Modeling the Distribution of Wild Cotton Gossypium aridum in Mexico Using Flowering Growing Degree Days and Annual Available Soil Water
Abstract
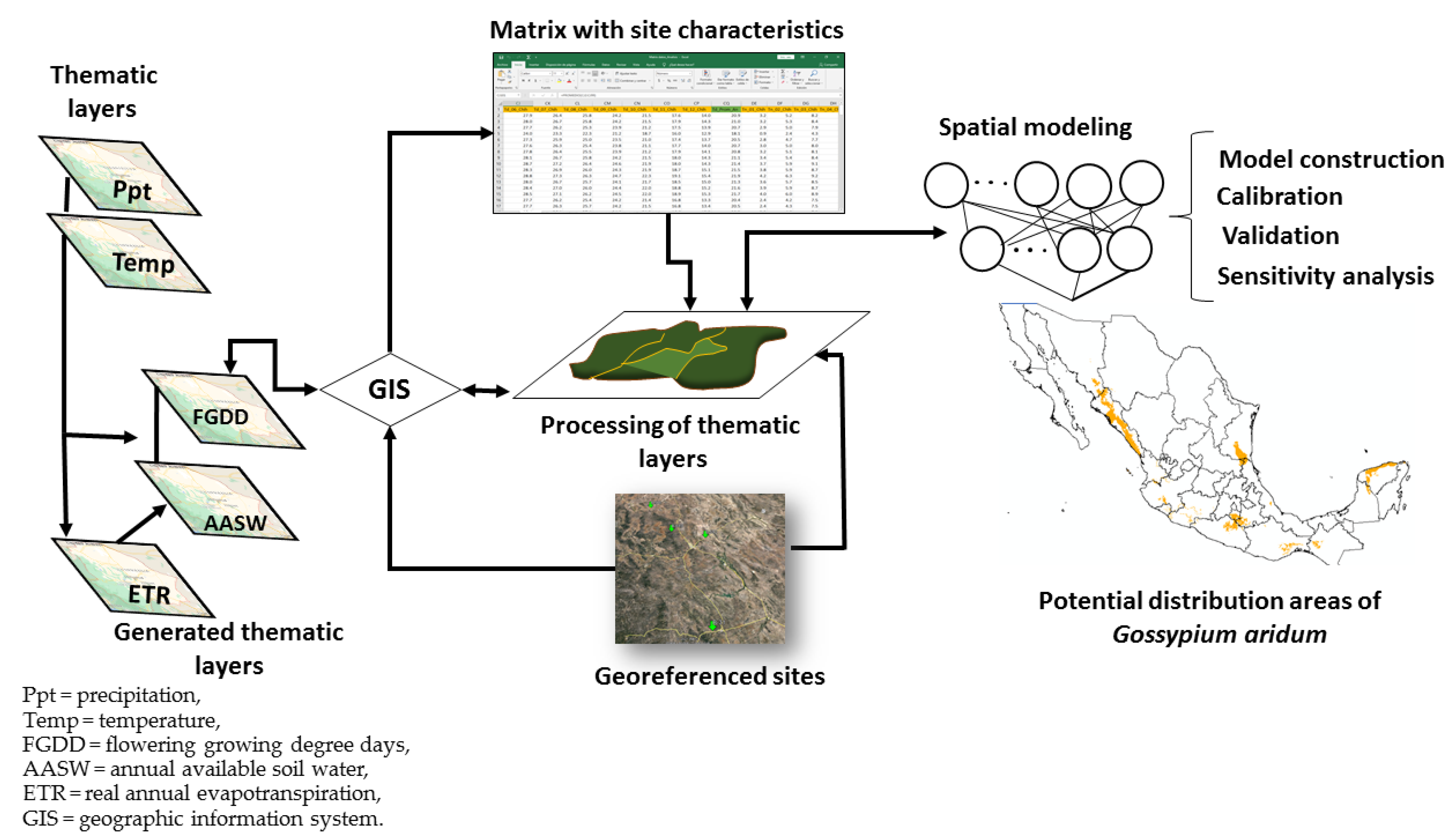
:1. Introduction
2. Materials and Methods
2.1. Study Area and Species under Study
2.2. Plant Databases
2.3. Climate Database and Parameters
2.3.1. Annual Available Soil Water
2.3.2. Growing Degree Days for Flowering
2.4. Characterization of the Sites
2.5. Model Construction
2.6. Model Calibration
2.7. Model Validation
2.8. Sensitivity Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Characterization of the Sites
3.2. Model Construction
3.3. Model Calibration
3.4. Model Validation
3.5. Sensitivity Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Contreras-Toledo, A.R.; Cortés-Cruz, M.A.; Costich, D.; de Lourdes Rico-Arce, M.; Brehm, J.M.; Maxted, N. A crop wild relative inventory for Mexico. Crop Sci. 2018, 58, 1292–1305. [Google Scholar] [CrossRef] [Green Version]
- Goettsch, B.; Urquiza-Haas, T.; Koleff, P.; Acevedo Gasman, F.; Aguilar-Meléndez, A.; Alavez, V.; Alejandre-Iturbide, G.; Aragon Cuevas, F.; Azurdia Pérez, C.; Carr, J.A.; et al. Extinction risk of Mesoamerican crop wild relatives. Plants People Planet 2021, 3, 775–795. [Google Scholar] [CrossRef]
- Hossain, A.; Maitra, S.; Pramanick, B.; Bhutia, K.L.; Ahmad, Z.; Moulik, D.; Syed, M.A.; Shankar, T.; Adeel, M.; Hassan, M.M.; et al. Wild relatives of plants as sources for the development of abiotic stress tolerance in plants. In Plant Perspectives to Global Climate Changes Developing Climate-Resillient Plants, 1st ed.; Aftab, T., Roychoudhury, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 471–518. [Google Scholar]
- EL Sabagh, A.; Hossain, A.; Islam, M.; Barutcular, C.; Ratnasekera, D.; Gormus, O.; Amanet, K.; Mubeen, M.; Nasim, W.; Fahad, S.; et al. Drought and heat stress in cotton (Gossypium hirsutum L.): Consequences and their possible mitigation strategies. In Agronomic Crops; Springer: Singapore, 2020; pp. 613–634. [Google Scholar] [CrossRef]
- Gechev, T.; Petrov, V. Reactive oxygen species and abiotic stress in plants. Int. J. Mol. Sci. 2020, 21, 7433. [Google Scholar] [CrossRef] [PubMed]
- Majeed, S.; Chaudhary, M.T.; Hulse-Kemp, A.M.; Azhar, M.T. Introduction: Crop Wild Relatives in Plant Breeding. In Wild Germplasm for Genetic Improvement in Crop Plants; Azhar, M.T., Wani, S.H., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2021; pp. 1–18. [Google Scholar]
- Burgeff, C.; Huerta, E.; Acevedo, F.; Sarukhán, J. How much can GMO and non-GMO cultivars coexist in a megadiverse country? AgBioForum 2014, 17, 90–101. [Google Scholar]
- Rocha-Munive, M.G.; Soberón, M.; Castañeda, S.; Niaves, E.; Scheinvar, E.; Eguiarte, L.E.; Mota-Sánchez, D.; Rosales-Robles, E.; Nava-Camberos, U.; Martínez-Carrillo, J.L.; et al. Evaluation of the impact of genetically modified cotton after 20 years of cultivation in Mexico. Front. Bioeng. Biotechnol. 2018, 6, 82. [Google Scholar] [CrossRef]
- Wendel, J.F.; Grover, C.E. Taxonomy and evolution of the cotton genus, Gossypium. Cotton 2015, 57, 25–44. [Google Scholar]
- Perez-Mendoza, C.; Tovar-Gomez, M.R.; Baez-Gonzalez, A.D.; Flores-Zarate, M. Recolección de germoplasma del género Gossypium en el estado de Guerrero. In Foro de Estudios Sobre Guerrero; COCYTIEG (Consejo de Ciencia y Tecnologia en Innovacion del Estado de Guerrero): Iguala, Mexico, 2016; pp. 1043–1047. [Google Scholar]
- Ulloa, M.; Stewart, J.M.; Acosta, N.S. Cotton genetic resources in the western states of Mexico: In situ conservation status and germplasm collection for ex situ preservation. Genet. Resour. Crop Evol. 2006, 5, 653–668. [Google Scholar] [CrossRef]
- Álvarez, I.; Wendel, J.F. Cryptic interspecific introgression and genetic differentiation within Gossypium aridum (Malvaceae) and its relatives. Evolution 2006, 60, 505–517. [Google Scholar] [CrossRef] [Green Version]
- CONABIO-SNIB. Comisión Nacional Para el Conocimiento y Uso de la Biodiversidad-Sistema Nacional de Información Sobre Biodiversidad. Gossypium. Available online: https://enciclovida.mx/especies/139343-gossypium (accessed on 17 September 2021).
- Zeng, L.; Stetina, S.R.; Erpelding, J.E.; Bechere, E.; Turley, R.B.; Scheffler, J. History and current research in the USDA-ARS cotton breeding program at Stoneville, MS. J. Cotton Sci. 2018, 22, 24–35. [Google Scholar]
- Shim, J.; Mangat, P.K.; Angeles-Shim, R.B. Natural variation in wild Gossypium species as a tool to broaden the genetic base of cultivated cotton. J. Plant Sci. Curr. Res 2018, 2. [Google Scholar] [CrossRef]
- Guo, Q.; Zhao, L.; Fan, X.; Xu, P.; Xu, Z.; Zhang, X.; Meng, S.; Shen, X. Transcription factor GarWRKY5 is involved in salt stress response in diploid cotton species (Gossypium aridum L.). Int. J. Mol. Sci. 2019, 20, 5244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, G.; Grover, C.E.; Yuan, D.; Dong, Y.; Miller, E.; Conover, J.L.; Wendel, J.F. Evolution and Diversity of the Cotton Genome. In Cotton Precision Breeding, 1st ed.; Rahman, M., Zafar, Y., Zhang, T., Eds.; Springer: Cham, Switzerland, 2021; pp. 25–78. [Google Scholar]
- Saleem, H.; Tanees, C.M.; Shakeel, A.; Hussain, W.S.; Du, X.; Tehseen, A.M. Wild Cotton Genepool: An Unopened Treasure. In Wild Germplasm for Genetic Improvement in Crop Plants, 1st ed.; Azhar, M.T., Wani, S.H., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2021; pp. 19–53. [Google Scholar]
- Frelichowski, J.; Jenderek, M.M.; Greene, S.; Hinze, L.L.; Love, J. Conservation of Crop Wild Relatives of Cotton (Gossypium hirsutum L.) Originating from the USA and Mexico. In Proceedings of the ASA, CSSA, and CSA International Annual Meeting, Baltimore, MD, USA, 4 November 2018. [Google Scholar]
- SINAREFI (Sistema Nacional de Recursos Fitogeneticos para la Alimentación y la Agricultura). Red de Algodón: Datos de Pasaporte de Accesiones de Algodon (2010–2015) para los Recursos Fitogenéticos para la Alimentación y la Agricultura (RFAA). D.F. México, 2015 SAGARPA/SNICS/SINAREFI.
- INEGI. Instituto Nacional de Informacion Estadistica y Geografía. Climatología. 2007. Available online: https://www.inegi.org.mx/temas/climatologia/ (accessed on 29 August 2021).
- Sharma, A.; Deepa, R.; Sankar, S.; Pryor, M.; Stewart, B.; Johnson, E.; Anandhi, A. Use of growing degree indicator for developing adaptive responses: A case study of cotton in Florida. Ecol. Indic. 2021, 124, 107383. [Google Scholar] [CrossRef]
- Fraisse, C.W.; Paula-Moraes, S.V. Degree Days: Heating, Cooling, and Growing. Agricultural and Biological Engineering Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. Available online: http://edis.ifas.ufl.edu (accessed on 3 September 2021).
- Gudadhe, N.N.; Kumar, N.; Pisal, R.R.; Mote, B.M.; Dhonde, M.B. Evaluation of agrometeorological indices in relation to crop phenology of cotton (Gossipium spp.) and chickpea (Cicer aritinum L.) at Rahuri region of Maharashtra. Trends Biosci. 2013, 6, 246–250. [Google Scholar]
- Casenave, E.C.; Toselli, M.E. Hydropriming as a pre-treatment for cotton germination under thermal and water stress conditions. Seed Sci. Technol. 2007, 35, 88–98. [Google Scholar] [CrossRef]
- Maeda, A.B.; Wells, L.W.; Sheehan, M.A.; Dever, J.K. Stories from the Greenhouse—A Brief on Cotton Seed Germination. Plants 2021, 10, 2807. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.A. Climate Change Impacts on Agriculture in Europe: Spatial Modelling of Winter Wheat. Ph.D. Thesis, University of Oxford, Oxford, UK, 1999. [Google Scholar]
- Mudassir, M.A.; Rasul, F.; Khaliq, T.; Yaseen, M. Conformance of sowing dates for maximizing heat use efficiency and seed cotton yield in arid to semi-arid cotton zone of Pakistan. Environ. Sci. Pollut. Res. 2021, 29, 11359–11373. [Google Scholar] [CrossRef]
- Pearson, R.G.; Dawson, T.P.; Berry, P.M.; Harrison, P.A. SPECIES: A Spatial Evaluation of Climate Impact on the Envelope of Species. Ecol. Model. 2002, 154, 289–300. [Google Scholar] [CrossRef]
- Andréassian, V.; Sari, T. Technical Note: On the puzzling similarity of two water balance formulas–Turc–Mezentsev vs. Tixeront–Fu. Hydrol. Earth Syst. Sci. 2019, 23, 2339–2350. [Google Scholar] [CrossRef] [Green Version]
- Sanchez San Roman, F.J. Evapotranspiracion.Depto. Geologia Universidad de Salamanca. Available online: https://hidrologia.usal.es/ (accessed on 22 November 2021).
- Reddy, K.R.; Hodges, H.F.; McKinion, J.M.; Wall, G.W. Temperature effects on Pima cotton growth and development. Agron. J. 1992, 84, 237–243. [Google Scholar] [CrossRef]
- Fraisse, C.W.; Bellow, J.; Brown, C. Degree Days: Heating, Cooling, and Growing; IFAS Extension; ABE 381; 1-7EDIS; University of Florida: Gainesville, FL, USA, 2007. [Google Scholar]
- Supak, J.R. Understanding and Using Heat Units in Cotton Production Systems. In Summary of Proceedings-Western Cotton Producers Conference (USA). Available online: http://cotton.tamu.edu/General%20Production/arch-understandingandusingheat.pdf (accessed on 12 December 2021).
- Tcach, N.; Paytas, M. Incidencia de altas temperaturas durante el reproductivo sobre el rendimiento de algodón (Gossypium hirsutum) cultivado en diferentes distanciamientos entre surcos. RIA. Rev. Investig. Agropecu. 2020, 46, 56–65. [Google Scholar]
- Reddy, K.R.; Kakanl, V.G.; Zhao, D.; Kotl, S.; Gao, W. Interactive effects of ultraviolet-B radiation and temperature on cotton physiology, growth, development and hyperspectral reflectance. Photochem. Photobiol. 2004, 79, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.A.; Porter, J.R.; Downing, T.E. Scaling-up the AFRCWHEAT2 model to assess phenological development for wheat in Europe. Agric. For. Meteorol. 2000, 101, 167–186. [Google Scholar] [CrossRef]
- Caldow, R.W.G.; Racey, P.A. Introduction: Largescale processes in ecology and hydrology. J. Appl. Ecol. 2000, 37, 6–12. [Google Scholar] [CrossRef]
- Pearson, R.G.; Dawson, T.P. Predicting the impacts of climate change on the distribution of species: Are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Guarino, L.; Jarvis, A.; Hijmans, R.J.; Maxted, N. Geographic Information Systems (GIS) and the Conservation and Use of Plant Genetic Resources. In Managing Plant Genetic Resources; Engels, J.M.M., Rao, V.R., Brown, A.H.D., Jackson, M.T., Eds.; IPGRI: Rome, Italy, 2002; pp. 387–404. [Google Scholar]
- Richardson, J.; Berish, C. Data and information issues in modeling for resource management decision making: Communication is the key. In Ecological Modeling for Resource Management, 1st ed.; Dale, V.H., Ed.; Springer: New York, NY, USA, 2003; pp. 167–179. [Google Scholar]
- Peterson, A.T.; Benz, B.W.; Papes, M. Highly pathogenic H5N1 avian influenza: Entry pathways into North America via bird migration. PLoS ONE 2007, 2, e261. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, S.; Watson, G.; Pearce, J.; Drielsma, M. Extended statistical approaches to modelling spatial pattern in biodiversity in northeast New South Wales. I. Species-level modelling. Biodivers. Conserv. 2002, 11, 2275–2307. [Google Scholar] [CrossRef]
- Engler, R.; Guisan, A.; Rechsteiner, L. An improved approach for predicting the distribution of rare and endangered species from occurrence and pseudo-absence data. J. Appl. Ecol. 2004, 41, 263–274. [Google Scholar] [CrossRef]
- Saltelli, A. What is sensitivity analysis? In Sensitivity Analysis, 1st ed.; Saltelli, A., Chan, K., Scott, E.M., Eds.; John Wiley and Sons: West Sussex, UK, 2005; pp. 3–14. [Google Scholar]
- Pearson, R.G.; Raxworthy, C.J.; Nakamura, M.; Peterson, T.A. Predicting species distributions from small numbers of occurrence records: A test case using cryptic geckos in Madagascar. J. Biogeogr. 2007, 34, 102–117. [Google Scholar] [CrossRef]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Buckland, S.T.; Elston, D.A. Empirical models for the spatial distribution of wildlife. J. Appl. Ecol. 1993, 30, 478–495. [Google Scholar] [CrossRef]
- Fielding, A. How should accuracy be measured? In Machine Learning Methods for Ecological Applications; Springer: Boston, MA, USA, 1999; pp. 209–223. [Google Scholar]
- Forbes, A.D. Classification-algorithm evaluation: Five performance measures based on confusion matrices. J. Clin. Monit. 1995, 11, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Sim, J.; Wright, C.C. The Kappa statistic in reliability studies: Use, interpretation, and sample size requirements. Phys. Ther. 2005, 85, 257–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manel, S.; Williams, H.C.; Ormerod, S.J. Evaluating presence–absence models in ecology: The need to account for prevalence. J. Appl. Ecol. 2001, 38, 921–931. [Google Scholar] [CrossRef]
- Advancing Cotton Education. Growth and Development of a Cotton Plant. National Cotton Council of America. Available online: https://www.cotton.org/tech/ace/growth-and-development.cfm (accessed on 18 February 2022).
- Fryxell, P.A. The Natural History of the Cotton Tribe (Malvaceae, Tribe Gossypieae); Texas A & M University Press: Collage Station, TX, USA, 1979; p. 264. [Google Scholar]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Gertseva, V.; Gertsev, V.; Ponomarev, N.Y. Integrative model of a population distribution in a habitat. Ecol. Model. 2005, 193, 575–588. [Google Scholar] [CrossRef]
- Diarra, A.; Barbier, B.; Zongo, B.; Yacouba, H. Impact of climate change on cotton production in Burkina Faso. Afr. J. Agric. Res. 2017, 12, 494–501. [Google Scholar]
- Reddy, K.R.; Doma, P.R.; Mearns, L.O.; Boone, M.Y.L.; Hodges, H.F.; Richardson, A.G.; Kakani, V.G. Simulating the impacts of climate change on cotton production in the Mississippi Delta. Clim. Res. 2002, 22, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Raxworthy, C.J. Introduction to the reptiles. In The Natural History of Madagascar; Goodman, S.M., Benstead, J.P., Eds.; University of Chicago Press: Chicago, IL, USA, 2003; pp. 934–949. [Google Scholar]
- Sánchez-Reyes, U.J.; Jones, R.W.; Raszick, T.J.; Ruiz-Arce, R.; Sword, G.A. Potential Distribution of Wild Host Plants of the Boll Weevil (Anthonomus grandis) in the United States and Mexico. Insects 2022, 30, 337. [Google Scholar] [CrossRef]
- Vaca, R.A. ‘Gossypium aridum. Distribución Potencial’, Escala: 1:5,000,000. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO). 2020. Available online: http://www.conabio.gob.mx/informacion/gis/ (accessed on 2 March 2022).
- Bourg, N.A.; McShea, W.J.; Gill, D.E. Putting a CART before the search: Successful habitat prediction for a rare forest herb. Ecology 2005, 86, 2793–2804. [Google Scholar] [CrossRef]







| MODEL | FGDD ¥ | AASW § |
|---|---|---|
| 1 | 330–580 | 4–110 |
| 2 | 460–766 | 4–77 |
| 3 | 330–766 | 4–103 |
| 4 | 460–860 | 0.0–77 |
| 5 | 275–460 | 150–210 |
| Models | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Kappa | 0.58 | 0.53 | 0.44 | 0.47 | 0.43 |
| Overall accuracy | 0.79 | 0.76 | 0.71 | 0.73 | 0.71 |
| Sensitivity | 0.85 | 0.85 | 0.82 | 0.79 | 0.82 |
| Specificity | 0.74 | 0.70 | 0.65 | 0.69 | 0.65 |
| Positive Predictive power | 0.73 | 0.66 | 0.58 | 0.67 | 0.58 |
| Negative Predictive power | 0.86 | 0.87 | 0.86 | 0.80 | 0.86 |
| Odds ratio | 16 | 13 | 9 | 8 | 8 |
| MODEL | FGDD ¥ | AASW § |
|---|---|---|
| A | 330–860 | 4–110, 150–210 |
| B | 460–860 | 4–110, 150–210 |
| C | 330–860 | 4–110 |
| D | 460–860 | 4–110 |
| Model | ||||
|---|---|---|---|---|
| Test | A | B | C | D |
| Kappa | 0.64 | 0.53 | 0.67 | 0.55 |
| Overall accuracy | 0.82 | 0.76 | 0.83 | 0.77 |
| Sensitivity | 0.80 | 0.81 | 0.85 | 0.84 |
| Specificity | 0.84 | 0.72 | 0.82 | 0.72 |
| Positive Predictive power | 0.87 | 0.71 | 0.83 | 0.70 |
| Negative Predictive power | 0.77 | 0.82 | 0.84 | 0.86 |
| Odds ratio | 21 | 11 | 26 | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baez-Gonzalez, A.D.; Melgoza-Castillo, A.; Royo-Marquez, M.H.; Kiniry, J.R.; Meki, M.N. Modeling the Distribution of Wild Cotton Gossypium aridum in Mexico Using Flowering Growing Degree Days and Annual Available Soil Water. Sustainability 2022, 14, 6383. https://doi.org/10.3390/su14116383
Baez-Gonzalez AD, Melgoza-Castillo A, Royo-Marquez MH, Kiniry JR, Meki MN. Modeling the Distribution of Wild Cotton Gossypium aridum in Mexico Using Flowering Growing Degree Days and Annual Available Soil Water. Sustainability. 2022; 14(11):6383. https://doi.org/10.3390/su14116383
Chicago/Turabian StyleBaez-Gonzalez, Alma Delia, Alicia Melgoza-Castillo, Mario Humberto Royo-Marquez, James R. Kiniry, and Manyowa N. Meki. 2022. "Modeling the Distribution of Wild Cotton Gossypium aridum in Mexico Using Flowering Growing Degree Days and Annual Available Soil Water" Sustainability 14, no. 11: 6383. https://doi.org/10.3390/su14116383
APA StyleBaez-Gonzalez, A. D., Melgoza-Castillo, A., Royo-Marquez, M. H., Kiniry, J. R., & Meki, M. N. (2022). Modeling the Distribution of Wild Cotton Gossypium aridum in Mexico Using Flowering Growing Degree Days and Annual Available Soil Water. Sustainability, 14(11), 6383. https://doi.org/10.3390/su14116383






