miR-21, miR-93, miR-191, miR-let-7b, and miR-499 Expression Level in Plasma and Cerebrospinal Fluid in Patients with Prolonged Disorders of Consciousness
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection and Preparation
2.3. Isolation of Total RNA
2.4. Reverse Transcription and qPCR
2.5. qPCR Data Analysis
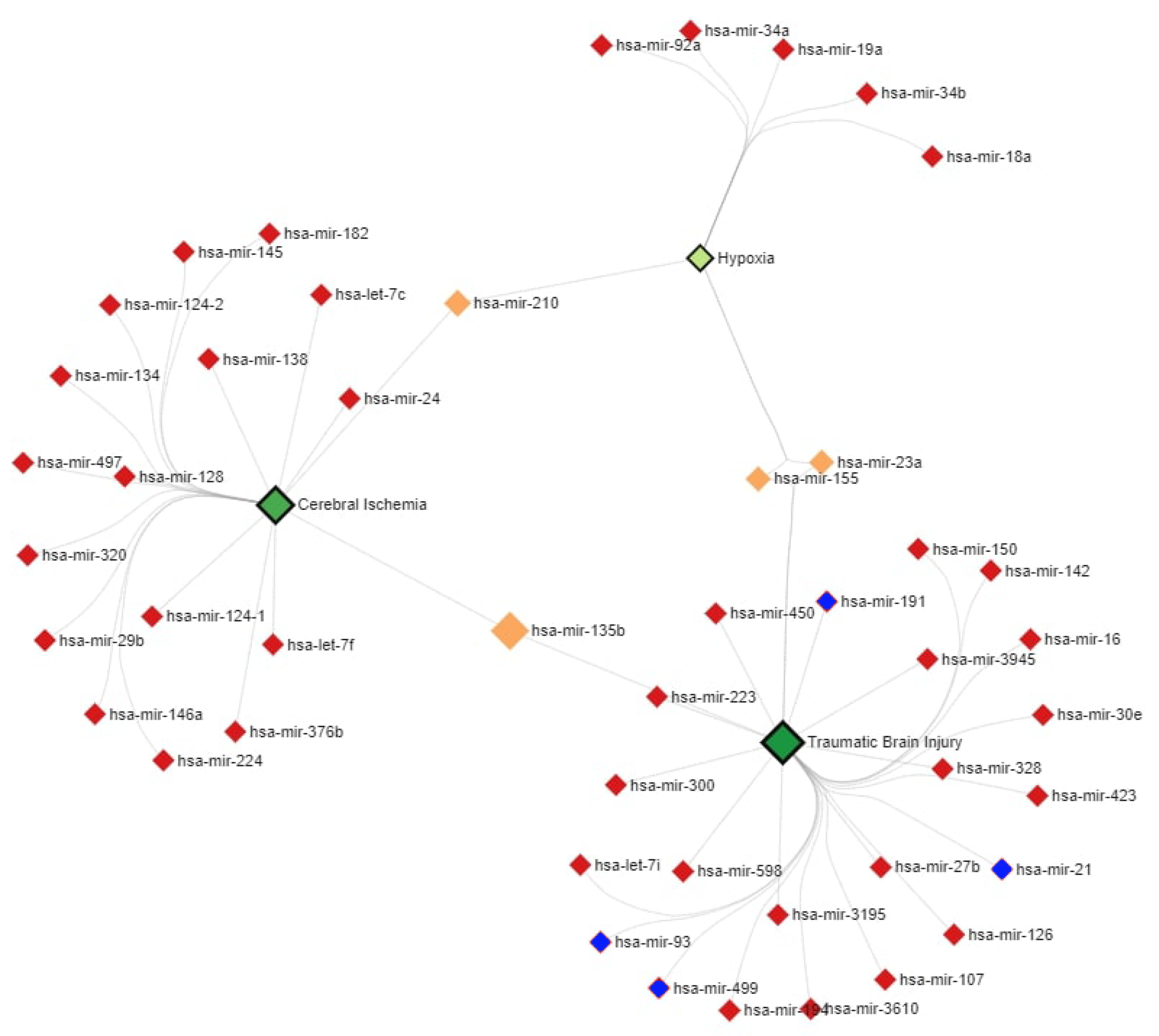
2.6. MiR–Disease Association Network Analysis
3. Results
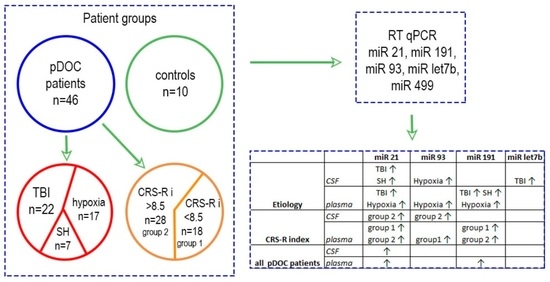
3.1. Description of Patient Groups
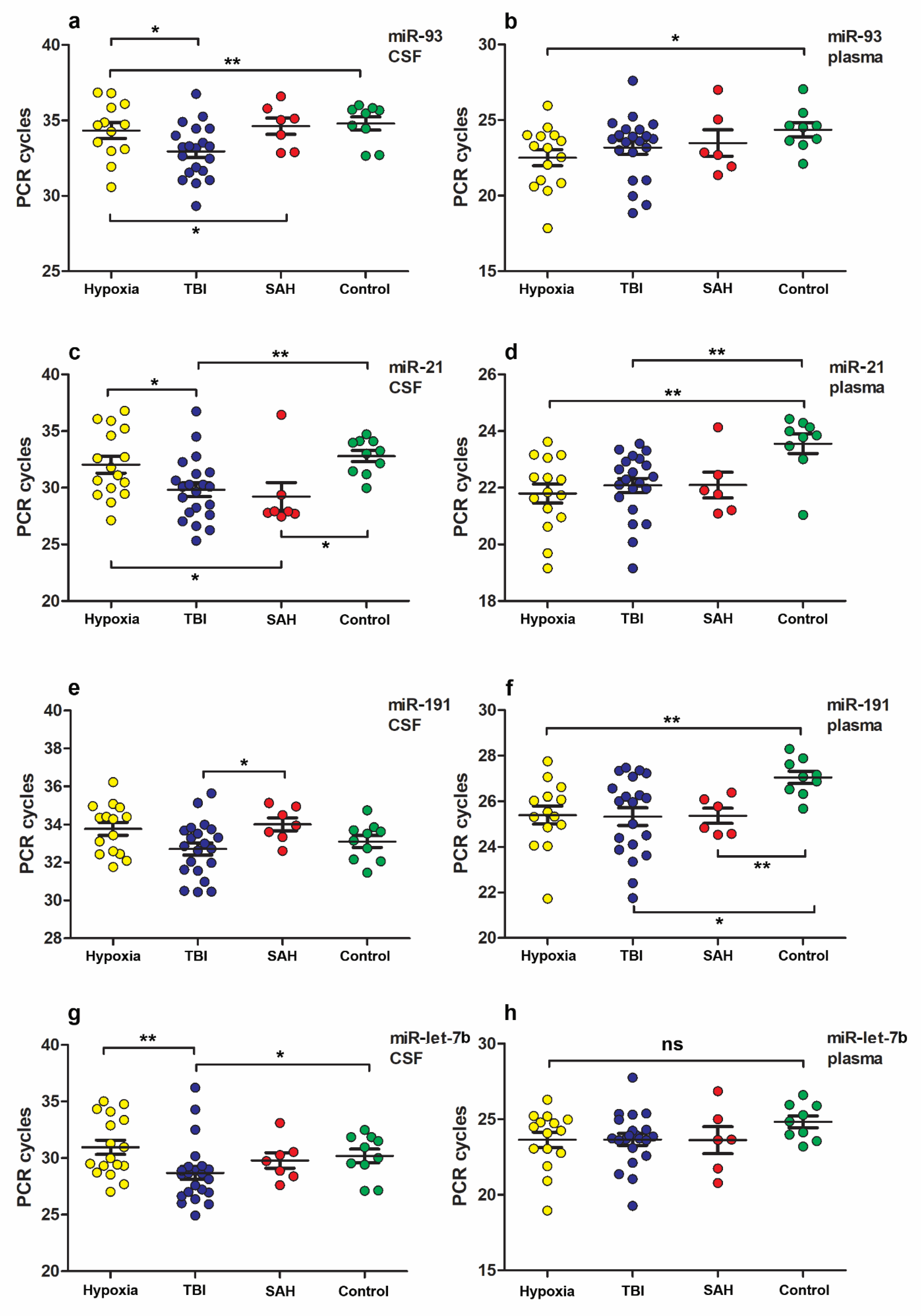
3.2. MiR Expression Levels
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giacino, J.T.; Kalmar, K. The Vegetative and Minimally Conscious States: A Comparision of Clinical Features and Functional Outcome. J. Head Trauma Rehabil. 1997, 12, 36–51. [Google Scholar] [CrossRef]
- Giacino, J.T.; Ashwal, S.; Childs, N.; Cranford, R.; Jennett, B.; Katz, D.I.; Kelly, J.P.; Rosenberg, J.H.; Whyte, J.; Zafonte, R.D.; et al. The Minimally Conscious State: Definition and Diagnostic Criteria. Neurology 2002, 58, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Giacino, J.T.; Kalmar, K. Diagnostic and Prognostic Guidelines for the Vegetative and Minimally Conscious States. Neuropsychol. Rehabil. 2005, 15, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Giacino, J.T.; Hirsch, J.; Schiff, N.; Laureys, S. Functional Neuroimaging Applications for Assessment and Rehabilitation Planning in Patients with Disorders of Consciousness. Arch. Phys. Med. Rehabil. 2006, 87, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Goldfine, A.M.; Victor, J.D.; Conte, M.M.; Bardin, J.C.; Schiff, N.D. Determination of Awareness in Patients with Severe Brain Injury Using EEG Power Spectral Analysis. Clin. Neurophysiol. 2011, 122, 2157–2168. [Google Scholar] [CrossRef] [Green Version]
- Edlow, B.L.; Claassen, J.; Schiff, N.D.; Greer, D.M. Recovery from Disorders of Consciousness: Mechanisms, Prognosis and Emerging Therapies. Nat. Rev. Neurol. 2021, 17, 135–156. [Google Scholar] [CrossRef]
- Schiff, N.D. Cognitive Motor Dissociation Following Severe Brain Injuries. JAMA Neurol. 2015, 72, 1413–1415. [Google Scholar] [CrossRef]
- Kim, N.; O’Sullivan, J.; Olafson, E.; Caliendo, E.; Nowak, S.; Voss, H.U.; Lowder, R.; Watson, W.D.; Ivanidze, J.; Fins, J.J.; et al. Cognitive-Motor Dissociation Following Pediatric Brain Injury: What About the Children? Neurol. Clin. Pract. 2022, 12, 248–257. [Google Scholar] [CrossRef]
- Redell, J.B.; Liu, Y.; Dash, P.K. Traumatic Brain Injury Alters Expression of Hippocampal MicroRNAs: Potential Regulators of Multiple Pathophysiological Processes. J. Neurosci. Res. 2009, 87, 1435–1448. [Google Scholar] [CrossRef]
- Mrozek, S.; Dumurgier, J.; Citerio, G.; Mebazaa, A.; Geeraerts, T. Biomarkers and Acute Brain Injuries: Interest and Limits. Crit. Care 2014, 18, 1–12. [Google Scholar] [CrossRef]
- Kawata, K.; Liu, C.Y.; Merkel, S.F.; Ramirez, S.H.; Tierney, R.T.; Langford, D. Blood Biomarkers for Brain Injury: What Are We Measuring? Neurosci. Biobehav. Rev. 2016, 68, 460–473. [Google Scholar] [CrossRef] [Green Version]
- Luppi, A.I.; Cain, J.; Spindler, L.R.B.; Górska, U.J.; Toker, D.; Hudson, A.E.; Brown, E.N.; Diringer, M.N.; Stevens, R.D.; Massimini, M.; et al. Mechanisms Underlying Disorders of Consciousness: Bridging Gaps to Move Toward an Integrated Translational Science. Neurocrit. Care 2021, 35, 37–54. [Google Scholar] [CrossRef]
- Zeiler, F.A.; McFadyen, C.; Newcombe, V.F.J.; Synnot, A.; Donoghue, E.L.; Ripatti, S.; Steyerberg, E.W.; Gruen, R.L.; McAllister, T.W.; Rosand, J.; et al. Genetic Influences on Patient-Oriented Outcomes in Traumatic Brain Injury: A Living Systematic Review of Non-Apolipoprotein E Single-Nucleotide Polymorphisms. J. Neurotrauma 2021, 38, 1107–1123. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V. The Functions of Animal MicroRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Im, H.I.; Kenny, P.J. MicroRNAs in Neuronal Function and Dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA Profiling: Approaches and Considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Pan, Y.B.; Sun, Z.L.; Feng, D.F. The Role of MicroRNA in Traumatic Brain Injury. Neuroscience 2017, 367, 189–199. [Google Scholar] [CrossRef]
- Wu, J.; Li, H.; He, J.; Tian, X.; Luo, S.; Li, J.; Li, W.; Zhong, J.; Zhang, H.; Huang, Z.; et al. Downregulation of MicroRNA-9–5p Promotes Synaptic Remodeling in the Chronic Phase after Traumatic Brain Injury. Cell Death Dis. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Atif, H.; Hicks, S.D. A Review of MicroRNA Biomarkers in Traumatic Brain Injury. J. Exp. Neurosci. 2019, 13, 117906951983228. [Google Scholar] [CrossRef] [Green Version]
- Bhalala, O.G. The Emerging Impact of MicroRNAs in Neurotrauma Pathophysiology and Therapy. In Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects; Kobeissy, F.H., Ed.; Taylor & Francis Group, LLC.: Washington, DC, USA, 2015; pp. 351–364. [Google Scholar]
- Najem, D.; Rennie, K.; Ribecco-Lutkiewicz, M.; Ly, D.; Haukenfrers, J.; Liu, Q.; Nzau, M.; Fraser, D.D.; Bani-Yaghoub, M. Traumatic Brain Injury: Classification, Models, and Markers. Biochem. Cell Biol. 2018, 96, 391–406. [Google Scholar] [CrossRef]
- Pasinetti, G.M.; Ho, L.; Dooley, C.; Abbi, B.; Lange, G. Select Non-Coding RNA in Blood Components Provide Novel Clinically Accessible Biological Surrogates for Improved Identification of Traumatic Brain Injury in OEF/OIF Veterans. J. Neurodegener. Dis. 2012, 1, 88–98. [Google Scholar]
- Yang, T.; Song, J.; Bu, X.; Wang, C.; Wu, J.; Cai, J.; Wan, S.; Fan, C.; Zhang, C.; Wang, J. Elevated Serum MiR-93, MiR-191, and MiR-499 Are Noninvasive Biomarkers for the Presence and Progression of Traumatic Brain Injury. J. Neurochem. 2016, 137, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Tanriverdi, F.; Zararsiz, G.; Elbuken, G.; Ulutabanca, H.; Karaca, Z.; Selcuklu, A.; Unluhizarci, K.; Tanriverdi, K.; Kelestimur, F. Circulating MicroRNAs as Potential Biomarkers for Traumatic Brain Injury-Induced Hypopituitarism. J. Neurotrauma 2016, 33, 1818–1825. [Google Scholar] [CrossRef]
- Mitra, B.; Rau, T.F.; Surendran, N.; Brennan, J.H.; Thaveenthiran, P.; Sorich, E.; Fitzgerald, M.C.; Rosenfeld, J.V.; Patel, S.A. Plasma Micro-RNA Biomarkers for Diagnosis and Prognosis after Traumatic Brain Injury: A Pilot Study. J. Clin. Neurosci. 2017, 38, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Peplow, P.V. MicroRNAs as Diagnostic Markers and Therapeutic Targets for Traumatic Brain Injury. Neural Regen. Res. 2017, 12, 1749–1761. [Google Scholar] [CrossRef]
- Redell, J.B.; Moore, A.N.; Ward, N.H.; Hergenroeder, G.W.; Dash, P.K. Human Traumatic Brain Injury Alters Plasma MicroRNA Levels. J. Neurotrauma 2010, 27, 2147–2156. [Google Scholar] [CrossRef]
- Schindler, C.R.; Woschek, M.; Vollrath, J.T.; Kontradowitz, K.; Lustenberger, T.; Störmann, P.; Marzi, I.; Henrich, D. MiR-142–3p Expression Is Predictive for Severe Traumatic Brain Injury (TBI) in Trauma Patients. Int. J. Mol. Sci. 2020, 21, 5381. [Google Scholar] [CrossRef]
- Bhomia, M.; Balakathiresan, N.S.; Wang, K.K.; Papa, L.; Maheshwari, R.K. A Panel of Serum MiRNA Biomarkers for the Diagnosis of Severe to Mild Traumatic Brain Injury in Humans. Sci. Rep. 2016, 6, 28148. [Google Scholar] [CrossRef]
- Liu, L.; Sun, T.; Liu, Z.; Chen, X.; Zhao, L.; Qu, G.; Li, Q. Traumatic Brain Injury Dysregulates MicroRNAs to Modulate Cell Signaling in Rat Hippocampus. PLoS ONE 2014, 9, e103948. [Google Scholar] [CrossRef] [Green Version]
- Herrold, A.; Kletzel, S.L.; Foecking, E.M.; Saban, K.L.; Przybycien-Szymanska, M.M.; Zilliox, M.; Bhaumik, D.; Lange, D.; Radke, J.R.; Salinas, I.; et al. MiRNAs as Potential Biomarkers for Traumatic Brain Injury: Pathway From Diagnosis to Neurorehabilitation. J. Head Trauma Rehabil. 2021, 36, E155–E169. [Google Scholar] [CrossRef]
- Qin, X.; Li, L.; Lv, Q.; Shu, Q.; Zhang, Y.; Wang, Y. Expression Profile of Plasma MicroRNAs and Their Roles in Diagnosis of Mild to Severe Traumatic Brain Injury. PLoS ONE 2018, 13, e0204051. [Google Scholar] [CrossRef]
- Zhou, Q.; Yin, J.; Wang, Y.; Zhuang, X.; He, Z.; Chen, Z.; Yang, X. MicroRNAs as Potential Biomarkers for the Diagnosis of Traumatic Brain Injury: A Systematic Review and Meta-Analysis. Int. J. Med. Sci 2021, 18, 128–136. [Google Scholar] [CrossRef]
- Hiskens, M.I.; Mengistu, T.S.; Li, K.M.; Fenning, A.S. Systematic Review of the Diagnostic and Clinical Utility of Salivary MicroRNAs in Traumatic Brain Injury (TBI). Int. J. Mol. Sci. 2022, 23, 13160. [Google Scholar] [CrossRef]
- Sun, P.; Liu, D.Z.; Jickling, G.C.; Sharp, F.R.; Yin, K.J. MicroRNA-Based Therapeutics in Central Nervous System Injuries. J. Cereb. Blood Flow Metab. 2018, 38, 1125–1148. [Google Scholar] [CrossRef]
- You, W.D.; Tang, Q.L.; Wang, L.; Lei, J.; Feng, J.F.; Mao, Q.; Gao, G.Y.; Jiang, J.Y. Alteration of MicroRNA Expression in Cerebrospinal Fluid of Unconscious Patients after Traumatic Brain Injury and a Bioinformatic Analysis of Related Single Nucleotide Polymorphisms. Chin. J. Traumatol. Engl. Ed. 2016, 19, 11–15. [Google Scholar] [CrossRef]
- Zilliox, M.; Foecking, E.; Kuffel, G.; Conneely, M.; Saban, K.; Herrold, A.; Kletzel, S.; Radke, J.; Walsh, E.; Guernon, A.; et al. An Initial MiRNA Profile of Persons with Persisting Neurobehavioral Impairments and States of Disordered Consciousness after Severe Traumatic Brain Injury. J. Head Trauma Rehabil. 2022. [Google Scholar] [CrossRef]
- Sanz, L.R.D.; Thibaut, A.; Edlow, B.L.; Laureys, S.; Gosseries, O. Update on Neuroimaging in Disorders of Consciousness. Curr. Opin. Neurol. 2021, 34, 488–496. [Google Scholar] [CrossRef]
- Di Pietro, V.; Ragusa, M.; Davies, D.; Su, Z.; Hazeldine, J.; Lazzarino, G.; Hill, L.J.; Crombie, N.; Foster, M.; Purrello, M.; et al. MicroRNAs as Novel Biomarkers for the Diagnosis and Prognosis of Mild and Severe Traumatic Brain Injury. J. Neurotrauma 2017, 34, 1948–1956. [Google Scholar] [CrossRef]
- Ji, W.; Jiao, J.; Cheng, C.; Shao, J. MicroRNA-21 in the Pathogenesis of Traumatic Brain Injury. Neurochem. Res. 2018, 43, 1863–1868. [Google Scholar] [CrossRef]
- Kondratov, K.; Nikitin, Y.; Fedorov, A.; Kostareva, A.; Mikhailovskii, V.; Isakov, D.; Ivanov, A.; Golovkin, A. Heterogeneity of the Nucleic Acid Repertoire of Plasma Extracellular Vesicles Demonstrated Using High-Sensitivity Fluorescence-Activated Sorting. J. Extracell. Vesicles 2020, 9, 1743139. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Cente, M.; Matyasova, K.; Csicsatkova, N.; Tomikova, A.; Porubska, S.; Niu, Y.; Majdan, M.; Filipcik, P.; Jurisica, I. Traumatic MicroRNAs: Deconvolving the Signal After Severe Traumatic Brain Injury. Cell Mol. Neurobiol. 2022, 2022, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.I.; Reis, R.M.; Calin, G.A. MicroRNA History: Discovery, Recent Applications, and next Frontiers. Mutat. Res. 2011, 717, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the MIR155 Host Gene in Physiological and Pathological Processes. Gene 2013, 532, 1–12. [Google Scholar] [CrossRef]



| Group | Total | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | ||||||
| N | % | N | % | N | % | ||
| Etiology (TBI, hypoxia, SAH) | Hypoxia | 9 | 50.0% | 8 | 28.6% | 17 | 37.0% |
| SAH | 5 | 27.8% | 2 | 7.1% | 7 | 15.2% | |
| TBI | 4 | 22.2% | 18 | 64.3% | 22 | 47.8% | |
| Total | 18 | 100.0% | 28 | 100.0% | 46 | 100.0% | |
| Gender | female | 7 | 38.9% | 10 | 35.7% | 17 | 37.0% |
| male | 11 | 61.1% | 18 | 64.3% | 29 | 63.0% | |
| Total | 18 | 100.0% | 28 | 100.0% | 46 | 100.0% | |
| Group | N | Average | Median | Min | Max | Q1 (25%) | Q3 (75%) | |
|---|---|---|---|---|---|---|---|---|
| Duration of pDOC | 1 | 18 | 10.2 | 3.5 | 1.0 | 54.0 | 1.0 | 11.0 |
| 2 | 28 | 6.2 | 2.5 | 1.0 | 37.0 | 1.0 | 5.5 | |
| Age | 1 | 18 | 42.1 | 44.0 | 18.0 | 63.0 | 33.0 | 54.0 |
| 2 | 28 | 35.2 | 32.5 | 21.0 | 61.0 | 25.0 | 44.0 | |
| CRS-R Score—at admission | 1 | 18 | 4.4 | 4.5 | 2.0 | 6.0 | 4.0 | 5.0 |
| 2 | 28 | 9.8 | 9.0 | 6.0 | 15.0 | 7.0 | 12.0 | |
| CRS-R Score after one month | 1 | 18 | 5.7 | 5.0 | 3.0 | 23.0 | 4.0 | 6.0 |
| 2 | 28 | 12.0 | 12.0 | 5.0 | 23.0 | 8.0 | 15.0 | |
| CRS-R index at admission | 1 | 18 | 3.41 | 3.80 | 0.33 | 4.84 | 2.75 | 4.50 |
| 2 | 28 | 26.41 | 20.80 | 12.13 | 64.57 | 14.22 | 39.56 | |
| CRS-R index after one month | 1 | 18 | 9.10 | 3.80 | 2.41 | 100.00 | 2.75 | 4.84 |
| 2 | 28 | 37.67 | 32.79 | 3.80 | 99.67 | 14.74 | 52.58 |
| Groups by Etiology CSF/Plasma | p-Value miR-93 | p-Value miR-21 | p-Value miR-191 | p-Value miR-let-7b | p-Value miR-499 |
|---|---|---|---|---|---|
| TBI vs. Hypoxia | 0.0487/0.3357 | 0.0357/0.5004 | 0.0542/0.9442 | 0.0034/0.7850 | 0.1193/0.6303 |
| TBI vs. SAH | 0.0434/0.8383 | 0.3396/0.5794 | 0.0411/0.9273 | 0.1201/0.7484 | 0.2515/0.9071 |
| Hypoxia vs. SAH | 0.9368/0.5593 | 0.0252/0.7853 | 0.5258/0.7728 | 0.3739/0.8457 | 0.7268/0.5333 |
| TBI vs. Control | 0.0083/0.1606 | 00038/0.0013 | 0.4220/0.0133 | 0.0328/0.0855 | 0.6113/0.6837 |
| Hypoxia vs. Control | 0.8412/0.0235 | 0.3990/0.0024 | 0.2330/0.0042 | 0.7632/0.1360 | 0.5468/0.3553 |
| SAH vs. Control | 0.9182/0.2721 | 0.0136/0.0768 | 0.1088/0.0048 | 0.6691/0.3277 | 0.7396/0.9061 |
| CSF/Plasma | p-Value miR-93 | p-Value miR-21 | p-Value miR-191 | p-Value miR-let-7 | p-Value miR-499 |
|---|---|---|---|---|---|
| Group 1vs. Group 2 | 0.6400/0.2334 | 0.7343/0.2334 | 0.5230/0.8481 | 0.7180/0.6912 | 0.3141/0.4344 |
| Group 1 vs. Control | 0.2799/0.0132 | 0.0564/0.0033 | 0.4666/0.0030 | 0.6488/0.0948 | 0.5814/0.3186 |
| Group 2 vs. Control | 0.0214/0.1977 | 0.0148/0.0012 | 0.7582/0.0057 | 0.1982/0.1095 | 0.8976/0.7696 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrova, T.A.; Kondratyev, S.A.; Kostareva, A.A.; Rutkovskiy, R.V.; Savvina, I.A.; Kondratyeva, E.A. miR-21, miR-93, miR-191, miR-let-7b, and miR-499 Expression Level in Plasma and Cerebrospinal Fluid in Patients with Prolonged Disorders of Consciousness. Neurol. Int. 2023, 15, 40-54. https://doi.org/10.3390/neurolint15010004
Petrova TA, Kondratyev SA, Kostareva AA, Rutkovskiy RV, Savvina IA, Kondratyeva EA. miR-21, miR-93, miR-191, miR-let-7b, and miR-499 Expression Level in Plasma and Cerebrospinal Fluid in Patients with Prolonged Disorders of Consciousness. Neurology International. 2023; 15(1):40-54. https://doi.org/10.3390/neurolint15010004
Chicago/Turabian StylePetrova, Tatiana A., Sergey A. Kondratyev, Anna A. Kostareva, Roman V. Rutkovskiy, Irina A. Savvina, and Ekaterina A. Kondratyeva. 2023. "miR-21, miR-93, miR-191, miR-let-7b, and miR-499 Expression Level in Plasma and Cerebrospinal Fluid in Patients with Prolonged Disorders of Consciousness" Neurology International 15, no. 1: 40-54. https://doi.org/10.3390/neurolint15010004
APA StylePetrova, T. A., Kondratyev, S. A., Kostareva, A. A., Rutkovskiy, R. V., Savvina, I. A., & Kondratyeva, E. A. (2023). miR-21, miR-93, miR-191, miR-let-7b, and miR-499 Expression Level in Plasma and Cerebrospinal Fluid in Patients with Prolonged Disorders of Consciousness. Neurology International, 15(1), 40-54. https://doi.org/10.3390/neurolint15010004