Molecular Biological Methods to Assess Different Botrytis cinerea Strains on Grapes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Strain Sampling and Cultivation
2.1.2. Preparation of Field Samples
2.2. Methods
2.2.1. DNA Extraction
2.2.2. qPCR—Preparation of Cultivated Botrytis Samples—Cross-Contamination
2.2.3. qPCR—Preparation of Standard Curves
2.2.4. qPCR—Run
2.2.5. qPCR—Early Detection Method—Limit of Detection
2.2.6. SSR-PCR—PCR Run
2.2.7. SSR-PCR—Agarose Gel Electrophoresis
2.2.8. SSR-PCR—Evaluation of the Gel
2.2.9. SSR-PCR—Capillary Sequencer
2.2.10. Statistical Analysis
3. Results
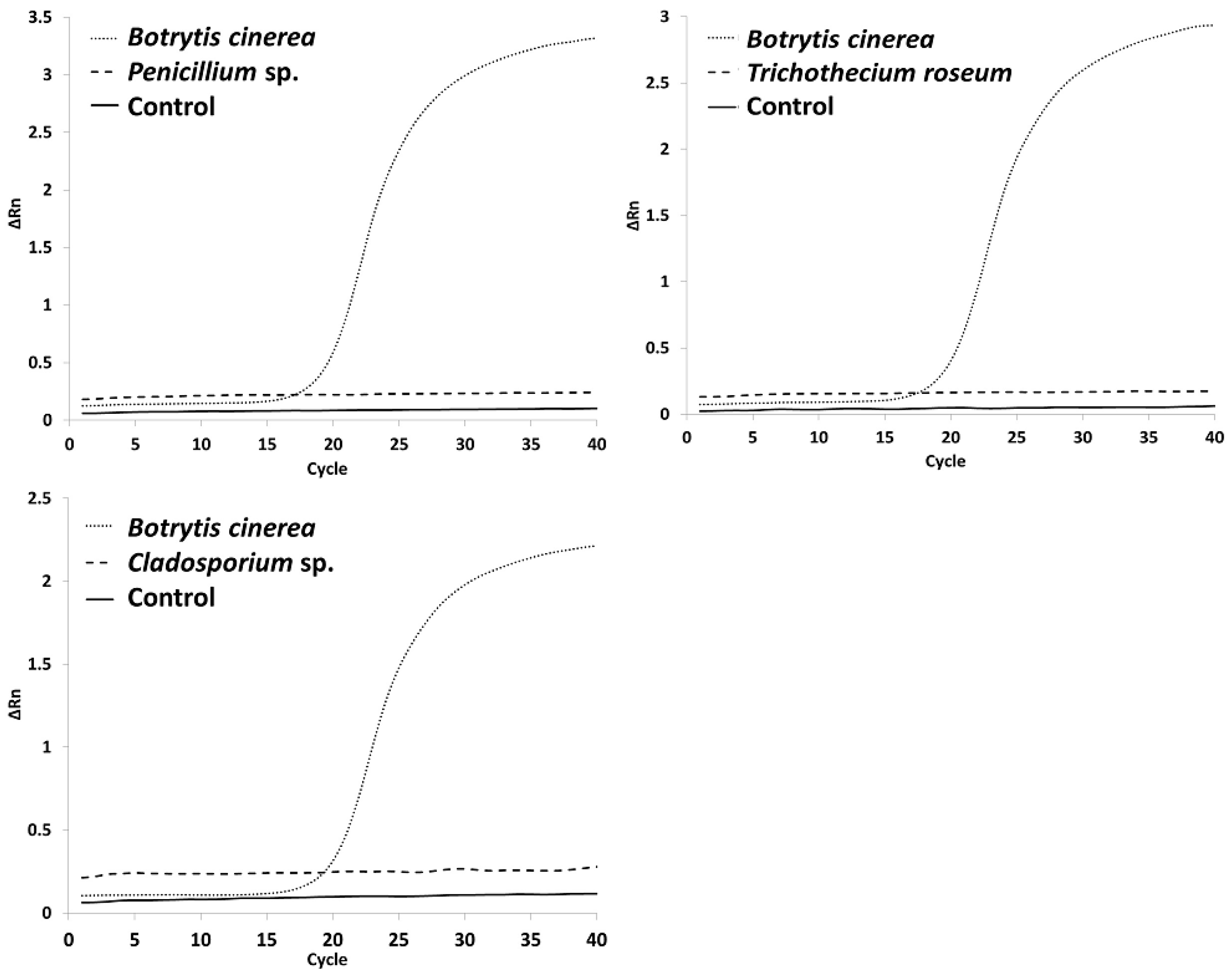
3.1. qPCR Cross-Contamination
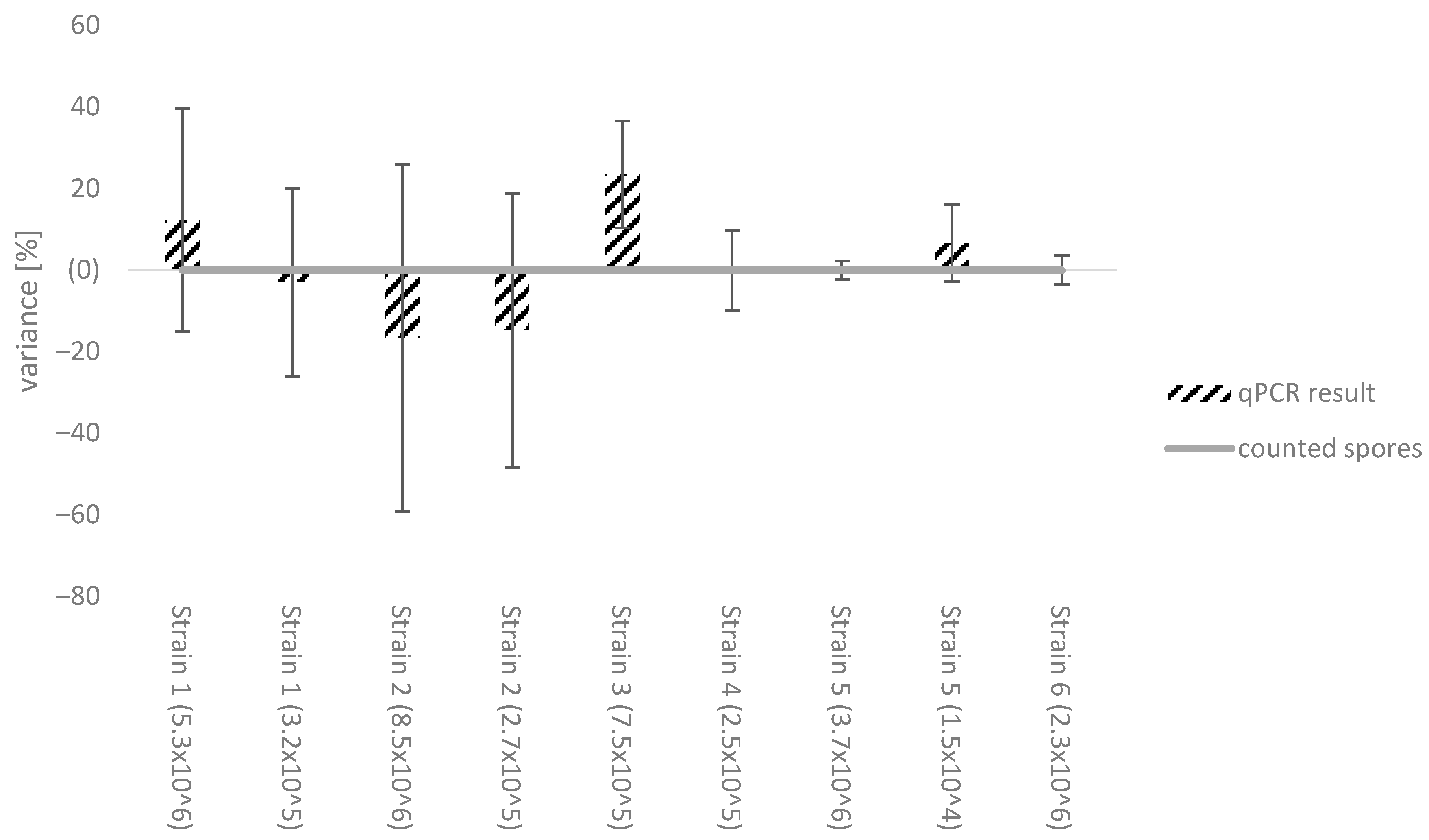
3.2. Effect of Different Botrytis Strains on the Standard Calibration Curve
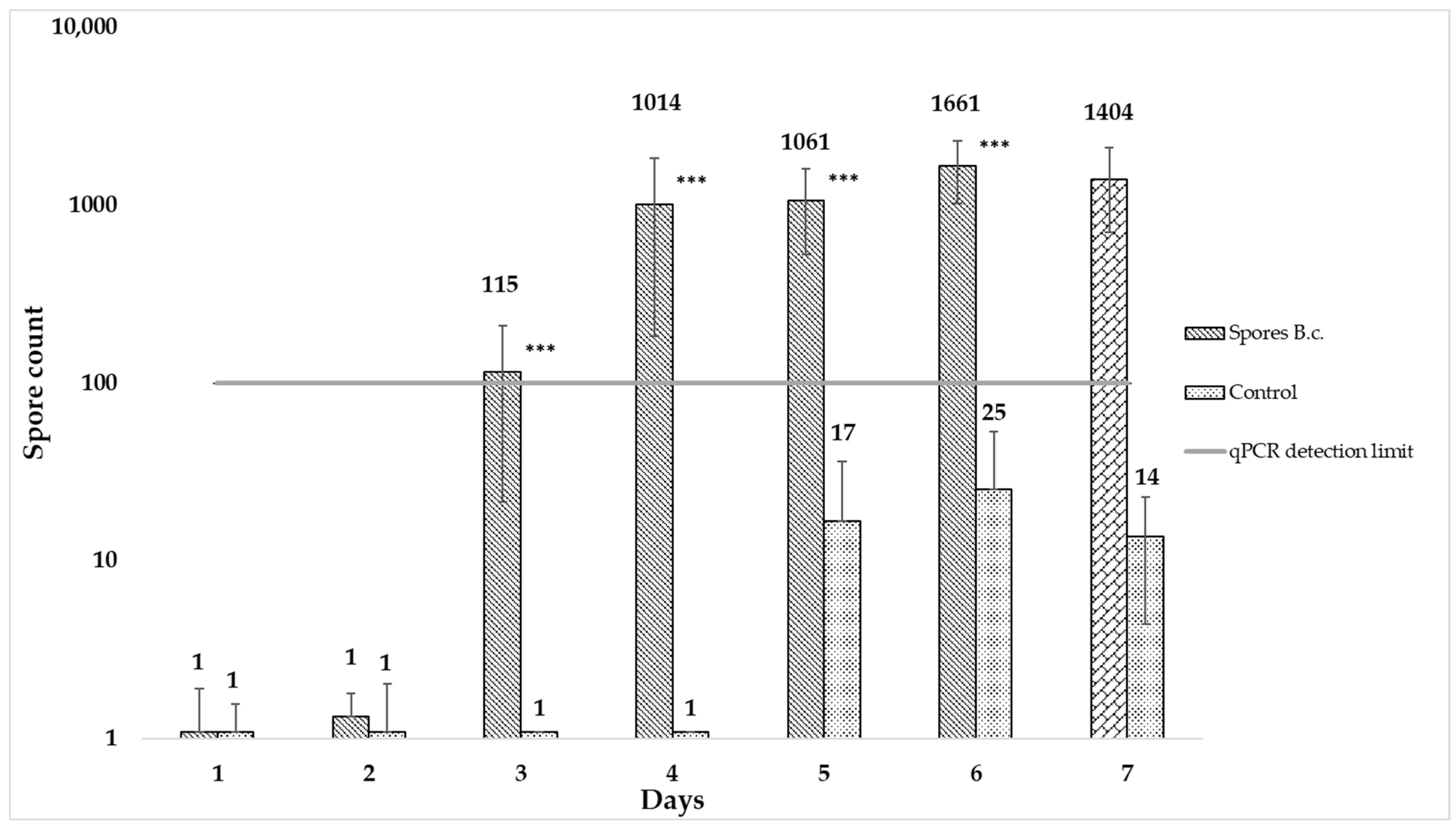
3.3. qPCR as an Early Detection Method—Limit of Detection
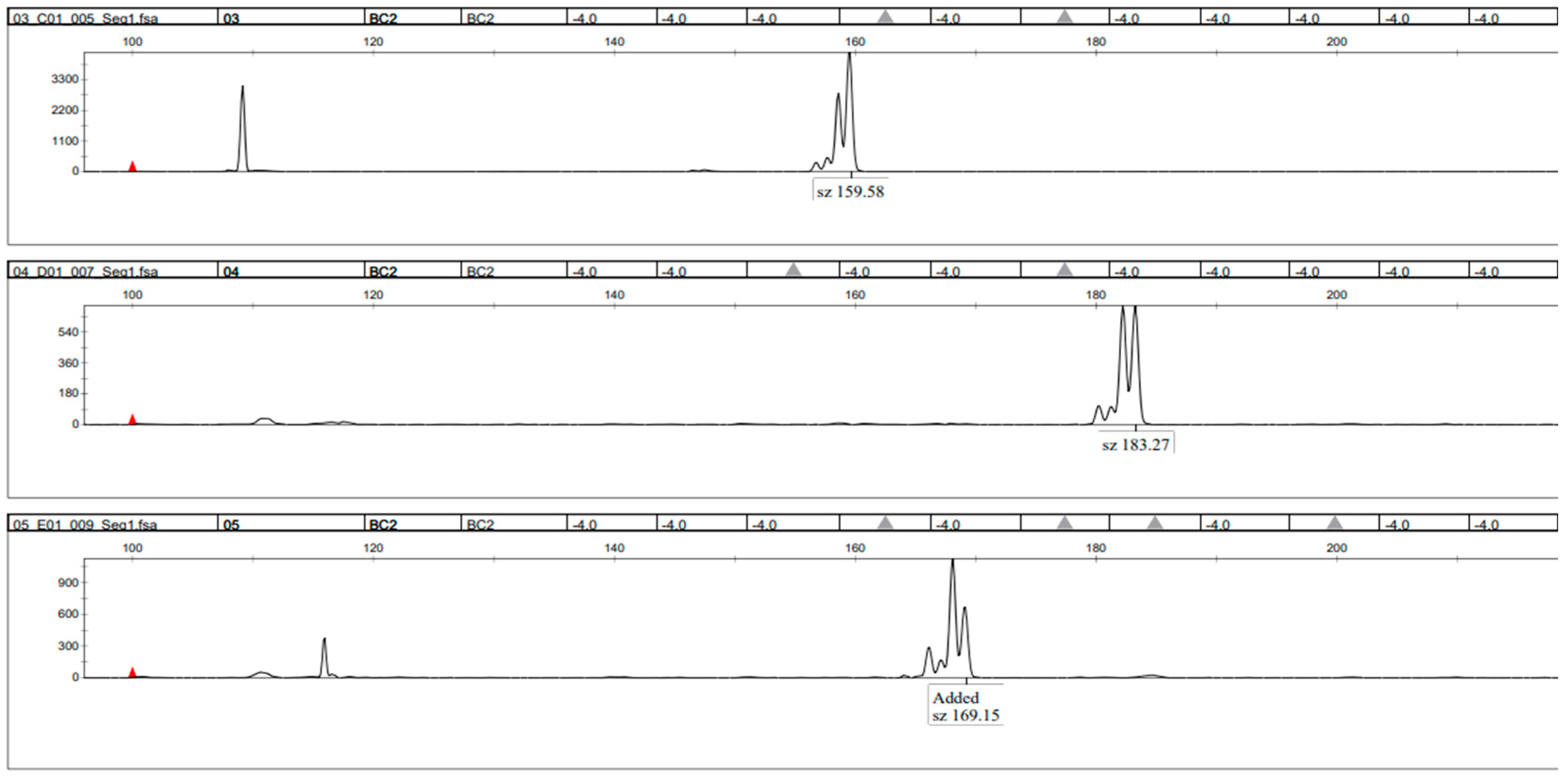
3.4. PCR Method Validation—Size Range of Primer Sets and Composition
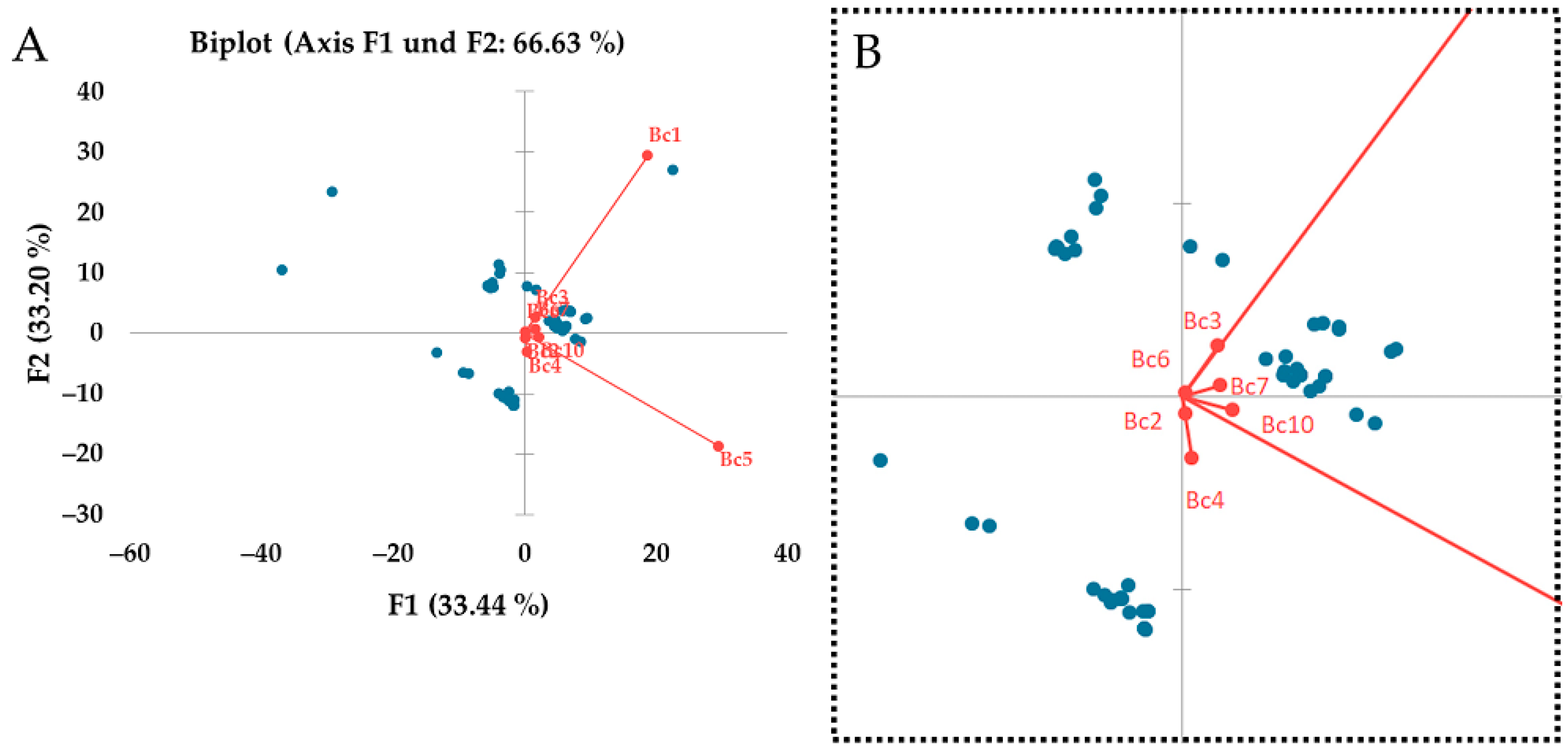
3.5. Regional Differences between Strains
4. Discussion
4.1. Cross-Contamination
4.2. Effect of Different Botrytis Strains on the Standard Calibration Curve
4.3. qPCR as an Early Detection Method against Botrytis cinerea
4.4. PCR Method Validation—Size Ranges of Primer Sets and Composition
4.5. Regional Differences between Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elad, Y.; Pertot, I.; Cotes Prado, A.M.; Stewart, A. Plant Hosts of Botrytis spp. In Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems, 1st ed.; Fillinger, S., Elad, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; Volume 1, pp. 413–486. [Google Scholar]
- Claus, H. Laccases of Botrytis cinerea. In Biology of Microorganisms on Grapes, in Must and in Wine, 1st ed.; König, H., Unden, G., Fröhlich, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1, pp. 339–356. [Google Scholar]
- Ferrada, E.E.; Latorre, B.A.; Zoffoli, J.P.; Castillo, A. Identification and Characterization of Botrytis Blossom blight of Japanese Plums Caused by Botrytis cinerea and B. prunorum sp. Nov, in Chile. Phytopathology 2016, 106, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Steel, C.C.; Blackman, J.W.; Schmidtke, L.M. Grapevine bunch rots: Impacts on wine composition, quality, and potential procedures for the removal of wine faults. J. Agric. Food Chem. 2013, 61, 5189–5206. [Google Scholar] [CrossRef] [PubMed]
- Rigling, D.; Robin, C.; Prospero, S. Mycovirus-Mediated Biological Control. In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 4, pp. 468–477. [Google Scholar]
- Poveda, J.; Barquero, M.; Gonzáles-Andrés, F. Insight into the Microbiological Control Strategies against Botrytis cinerea Using Systemic Plant Resistance Activation. Agronomy 2020, 10, 1822. [Google Scholar] [CrossRef]
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; Van Kan, J.A.L. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef] [PubMed]
- Ribéreau-Gayon, P.; Glories, Y.; Maujean, A.; Dubourdieu, D. Handbook of Enology—The Chemistry of Wine Stabilization and Treatments, 2nd ed.; Jon Wiley & Sons Ltd.: Chichester, UK, 2006; pp. 277–278. [Google Scholar]
- Armijo, G.; Schlechter, R.; Agurto, M.; Munos, D.; Nunez, C.; Arce-Johnson, P. Grapevine Pathogenic Microorganisms: Understanding Infection Strategies and Host Response Scenarios. Front. Plant Sci. 2016, 7, 382. [Google Scholar] [CrossRef]
- Gil-ad, N.L.; Bar-Nun, N.; Mayer, A.M. The possible function of the glucan sheath of Botrytis cinerea: Effects on the distribution of enzyme activities. FEMS Microbiol. Lett. 2001, 199, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Claus, H. How to deal with Uninvited Guets in Wine: Copper and Copper-containing Oxidases. Fermentation 2020, 6, 38. [Google Scholar] [CrossRef]
- Thurston, C.F. The structure and function of fungal laccases. Microbiology 1994, 140, 19–26. [Google Scholar] [CrossRef]
- Vignault, A.; Pascual, O.; Jourdes, M.; Moine, V.; Fermaud, M.; Roudet, J.; Canals, J.M.; Teissedre, P.-L.; Zamora, F. Impact of enological tannins on laccase activity. Oeno One 2019, 53, 27–38. [Google Scholar] [CrossRef]
- La Guerche, S.; Chamont, S.; Blancard, D.; Dubourdieu, D.; Darriet, P. Origin of (-)-geosmin on grapes: On the complementary action of two fungi, Botrytis cinerea and Penicillium expansum. Antonie Van Leeuwenhoek 2005, 88, 131–139. [Google Scholar] [CrossRef]
- La Guerche, S.; De Senneville, L.; Blancard, D. Impact of the Botrytis cinerea strain and metabolism on (−)-geosmin production by Penicillium expansum in grape juice. Antonie Van Leeuwenhoek 2007, 92, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Ky, I.; Lorrain, B.; Jourdes, M.; Pasquier, G.; Fermaud, M.; Gény, L.; Rey, P.; Doneche, B.; Teissedre, P.-L. Assessment of grey mould (Botrytis cinerea) impact on phenolic and sensory quality of Bordeaux grapes, musts and wines for two consecutive vintages. Aust. J. Agric. Res. 2012, 18, 215–226. [Google Scholar] [CrossRef]
- Marchal, R.; Salmon, T.; Gonzalez, R.; Kemp, B.; Vrigneau, C.; Willams, P.; Doco, T. Impact of Botrytis cinerea Contamination on the Characteristics and Foamability of Yeast Macromolecules Released during the Alcoholic Fermentation of a Model Grape Juice. Molecules 2020, 25, 472. [Google Scholar] [CrossRef] [PubMed]
- Thieme Römpp—Botrytizide. Available online: https://roempp.thieme.de/lexicon/RD-02-02370 (accessed on 22 November 2023).
- Caseys, C.; Shi, G.; Soltis, N.E.; Gwinner, R.; Corwin, J.A.; Atwell, S.; Kliebenstein, D.J. Quantitative interactions: The disease outcome of Botrytis cinerea across the plant kingdom. G3 2018, 11, jkab175. [Google Scholar] [CrossRef] [PubMed]
- Mercier, J.; Roussel, D.; Charles, M.-T.; Arul, J. Systemic and Local Responses Associated with UV- and Pathogen-Induced Resistance to Botrytis cinerea in Stored Carrot. Phytopathology 2000, 90, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Mercier, A.; Carpentier, F.; Duplaix, C.; Auger, A.; Pradier, J.M.; Viaud, M.; Gladieux, P.; Walker, A.S. The polyphagous plant pathogenic fungus Botrytis cinerea encompasses host-specialized and generalist populations. Environ. Microbiol. 2019, 21, 4808–4821. [Google Scholar] [CrossRef] [PubMed]
- Shuping, D.S.S.; Eloff, J.N. The Use of Plants to Protect Plants and Food against Fungal Pathogens: A Review. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 120–127. [Google Scholar] [CrossRef]
- DLR Rheinpfalz 2020. Rebschutz 2020. Available online: https://www.dlr.rlp.de/Internet/global/themen.nsf/ (accessed on 2 June 2020).
- European Commission. Available online: https://commission.europa.eu/strategy-and-policy/priorities-2019-2024/european-green-deal_de (accessed on 6 November 2023).
- Li, H.; Chen, Y.; Zhang, Z.; Li, B.; Qin, G.; Tian, S. Pathogenic mechanisms and control strategies of Botrytis cinerea causing post-harvest decay in fruits and vegetables. Food Qual. Saf. 2018, 2, 111–119. [Google Scholar]
- Elad, Y.; Yunis, H.; Katan, T. Multiple fungicide resistance to benzimidazoles, dicarboximides and diethofencarb in field isolates of Botrytis cinerea in Israel. Plant Pathol. 1992, 41, 41–46. [Google Scholar] [CrossRef]
- Grabke, A.; Fernández-Ortuño, D.; Schnabel, G. Fenhexamid resistance in Botrytis cinerea from strawberry fields in the Carolinas is associated with four target gene mutations. Plant Dis. 2012, 97, 271–276. [Google Scholar] [CrossRef]
- Jiang, J.; Ding, L.; Michailides, T.J.; Li, H.; Ma, Z. Molecular characterization of field azoxystrobin-resistant isolates of Botrytis cinerea. Pestic. Biochem. Physiol. 2009, 93, 72–76. [Google Scholar] [CrossRef]
- Fournier, E.; Giraud, T.; Loiseau, A.; Vautrin, D.; Estoup, A.; Solignac, M.; Cornuet, J.M.; Brygoo, Y. Characterization of nine polymorphic microsatellite loci in the fungus Botrytis cinerea (Ascomycota). Mol. Ecol. Notes 2002, 2, 253–255. [Google Scholar] [CrossRef]
- Fournier, E.; Gladieux, P.; Giraud, T. The ‘Dr Jekyll and Mr Hyde fungus’: Noble rot versus gray mold symptoms of Botrytis cinerea on grapes. Evol. Appl. 2013, 6, 960–969. [Google Scholar] [CrossRef]
- Diguta, C.F.; Rousseaux, S.; Weidmann, S.; Bretin, N.; Vincent, B.; Guilloux-Benatier, M.; Alexandre, H. Development of a qPCR assay for specific quantification of Botrytis cinerea on grapes. FEMS Microbiol. Lett. 2010, 313, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.B.; Walsh, K.; Boonham, N.; O’Neill, T.; Pearson, S.; Barker, I. Development of real-time PCR (TaqMan) assays for the detection and quantification of Botrytis cinerea in planta. Plant Physiol. Biochem. 2005, 43, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Huber, F.; Röckel, F.; Schwander, F.; Maul, E.; Eibach, R.; Cousins, P.; Töpfer, R. A view into American grapevine history: Vitis vinifera cv. ‘Sémillon’ is an ancestor of ‘Catawba’ and ‘Concord’. J. Grapevine Res. 2016, 55, 53–56. [Google Scholar]
- Jayawardena, R.S.; Purahong, W.; Zhang, W.; Wubet, T.; Li, X.; Liu, M.; Zhao, W.; Hyde, K.D.; Liu, J.; Yan, J. Biodervisity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. 2018, 90, 1–84. [Google Scholar] [CrossRef]
- Ammour, M.S.; Fedele, G.; Morcia, C.; Terzi, V.; Rossi, V. Quantification of Botrytis cinerea in Grapevine Bunch Trash by Real-Time PCR. Phytopathology 2019, 109, 1312–1319. [Google Scholar] [CrossRef]
- Zhang, M.; Gu, L.; Zheng, P.; Chen, Z.; Dou, X.; Qin, Q.; Cai, X. Improvement of cell counting method for Neubauer counting chamber. J. Clin. Lab. Anal. 2020, 1, e23024. [Google Scholar] [CrossRef]
- Alitieri, V.; Rossi, V.; Fedele, G. Biocontrol of Botrytis cinerea as Influenced by Grapevine Growth Stages and Environmental Conditions. Plants 2023, 12, 3430. [Google Scholar] [CrossRef] [PubMed]
- Pezet, R.; Viret, O.; Perret, C.; Tabachhi, R. Latency of Botrytis cinerea Pers.: Fr. and Biochemical Studies During Growth and Ripening of Two Grape Berry Cultivars, Respectively Susceptible and Resistant to Grey Mould. J. Phytopathol. 2003, 151, 208–214. [Google Scholar] [CrossRef]
- Kretschmer, M.; Hahn, M. Fungicide resistance and genetic diversity of Botrytis cinerea isolates from a vineyard in Germany. J. Plant Dis. Protect. 2008, 115, 214–219. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Shen, F.; Xu, H.; Li, Y.; Liu, D. Characterization of Botrytis cinerea Isolates from grape vineyards in China. Plant Dis. 2018, 102, 40–48. [Google Scholar] [CrossRef] [PubMed]

| Strain | Date of Collection | Location (°N, °E) | Region |
|---|---|---|---|
| 1 | 7 October 2022 | 49.285899, 8.194810 | Edenkoben |
| 2 | 7 October 2022 | 49.285899, 8.194810 | Edenkoben |
| 3 | - | DSMZ Strain | - |
| 4 | 11 October 2013 | 49.553507, 8.154937 | Grünstadt |
| 5 | 7 September 2022 | 49.153390, 8.022599 | Göcklingen |
| 6 | 7 September 2022 | 49.153390, 8.022599 | Göcklingen |
| 7 | 25 September 2022 | 49.368001, 8.186013 | Mußbach |
| 8 | 25 September 2022 | 49.368001, 8.186013 | Mußbach |
| 9 | 6 October 2022 | 50.729375, 7.067771 | Bonn |
| 10 | 25 October 2022 | 49.293299, 8.208207 | Edenkoben |
| 11 | 25 October 2022 | 49.293299, 8.208207 | Edenkoben |
| 12 | 25 October 2022 | 49.293299, 8.208207 | Edenkoben |
| 13 | 25 October 2022 | 49.285899, 8.194810 | Edenkoben |
| 14 | 25 October 2022 | 49.285899, 8.194810 | Edenkoben |
| 15 | 25 October 2022 | 49.285899, 8.194810 | Edenkoben |
| 16 | 28 October 2022 | 49.222630, 8.112190 | Mußbach |
| 17 | 28 October 2022 | 49.222630, 8.112190 | Mußbach |
| 18 | 28 October 2022 | 49.222630, 8.112190 | Mußbach |
| 19 | 4 October 2022 | 48.539542, 7.472217 | Mutzig |
| 20 | 4 October 2022 | 48.539542, 7.472217 | Mutzig |
| 21 | 4 October 2022 | 48.539542, 7.472217 | Mutzig |
| 22 | 25 September 2022 | 49.368001, 8.186013 | Mußbach |
| 23 | 28 October 2022 | 44.612330, −0.321527 | Barsac |
| 24 | 7 September 2022 | 48.054682, 7.624740 | Ihrlingen |
| 25 | 7 September 2022 | 48.054682, 7.624740 | Ihrlginen |
| 26 | 7 September 2022 | 48.054682, 7.624740 | Ihrlingen |
| 27 | 7 September 2022 | 47.979199, 7.833400 | Freiburg |
| 28 | 7 September 2022 | 47.979199, 7.833400 | Freiburg |
| 29 | 7 September 2022 | 47.979199, 7.833400 | Freiburg |
| 30 | 19 September 2022 | 49.625441, 8.647692 | Heppenheim |
| 31 | 19 September 2022 | 49.625797, 8.648443 | Heppenheim |
| 32 | 23 September 2022 | 49.047806, 8.842546 | Heppenheim |
| 33 | 23 September 2022 | 49.047806, 8.842546 | Sternenfels |
| 34 | 23 September 2022 | 49.047806, 8.842546 | Sternenfels |
| 35 | 23 September 2022 | 49.043591, 8.850927 | Sternenfels |
| 36 | 23 September 2022 | 49.043591, 8.850927 | Sternenfels |
| 37 | 15 September 2022 | 50.006921, 8.001069 | Oestrich-Winkel |
| 38 | 15 September 2022 | 50.006921, 8.001069 | Oestrich-Winkel |
| 39 | 15 September 2022 | 50.006921, 8.001069 | Oestrich-Winkel |
| 40 | 15 September 2022 | 50.006921, 8.001069 | Oestrich-Winkel |
| 41 | 15 September 2022 | 50.006921, 8.001069 | Oestrich-Winkel |
| 42 | 7 September 2022 | 49.183386, 8.089927 | Wollmesheim |
| 43 | 7 September 2022 | 49.183386, 8.089927 | Wollmesheim |
| 44 | 7 September 2022 | 49.183386, 8.089927 | Wollmesheim |
| 45 | 25 September 2022 | 49.368001, 8.186013 | Mußbach |
| 46 | 25 September 2022 | 49.368001, 8.186013 | Mußbach |
| 47 | 15 September 2022 | 49.735226, 8.327686 | Mettenheim |
| 48 | 26 September 2022 | 49.952237, 7.026950 | Zeltingen-Rachtig |
| 49 | 26 September 2022 | 49.952237, 7.026950 | Zeltingen-Rachtig |
| 50 | 30 October 2022 | 49.225930, 8.104260 | Mußbach |
| 51 | 30 October 2022 | 49.225930, 8.104260 | Mußbach |
| 52 | 30 October 2022 | 49.225930, 8.104260 | Mußbach |
| 53 | 30 October 2022 | 49.225930, 8.104260 | Mußbach |
| 54 | 15 September 2022 | 50.014349, 8.002587 | Oestrich-Winkel |
| 55 | 15 September 2022 | 50.014349, 8.002587 | Mörzheim |
| 56 | 7 September 2022 | 49.155096, 8.071153 | Mörzheim |
| 57 | 7 September 2022 | 49.155096, 8.071153 | Mörzheim |
| 58 | 7 September 2022 | 49.155096, 8.071153 | Mörzheim |
| 59 | 26 September 2022 | 49.913841, 7.049492 | Zeltingen-Rachtig |
| Strain | Location | Date of Collection |
|---|---|---|
| Strain 1 | Rupperstberg, Germany | 9 September 2008 |
| Strain 2 | Göcklingen, Germany | 17 September 2021 |
| Strain 3 | Italy | 2 February 2011 |
| Strain 4 | Wachenheim, Germany | 16 September 2021 |
| Strain 5 | Geinsheim, Germany | 31 August 2021 |
| Strain 6 | Laumersheim, Germany | 30 August 2021 |
| Strain 7 | Deidesheim, Germany | 22 September 2009 |
| Primer | Sequence |
|---|---|
| QBc | F: GCTGTAATTTCAATGTGCAGAATCC R: GGAGCAACAATTAATCGCATTTC |
| QBc10 | F: GCTGTAATTTCAATGTGCAGAATCCTGTCCCCGGT R: GGAGCAACAATTAATCGCATTTCAAACATGCTG |
| Primer | Sequence | Temperature and Primer Mix | Expected Size Range (Fournier et al. [29]) |
|---|---|---|---|
| Bc1 | F: AGGGAGGGTATGAGTGTGTA R: TTGAGGAGGTGGAAGTTGTA | Primer Mix 1 (50 °C) | 245–281 |
| Bc2 | F: CATACACGTATTTCTTCCAA R: TTTACGAGTGTTTTTGTTAG | Primer Mix 1 (50 °C) | 161–205 |
| Bc3 | F: GGATGAATCAGTTGTTTGTG R: CACCTAGGTATTTCCTGGTA | Primer Mix 2 (50 °C) | 197–229 |
| Bc4 | F: CATCTTCTGGGAACGCACAT R: ATCCACCCCCAAACGATTGT | Primer Mix 1 (50 °C) | 98–125 |
| Bc5 | F: CGTTTTCCAGCATTTCAAGT R: CATCTCATATTCGTTCCTCA | Primer Mix 2 (50 °C) | 143–163 |
| Bc6 | F: ACTAGATTCGAGATTCAGTT R: AAGGTGGTATGAGCGGTTTA | Primer Mix 2 (50 °C) | 88–158 |
| Bc7 | F: CCAGTTTCGAGGAGGTCCAC R: GCCTTAGCGGATGTGAGGTA | Primer Mix 3 (60 °C) | 113–131 |
| Bc10 | F: TCCTCTTCCCTCCCATCAAC R: GGATCTGCGTGGTTATGA | Primer Mix 3 (60 °C) | 158–189 |
| Primer | Sequence | Label | Primer Mix |
|---|---|---|---|
| Bc1 | F: AGGGAGGGTATGAGTGTGTA R: TTGAGGAGGTGGAAGTTGTA | Rox - | Primer Mix 1 |
| Bc2 | F: CATACACGTATTTCTTCCAA R: TTTACGAGTGTTTTTGTTAG | Tamra - | Primer Mix 2 |
| Bc3 | F: GGATGAATCAGTTGTTTGTG R: CACCTAGGTATTTCCTGGTA | 6-Fam - | Primer Mix 2 |
| Bc4 | F: CATCTTCTGGGAACGCACAT R: ATCCACCCCCAAACGATTGT | Rox - | Primer Mix 3 |
| Bc5 | F: CGTTTTCCAGCATTTCAAGT R: CATCTCATATTCGTTCCTCA | 6-Fam - | Primer Mix 1 |
| Bc6 | F: ACTAGATTCGAGATTCAGTT R: AAGGTGGTATGAGCGGTTTA | 6-Fam - | Primer Mix 2 |
| Bc7 | F: CCAGTTTCGAGGAGGTCCAC R: GCCTTAGCGGATGTGAGGTA | Hex - | Primer Mix 3 |
| Bc9 | F: CTCGTCATAACCACGCAGAT R: GCAAGGTCTCGATGTCGATC | 6-Fam - | Primer Mix 3 |
| Bc10 | F: TCCTCTTCCCTCCCATCAAC R: GGATCTGCGTGGTTATGA | Hex - | Primer Mix 1 |
| Primer | Capillary Sequencer | Agarose Gel | Fournier et al. [29] |
|---|---|---|---|
| Bc1 | 219–265 | 195–256 | 245–281 |
| Bc2 | 144–183 | 132–180 | 161–205 |
| Bc3 | 213–221 | 189–240 | 197–229 |
| Bc4 | 118–127 | 107–132 | 98–125 |
| Bc5 | 117–162 | 135–169 | 143–163 |
| Bc6 | 109–125 | 105–130 | 88–158 |
| Bc7 | 111–119 | 105–120 | 113–131 |
| Bc9 | 146–147 | / | 150–194 |
| Bc10 | 178–189 | 165–200 | 158–189 |
| Location | Difference from All Strains |
|---|---|
| Bonn (n = 1) | 85.8% |
| (DSMZ) (n = 1) | 73.5% |
| Ihrlingen (n = 3) | 66.5% ± 6.2% |
| Zeltingen-Rachtig (n = 3) | 62.4% ± 9.0% |
| Mörzheim (n = 4) | 62.4% ± 12.1% |
| Freiburg (n = 3) | 58.9% ± 4.1% |
| Grünstadt (n = 1) | 58.3% |
| Edenkoben (n = 8) | 57.0% ± 8.4% |
| Barsac (n = 1) | 55.3% |
| Wollmesheim (n = 3) | 53.5% ± 2.3% |
| Mußbach (n = 12) | 53.2% ± 5.6% |
| Oestrich-Winkel (n = 6) | 52.3% ± 5.2% |
| Sternenfels (n = 5) | 52.0% ± 8.8% |
| Mutzig (n = 3) | 48.2% ± 2.9% |
| Mettenheim (n = 1) | 48.1% |
| Heppenheim (n = 2) | 46.4% ± 1.7% |
| Göcklingen (n = 2) | 45.6% ± 0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Backmann, L.; Schmidtmann, K.; Wegmann-Herr, P.; Jürgens, A.; Scharfenberger-Schmeer, M. Molecular Biological Methods to Assess Different Botrytis cinerea Strains on Grapes. Microbiol. Res. 2024, 15, 567-581. https://doi.org/10.3390/microbiolres15020037
Backmann L, Schmidtmann K, Wegmann-Herr P, Jürgens A, Scharfenberger-Schmeer M. Molecular Biological Methods to Assess Different Botrytis cinerea Strains on Grapes. Microbiology Research. 2024; 15(2):567-581. https://doi.org/10.3390/microbiolres15020037
Chicago/Turabian StyleBackmann, Louis, Katharina Schmidtmann, Pascal Wegmann-Herr, Andreas Jürgens, and Maren Scharfenberger-Schmeer. 2024. "Molecular Biological Methods to Assess Different Botrytis cinerea Strains on Grapes" Microbiology Research 15, no. 2: 567-581. https://doi.org/10.3390/microbiolres15020037
APA StyleBackmann, L., Schmidtmann, K., Wegmann-Herr, P., Jürgens, A., & Scharfenberger-Schmeer, M. (2024). Molecular Biological Methods to Assess Different Botrytis cinerea Strains on Grapes. Microbiology Research, 15(2), 567-581. https://doi.org/10.3390/microbiolres15020037






