-associated macrophages |
| MDSCs | Myeloid-derived suppressor cells |
| TGF-β | Transforming growth factor beta |
| VEGF | Vascular endothelial growth factor |
| TILs | Tumor-infiltrating lymphocytes |
| HIFs | Hypoxia-inducible factors |
| PD-L1 | Programmed-death ligand |
| CTLA-4 | Cytotoxic T-lymphocyte-associated protein-4 |
| NKG-2A | Natural killer group 2A |
| EDC | 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride |
| NHS | N-hydroxy succinimide |
| DMAP | 4-(Dimethyl amino) pyridine |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| DAPI | 4′,6-diamidino-2-phenylindole dihydrochloride |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal bovine serum |
| DCM | Dichloromethane |
| PBS | Phosphate buffered saline |
| RSM | Response surface methodology |
| DOE | Design of experiments |
| BBD | Box–Behnken design |
| R2 | Correlation coefficient |
| RMSE | Root mean square error |
| SEM | Scanning electron microscopy |
| TEM | Transmission electron microscopy |
| XRD | X-ray diffraction |
| FTIR | Fourier transform infrared spectroscopy |
| EE | Entrapment efficiency |
| DL | Drug loading |
| RP-HPLC | Reverse-phase high-performance liquid chromatography |
| ICH | International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use |
| ACN | Acetonitrile |
| TFA | Trifluoroacetic acid |
| NCCS | National Centre for Cell Science |
| DMSO | Dimethyl sulfoxide |
| CLSM | Confocal laser scanning microscopy |
| MFI | Mean fluorescence intensity |
References
- Chen, G.; Wang, Y.; Wu, P.; Zhou, Y.; Yu, F.; Zhu, C.; Li, Z.; Hang, Y.; Wang, K.; Li, J.; et al. Reversibly stabilized polycation nanoparticles for combination treatment of early-and late-stage metastatic breast cancer. ACS Nano 2018, 12, 6620–6636. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, C.; Kim, I.; Ro, J.; Kim, J.; Min, Y.; Park, J.; Sunkara, V.; Park, Y.-S.; Michael, I.; et al. Three-dimensional human liver-chip emulating premetastatic niche formation by breast cancer-derived extracellular vesicles. ACS Nano 2020, 14, 14971–14988. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Dan, Z.; He, X.; Zhang, Z.; Yu, H.; Yin, Q.; Li, Y. Liposomes coated with isolated macrophage membrane can target lung metastasis of breast cancer. ACS Nano 2016, 10, 7738–7748. [Google Scholar] [CrossRef]
- Pandey, P.; Arya, D.K.; Ramar, M.K.; Chidambaram, K.; Rajinikanth, P. Engineered nanomaterials as an effective tool for HER2+ breast cancer therapy. Drug Discov. Today 2022, 27, 2526–2540. [Google Scholar] [CrossRef] [PubMed]
- Zi, Y.; Yang, K.; He, J.; Wu, Z.; Liu, J.; Zhang, W. Strategies to enhance drug delivery to solid tumors by harnessing the EPR effects and alternative targeting mechanisms. Adv. Drug Deliv. Rev. 2022, 188, 114449. [Google Scholar] [CrossRef]
- Huang, D.; Sun, L.; Huang, L.; Chen, Y. Nanodrug delivery systems modulate tumor vessels to increase the enhanced permeability and retention effect. J. Pers. Med. 2021, 11, 124. [Google Scholar] [CrossRef]
- Piccart-Gebhart, M.J.; Procter, M.; Leyland-Jones, B.; Goldhirsch, A.; Untch, M.; Smith, I.; Gianni, L.; Baselga, J.; Bell, R.; Jackisch, C.; et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1659–1672. [Google Scholar] [CrossRef]
- Loibl, S.; Gianni, L. HER2-positive breast cancer. Lancet 2017, 389, 2415–2429. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Gelber, R.D.; Piccart-Gebhart, M.J.; De Azambuja, E.; Procter, M.; Suter, T.M.; Jackisch, C.; Cameron, D.; Weber, H.A.; Heinzmann, D.; et al. 2 years versus 1 year of adjuvant trastuzumab for HER2-positive breast cancer (HERA): An open-label, randomised controlled trial. Lancet 2013, 382, 1021–1028. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Irudayaraj, J. Nuclear targeting dynamics of gold nanoclusters for enhanced therapy of HER2+ breast cancer. ACS Nano 2011, 5, 9718–9725. [Google Scholar] [CrossRef]
- Slamon, D.; Eiermann, W.; Robert, N.; Pienkowski, T.; Martin, M.; Press, M.; Mackey, J.; Glaspy, J.; Chan, A.; Pawlicki, M.; et al. Adjuvant trastuzumab in HER2-positive breast cancer. N. Engl. J. Med. 2011, 365, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Xiao, Y.; Pan, M.; Li, F.; Duan, W.; Meng, L.; Liu, X.; Yan, F.; Zheng, H. Hyperthermia-triggered drug delivery from iRGD-modified temperature-sensitive liposomes enhances the anti-tumor efficacy using high intensity focused ultrasound. J. Control. Release 2016, 243, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Sonju, J.J.; Dahal, A.; Singh, S.S.; Jois, S.D. Peptide-functionalized liposomes as therapeutic and diagnostic tools for cancer treatment. J. Control. Release 2021, 329, 624–644. [Google Scholar] [CrossRef]
- Deepak, P.; Kumar, P.; Pandey, P.; Arya, D.K.; Jaiswal, S.; Kumar, A.; Sonkar, A.B.; Ali, D.; Alarifi, S.; Ramar, M.; et al. Pentapeptide cRGDfK-Surface Engineered Nanostructured Lipid Carriers as an Efficient Tool for Targeted Delivery of Tyrosine Kinase Inhibitor for Battling Hepatocellular Carcinoma. Int. J. Nanomed. 2023, 18, 7021–7046. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Zhang, Q.; Fu, Y.; Sun, X.; Gong, T.; Zhang, Z. Coadministration of oligomeric hyaluronic acid-modified liposomes with tumor-penetrating peptide-iRGD enhances the antitumor efficacy of doxorubicin against melanoma. ACS Appl. Mater. Interfaces 2017, 9, 1280–1292. [Google Scholar] [CrossRef]
- Guan, J.; Guo, H.; Tang, T.; Wang, Y.; Wei, Y.; Seth, P.; Li, Y.; Dehm, S.M.; Ruoslahti, E.; Pang, H. iRGD-liposomes enhance tumor delivery and therapeutic efficacy of antisense oligonucleotide drugs against primary prostate cancer and bone metastasis. Adv. Funct. Mater. 2021, 31, 2100478. [Google Scholar] [CrossRef]
- Sun, Y.; Gu, Y.; Gao, X.; Jin, X.; Wink, M.; Sharopov, F.S.; Yang, L.; Sethi, G. Lycorine suppresses the malignancy of breast carcinoma by modulating epithelial mesenchymal transition and β-catenin signaling. Pharmacol. Res. 2023, 195, 106866. [Google Scholar] [CrossRef]
- Zhang, P.; Yuan, X.; Yu, T.; Huang, H.; Yang, C.; Zhang, L.; Yang, S.; Luo, X.; Luo, J. Lycorine inhibits cell proliferation, migration and invasion, and primarily exerts in vitro cytostatic effects in human colorectal cancer via activating the ROS/p38 and AKT signaling pathways. Oncol. Rep. 2021, 45, 19. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, J.; Chen, Y.; Xue, R.; Mao, Z.; Lu, W.; Jiang, Y. Lycorine hydrochloride suppresses stress-induced premature cellular senescence by stabilizing the genome of human cells. Aging Cell 2021, 20, e13307. [Google Scholar] [CrossRef]
- Shi, S.; Li, C.; Zhang, Y.; Deng, C.; Tan, M.; Pan, G.; Du, J.; Ji, Y.; Li, Q.; Liang, H.; et al. Lycorine hydrochloride inhibits melanoma cell proliferation, migration and invasion via down-regulating p21Cip1/WAF1. Am. J. Cancer Res. 2021, 11, 1391. [Google Scholar]
- Li, M.-H.; Liao, X.; Li, C.; Wang, T.-T.; Sun, Y.-S.; Yang, K.; Jiang, P.-W.; Shi, S.-T.; Zhang, W.-X.; Zhang, K.; et al. Lycorine hydrochloride induces reactive oxygen species-mediated apoptosis via the mitochondrial apoptotic pathway and the JNK signaling pathway in the oral squamous cell carcinoma HSC-3 cell line. Oncol. Lett. 2021, 21, 236. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Deng, C.; Pan, G.; Wang, X.; Zhang, K.; Dong, Z.; Zhao, G.; Tan, M.; Hu, X.; Shi, S.; et al. Lycorine hydrochloride inhibits cell proliferation and induces apoptosis through promoting FBXW7-MCL1 axis in gastric cancer. J. Exp. Clin. Cancer Res. 2020, 39, 230. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, J.; Xing, G. Lycorine inhibits the growth and metastasis of breast cancer through the blockage of STAT3 signaling pathway. Acta Biochim. Biophys. Sin. 2017, 49, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Nayek, S.; Raghavendra, N.; Kumar, B.S. Development of novel S PC-3 gefitinib lipid nanoparticles for effective drug delivery in breast cancer. Tissue distribution studies and cell cytotoxicity analysis. J. Drug Deliv. Sci. Technol. 2021, 61, 102073. [Google Scholar] [CrossRef]
- Lin, K.-H.; Hong, S.-T.; Wang, H.-T.; Lo, Y.-L.; Lin, A.M.-Y.; Yang, J.C.-H. Enhancing anticancer effect of gefitinib across the blood–brain barrier model using liposomes modified with one α-helical cell-penetrating peptide or glutathione and tween 80. Int. J. Mol. Sci. 2016, 17, 1998. [Google Scholar] [CrossRef]
- Takenaka, T.; Nakai, S.; Katayama, M.; Hirano, M.; Ueno, N.; Noguchi, K.; Takatani-Nakase, T.; Fujii, I.; Kobayashi, S.S.; Nakase, I. Effects of gefitinib treatment on cellular uptake of extracellular vesicles in EGFR-mutant non-small cell lung cancer cells. Int. J. Pharm. 2019, 572, 118762. [Google Scholar] [CrossRef]
- Sitia, L.; Sevieri, M.; Signati, L.; Bonizzi, A.; Chesi, A.; Mainini, F.; Corsi, F.; Mazzucchelli, S. HER-2-targeted nanoparticles for breast cancer diagnosis and treatment. Cancers 2022, 14, 2424. [Google Scholar] [CrossRef]
- Hu, C.; Liang, K.; An, R.; Wang, X.; You, L. The characterization, pharmacokinetic, and tissue distribution studies of TPGS-modified artesunate liposome in rats. Drug Dev. Ind. Pharm. 2018, 44, 1528–1535. [Google Scholar] [CrossRef]
- Raju, A.; Muthu, M.S.; Feng, S.-S. Trastuzumab-conjugated vitamin E TPGS liposomes for sustained and targeted delivery of docetaxel. Expert Opin. Drug Deliv. 2013, 10, 747–760. [Google Scholar] [CrossRef]
- Muthu, M.S.; Kulkarni, S.A.; Raju, A.; Feng, S.-S. Theranostic liposomes of TPGS coating for targeted co-delivery of docetaxel and quantum dots. Biomaterials 2012, 33, 3494–3501. [Google Scholar] [CrossRef]
- Assanhou, A.G.; Li, W.; Zhang, L.; Xue, L.; Kong, L.; Sun, H.; Mo, R.; Zhang, C. Reversal of multidrug resistance by co-delivery of paclitaxel and lonidamine using a TPGS and hyaluronic acid dual-functionalized liposome for cancer treatment. Biomaterials 2015, 73, 284–295. [Google Scholar] [CrossRef]
- Vijayakumar, M.R.; Vajanthri, K.Y.; Balavigneswaran, C.K.; Mahto, S.K.; Mishra, N.; Muthu, M.S.; Singh, S. Pharmacokinetics, biodistribution, in vitro cytotoxicity and biocompatibility of Vitamin E TPGS coated trans resveratrol liposomes. Colloids Surf. B Biointerfaces 2016, 145, 479–491. [Google Scholar] [CrossRef]
- Farooq, M.A.; Xinyu, H.; Jabeen, A.; Ahsan, A.; Seidu, T.A.; Kutoka, P.T.; Wang, B. Enhanced cellular uptake and cytotoxicity of vorinostat through encapsulation in TPGS-modified liposomes. Colloids Surf. B Biointerfaces 2021, 199, 111523. [Google Scholar] [CrossRef]
- Muthu, M.S.; Kulkarni, S.A.; Xiong, J.; Feng, S.-S. Vitamin E TPGS coated liposomes enhanced cellular uptake and cytotoxicity of docetaxel in brain cancer cells. Int. J. Pharm. 2011, 421, 332–340. [Google Scholar] [CrossRef]
- Pan, J.; Feng, S.-S. Targeted delivery of paclitaxel using folate-decorated poly (lactide)–vitamin E TPGS nanoparticles. Biomaterials 2008, 29, 2663–2672. [Google Scholar] [CrossRef]
- Kaddah, S.; Khreich, N.; Kaddah, F.; Charcosset, C.; Greige-Gerges, H. Cholesterol modulates the liposome membrane fluidity and permeability for a hydrophilic molecule. Food Chem. Toxicol. 2018, 113, 40–48. [Google Scholar] [CrossRef]
- Guimarães, D.; Cavaco-Paulo, A.; Nogueira, E. Design of liposomes as drug delivery system for therapeutic applications. Int. J. Pharm. 2021, 601, 120571. [Google Scholar] [CrossRef]
- Choi, S.; Kang, B.; Yang, E.; Kim, K.; Kwak, M.K.; Chang, P.-S.; Jung, H.-S. Precise control of liposome size using characteristic time depends on solvent type and membrane properties. Sci. Rep. 2023, 13, 4728. [Google Scholar] [CrossRef]
- Song, T.; Wang, H.; Liu, Y.; Cai, R.; Yang, D.; Xiong, Y. TPGS-modified long-circulating liposomes loading ziyuglycoside I for enhanced therapy of myelosuppression. Int. J. Nanomed. 2021, 16, 6281–6295. [Google Scholar] [CrossRef]
- Saraf, S.; Jain, A.; Tiwari, A.; Verma, A.; Panda, P.K.; Jain, S.K. Advances in liposomal drug delivery to cancer: An overview. J. Drug Deliv. Sci. Technol. 2020, 56, 101549. [Google Scholar] [CrossRef]
- Almeida, B.; Nag, O.K.; Rogers, K.E.; Delehanty, J.B. Recent progress in bioconjugation strategies for liposome-mediated drug delivery. Molecules 2020, 25, 5672. [Google Scholar] [CrossRef] [PubMed]
- Nel, J.; Elkhoury, K.; Velot, É.; Bianchi, A.; Acherar, S.; Francius, G.; Tamayol, A.; Grandemange, S.; Arab-Tehrany, E. Functionalized liposomes for targeted breast cancer drug delivery. Bioact. Mater. 2023, 24, 401–437. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, J.; Qian, H.; Zhuang, Q. Cancer-associated fibroblasts: From basic science to anticancer therapy. Exp. Mol. Med. 2023, 55, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Agostinetto, E.; Curigliano, G.; Piccart, M. Emerging treatments in HER2-positive advanced breast cancer: Keep raising the bar. Cell Rep. Med. 2024, 5, 101575. [Google Scholar] [CrossRef]
- Singh, R.P.; Sharma, G.; Kumari, L.; Koch, B.; Singh, S.; Bharti, S.; Rajinikanth, P.S.; Pandey, B.L.; Muthu, M.S. RGD-TPGS decorated theranostic liposomes for brain targeted delivery. Colloids Surf. B Biointerfaces 2016, 147, 129–141. [Google Scholar]
- Ong, T.H.; Chitra, E.; Ramamurthy, S.; Siddalingam, R.P.; Yuen, K.H.; Ambu, S.P.; Davamani, F. Chitosan-propolis nanoparticle formulation demonstrates anti-bacterial activity against Enterococcus faecalis biofilms. PLoS ONE 2017, 12, e0174888. [Google Scholar]
- Kumar, R.; Kumar, A.; Kumar, D.; Yadav, S.; Shrivastava, N.K.; Singh, J.; Sonkar, A.B.; Verma, P.; Arya, D.K.; Kaithwas, G.; et al. Harnessing Potential of ω-3 Polyunsaturated Fatty Acid with Nanotechnology for Enhanced Breast Cancer Therapy: A Comprehensive Investigation into ALA-Based Liposomal PTX Delivery. Pharmaceutics 2024, 16, 913. [Google Scholar] [CrossRef] [PubMed]
- Torres-Flores, G.; Gonzalez-Horta, A.; Vega-Cantu, Y.I.; Rodriguez, C.; Rodriguez-Garcia, A. Preparation and Characterization of Liposomal Everolimus by Thin-Film Hydration Technique. Adv. Polym. Technol. 2020, 2020, 5462949. [Google Scholar] [CrossRef]
- Jaiswal, S.; Anjum, M.M.; Arya, D.K.; Thakur, S.; Pandey, P.; Deepak, P.; Kanaujiya, S.; Anand, S.; Kaushik, A.S.; Mishra, V.; et al. Surface entrenched β-sitosterol niosomes for enhanced cardioprotective activity against isoproterenol induced cardiotoxicity in rats. Int. J. Pharm. 2024, 653, 123872. [Google Scholar] [CrossRef] [PubMed]
- Rajinikanth, P.; Sankar, C.; Mishra, B. Sodium alginate microspheres of metoprolol tartrate for intranasal systemic delivery: Development and evaluation. Drug Deliv. 2003, 10, 21–28. [Google Scholar] [CrossRef]
- Balasubramaniam, J.; Rao, V.U.; Vasudha, M.; Babu, J.; Rajinikanth, P. Sodium alginate microspheres of metformin HCl: Formulation and in vitro evaluation. Curr. Drug Deliv. 2007, 4, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Ruozi, B.; Belletti, D.; Tombesi, A.; Tosi, G.; Bondioli, L.; Forni, F.; Vandelli, M.A. AFM, ESEM, TEM, and CLSM in liposomal characterization: A comparative study. Int. J. Nanomed. 2011, 6, 557–563. [Google Scholar] [CrossRef]
- Anjum, S.; Wang, Y.; Xin, Y.; Li, X.; Li, T.; Zhang, H.; Quan, L.; Li, Y.; Arya, D.K.; Rajinikanth, P.; et al. Bioinspired core-shell nanofiber drug-delivery system modulates osteogenic and osteoclast activity for bone tissue regeneration. Mater. Today Bio 2024, 26, 101088. [Google Scholar] [CrossRef]
- Lim, W.M.; Rajinikanth, P.S.; Mallikarjun, C.; Kang, Y.B. Formulation and delivery of itraconazole to the brain using a nanolipid carrier system. Int. J. Nanomed. 2014, 9, 2117–2126. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.; Li, T.; Arya, D.K.; Ali, D.; Alarifi, S.; Yulin, W.; Hengtong, Z.; Rajinikanth, P.; Ao, Q. Biomimetic electrospun nanofibrous scaffold for tissue engineering: Preparation, optimization by design of experiments (DOE), in-vitro and in-vivo characterization. Front. Bioeng. Biotechnol. 2023, 11, 1288539. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.; Arya, D.K.; Saeed, M.; Ali, D.; Athar, M.S.; Yulin, W.; Alarifi, S.; Wu, X.; Rajinikanth, P.; Ao, Q. Multifunctional electrospun nanofibrous scaffold enriched with alendronate and hydroxyapatite for balancing osteogenic and osteoclast activity to promote bone regeneration. Front. Bioeng. Biotechnol. 2023, 11, 1302594. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Girotti, J.; Fan, Y.; Levy, E.S.; Zang, N.; Sethuraman, V.; Kou, P.; Zhang, K.; Gruenhagen, J.; Lin, J. Fast and versatile analysis of liposome encapsulation efficiency by nanoParticle exclusion chromatography. J. Chromatogr. A 2022, 1662, 462688. [Google Scholar] [CrossRef]
- Ohnishi, N.; Yamamoto, E.; Tomida, H.; Hyodo, K.; Ishihara, H.; Kikuchi, H.; Tahara, K.; Takeuchi, H. Rapid determination of the encapsulation efficiency of a liposome formulation using column-switching HPLC. Int. J. Pharm. 2013, 441, 67–74. [Google Scholar] [CrossRef]
- Rajinikanth, P.; Keat, N.W.; Garg, S. Self-nanoemulsifying drug delivery systems of Valsartan: Preparation and in-vitro characterization. Int. J. Drug Deliv. 2012, 4, 153. [Google Scholar]
- Rajinikanth, P.S.; Karunagaran, L.N.; Balasubramaniam, J.; Mishra, B. Formulation and evaluation of clarithromycin microspheres for eradication of Helicobacter pylori. Chem. Pharm. Bull. 2008, 56, 1658–1664. [Google Scholar] [CrossRef] [PubMed]
- Muppidi, K.; Pumerantz, A.S.; Wang, J.; Betageri, G. Development and Stability Studies of Novel Liposomal Vancomycin Formulations. ISRN Pharm. 2012, 2012, 636743. [Google Scholar] [CrossRef]
- Taira, M.C.; Chiaramoni, N.S.; Pecuch, K.M.; Alonso-Romanowski, S. Stability of Liposomal Formulations in Physiological Conditions for Oral Drug Delivery. Drug Deliv. 2004, 11, 123–128. [Google Scholar] [CrossRef]
- Rajinikanth, P.S.; Mishra, B. Stomach-site specific drug delivery system of clarithromycin for eradication of Helicobacter pylori. Chem. Pharm. Bull. 2009, 57, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Panwar, P.; Pandey, B.; Lakhera, P.C.; Singh, K.P. Preparation, characterization, and in vitro release study of albendazole-encapsulated nanosize liposomes. Int. J. Nanomed. 2010, 5, 101–108. [Google Scholar] [CrossRef]
- Rajinikanth, P.S.; Chellian, J. Development and evaluation of nanostructured lipid carrier-based hydrogel for topical delivery of 5-fluorouracil. Int. J. Nanomed. 2016, 11, 5067–5077. [Google Scholar] [CrossRef]
- Jaiswal, S.; Anjum, M.M.; Thakur, S.; Pandey, P.; Arya, D.K.; Kumar, A.; Kaushik, A.S.; Rajinikanth, P.S. Evaluation of cardioprotective effect of naringin loaded lignin nanoparticles against isoproterenol induced myocardial infarction. J. Drug Deliv. Sci. Technol. 2023, 89, 105076. [Google Scholar] [CrossRef]
- Maurelli, A.M.; De Leo, V.; Daniello, V.; Calvano, C.D.; Ciriaco, F.; Milano, F.; Ingrosso, C.; Cataldi, T.R.; Di Gioia, S.; Conese, M.; et al. In depth study of the polydopamine coating of liposomes as a potential alternative to PEGylation for the stabilization of nanocarriers in biological fluids. Mater. Today Chem. 2024, 37, 101994. [Google Scholar] [CrossRef]
- Wang, P.; Zhong, W.; Huang, Q.; Zhu, Y.; Chen, L.; Ye, K. Liposome Nanomedicine Based on Tumor Cell Lysate Mitigates the Progression of Lynch Syndrome-Associated Colon Cancer. ACS Biomater. Sci. Eng. 2024, 10, 3136–3147. [Google Scholar] [CrossRef]
- Kanaujiya, S.; Arya, D.K.; Pandey, P.; Singh, S.; Pandey, G.; Anjum, S.; Anjum, M.M.; Ali, D.; Alarifi, S.; Vijayakumar, M.R.; et al. Resveratrol-Ampicillin Dual-Drug Loaded Polyvinylpyrrolidone/Polyvinyl Alcohol Biomimic Electrospun Nanofiber Enriched with Collagen for Efficient Burn Wound Repair. Int. J. Nanomed. 2024, 19, 5397–5418. [Google Scholar] [CrossRef]
- Angius, F.; Floris, A. Liposomes and MTT cell viability assay: An incompatible affair. Toxicol. In Vitro 2015, 29, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Deepak, P.; Kumar, P.; Arya, D.K.; Pandey, P.; Kumar, S.; Parida, B.P.; Narayan, G.; Singh, S.; Rajinikanth, P.S. c(RGDfK) anchored surface manipulated liposome for tumor-targeted Tyrosine Kinase Inhibitor (TKI) delivery to potentiate liver anticancer activity. Int. J. Pharm. 2023, 642, 123160. [Google Scholar] [CrossRef] [PubMed]
- Bulut, G.; Atmaca, H.; Karaca, B. Trastuzumab in combination with AT-101 induces cytotoxicity and apoptosis in Her2 positive breast cancer cells. Future Oncol. 2020, 16, 4485–4495. [Google Scholar] [CrossRef]
- Hadisi, Z.; Farokhi, M.; Bakhsheshi-Rad, H.R.; Jahanshahi, M.; Hasanpour, S.; Pagan, E.; Dolatshahi-Pirouz, A.; Zhang, Y.S.; Kundu, S.C.; Akbari, M. Hyaluronic acid (HA)-based silk fibroin/zinc oxide core–shell electrospun dressing for burn wound management. Macromol. Biosci. 2020, 20, 1900328. [Google Scholar] [CrossRef]
- Mouritzen, M.V.; Jenssen, H. Optimized Scratch Assay for In Vitro Testing of Cell Migration with an Automated Optical Camera. J. Vis. Exp. JoVE 2018, 138, 57691. [Google Scholar] [CrossRef]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Hutterer, K.M.; Polozova, A.; Kuhns, S.; McBride, H.J.; Cao, X.; Liu, J. Assessing analytical and functional similarity of proposed Amgen biosimilar ABP 980 to trastuzumab. BioDrugs 2019, 33, 321–333. [Google Scholar] [CrossRef]
| Variables | Levels | ||
|---|---|---|---|
| −1 | 0 | +1 | |
| Independent variables | |||
| X1 = E-80 (mg) | 30 | 45 | 60 |
| X2 = Cholesterol (mg) | 10 | 15 | 20 |
| X3 = TPGS (mg) | 5 | 7.5 | 10 |
| Dependent variables | Desired response | ||
| Y1 = PS (nm) | Minimum | ||
| Y2 = PDI | Minimum Maximum | ||
| Y3 = ZP (±) | |||
| Independent Variable (Factors) | Dependent Variable (Response) | |||||
|---|---|---|---|---|---|---|
| Formulation Code | Factor 1 (X1) | Factor 2 (X2) | Factor 3 (X3) | PS (nm) Y1 | PDI Y2 | ZP (±mV) Y3 |
| LP1 | 45 | 15 | 7.5 | 152.8 ± 1.54 | 0.375 ± 0.046 | −7.36 |
| LP 2 | 60 | 15 | 5 | 189.4 ± 2.06 | 0.423 ± 0.068 | −11.32 |
| LP 3 | 30 | 15 | 5 | 135.5 ± 1.36 | 0.355 ± 0.034 | −9.1 |
| LP 4 | 30 | 15 | 10 | 143.5 ± 1.48 | 0.362 ± 0.081 | −6.31 |
| LP 5 | 30 | 20 | 7.5 | 149.2 ± 1.59 | 0.348 ± 0.059 | −8.26 |
| LP 6 | 45 | 20 | 5 | 164.3 ± 1.85 | 0.318 ± 0.047 | −9.69 |
| LP 7 | 60 | 15 | 10 | 194.6 ± 2.18 | 0.463 ± 0.065 | −9.2 |
| LP 8 | 60 | 10 | 7.5 | 180.6 ± 2.03 | 0.494 ± 0.072 | −10.3 |
| LP 9 | 45 | 10 | 5 | 147.9 ± 1.47 | 0.395 ± 0.061 | −8.19 |
| LP 10 | 45 | 15 | 7.5 | 152.8 ± 1.54 | 0.375 ± 0.058 | −7.669 |
| LP 11 | 30 | 10 | 7.5 | 114.7 ± 1.16 | 0.385 ± 0.055 | −8.88 |
| LP 12 | 45 | 10 | 10 | 151.1 ± 1.54 | 0.378 ± 0.053 | −6.8 |
| LP 13 | 45 | 15 | 7.5 | 152.8 ± 1.54 | 0.375 ± 0.062 | −7.36 |
| LP 14 | 45 | 20 | 10 | 183.6 ± 1.97 | 0.325 ± 0.048 | −9.16 |
| LP 15 | 60 | 20 | 7.5 | 198.4 ± 2.38 | 0.338 ± 0.056 | −15.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arya, D.K.; Deshpande, H.; Kumar, A.; Chidambaram, K.; Pandey, P.; Anjum, S.; Deepak, P.; Kumar, V.; Kumar, S.; Pandey, G.; et al. HER-2 Receptor and αvβ3 Integrin Dual-Ligand Surface-Functionalized Liposome for Metastatic Breast Cancer Therapy. Pharmaceutics 2024, 16, 1128. https://doi.org/10.3390/pharmaceutics16091128
Arya DK, Deshpande H, Kumar A, Chidambaram K, Pandey P, Anjum S, Deepak P, Kumar V, Kumar S, Pandey G, et al. HER-2 Receptor and αvβ3 Integrin Dual-Ligand Surface-Functionalized Liposome for Metastatic Breast Cancer Therapy. Pharmaceutics. 2024; 16(9):1128. https://doi.org/10.3390/pharmaceutics16091128
Chicago/Turabian StyleArya, Dilip Kumar, Hemali Deshpande, Ashish Kumar, Kumarappan Chidambaram, Prashant Pandey, Shabnam Anjum, Payal Deepak, Vikas Kumar, Santosh Kumar, Giriraj Pandey, and et al. 2024. "HER-2 Receptor and αvβ3 Integrin Dual-Ligand Surface-Functionalized Liposome for Metastatic Breast Cancer Therapy" Pharmaceutics 16, no. 9: 1128. https://doi.org/10.3390/pharmaceutics16091128
APA StyleArya, D. K., Deshpande, H., Kumar, A., Chidambaram, K., Pandey, P., Anjum, S., Deepak, P., Kumar, V., Kumar, S., Pandey, G., Srivastava, S., & Rajinikanth, P. S. (2024). HER-2 Receptor and αvβ3 Integrin Dual-Ligand Surface-Functionalized Liposome for Metastatic Breast Cancer Therapy. Pharmaceutics, 16(9), 1128. https://doi.org/10.3390/pharmaceutics16091128









