Soil Microbial Communities in Desert Grassland around Rare Earth Mine: Diversity, Variation, and Response Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Setting up Sampling Sites and Collection of Soil Samples
2.3. Investigation of Plants
2.4. Soil Physical and Chemical Properties
2.5. Microbial DNA Extraction, Amplification, and Sequencing
2.6. Bioinformatics Analyses
2.7. Statistic Analyses
3. Results
3.1. REE, Soil Properties and Plant Communities
3.2. Diversity of Soil Microbial Communities
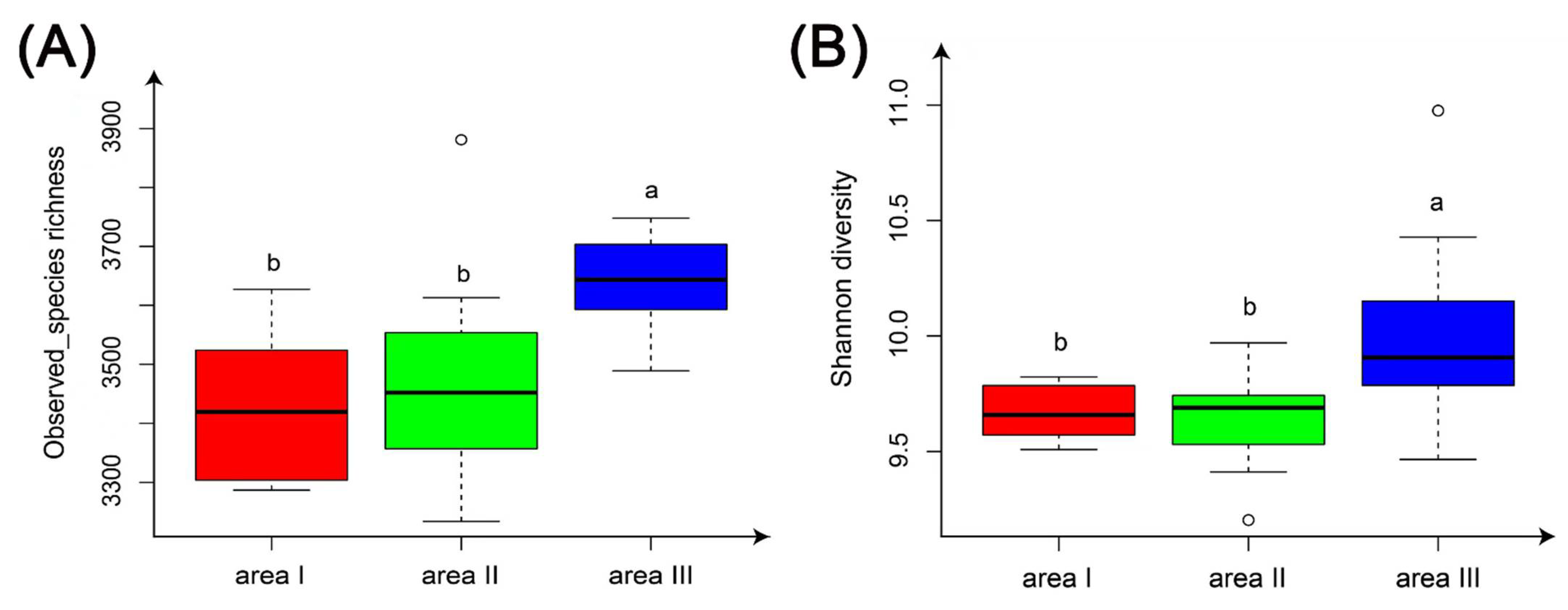
3.2.1. Alpha Diversity
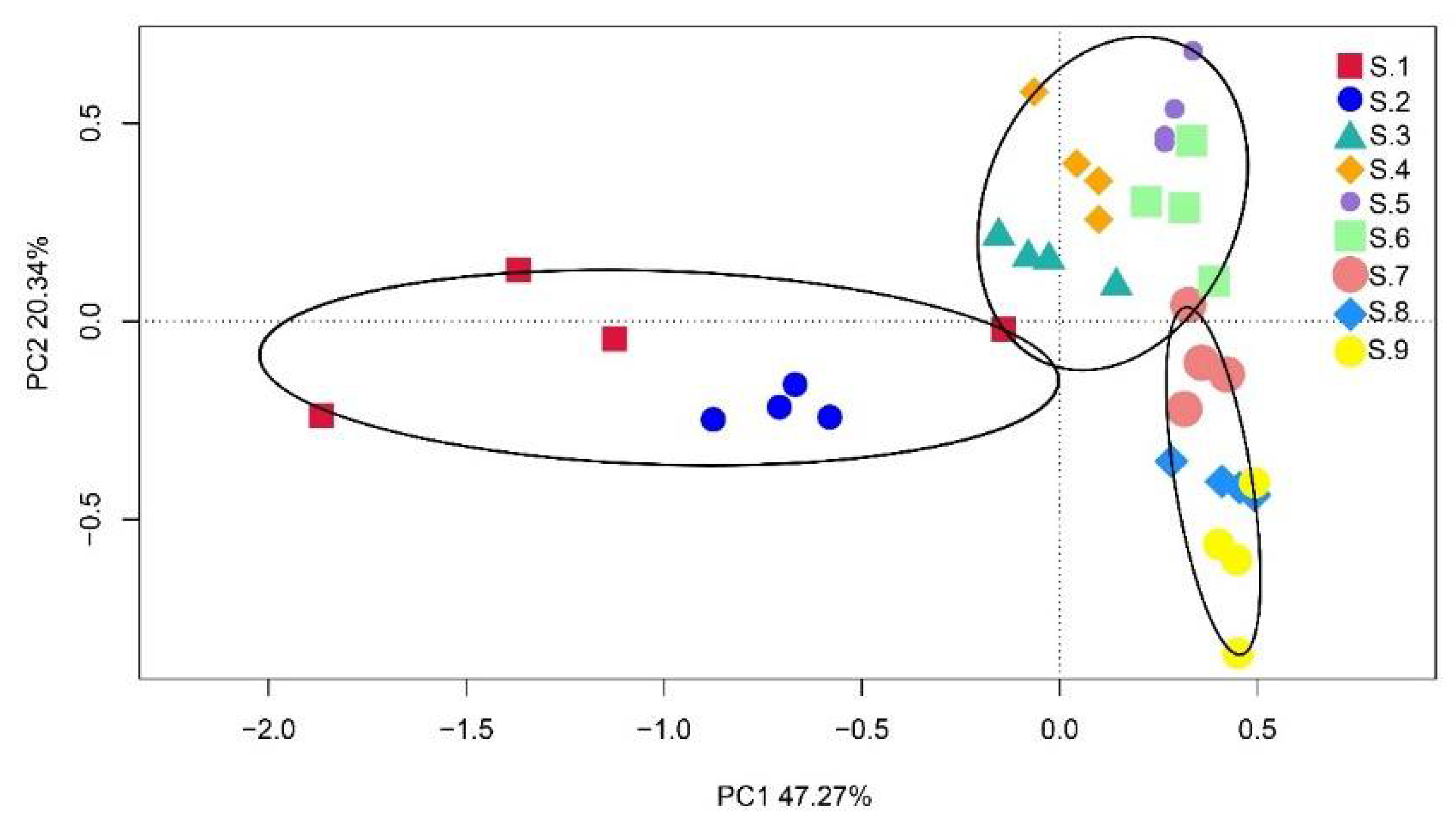
3.2.2. Beta Diversity
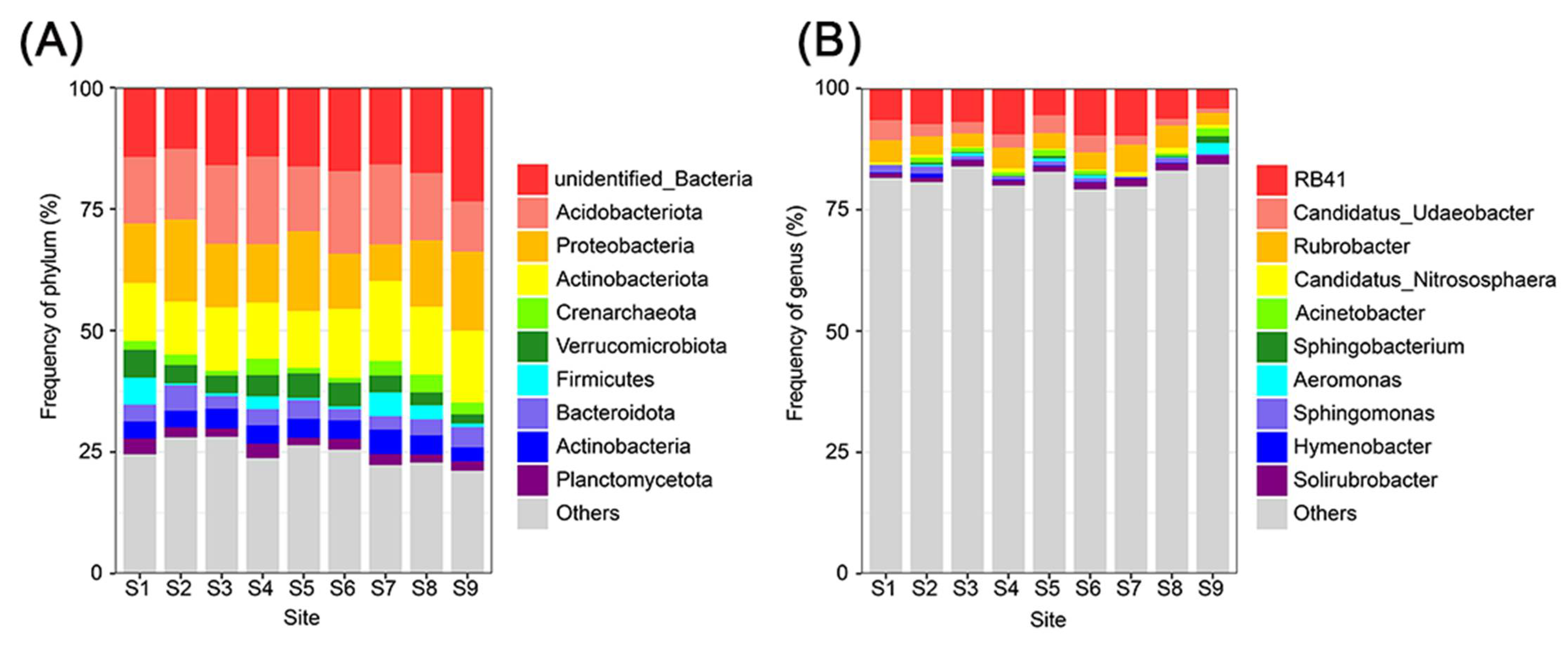
3.3. Taxonomic Profiles of the Soil Microbial Communities
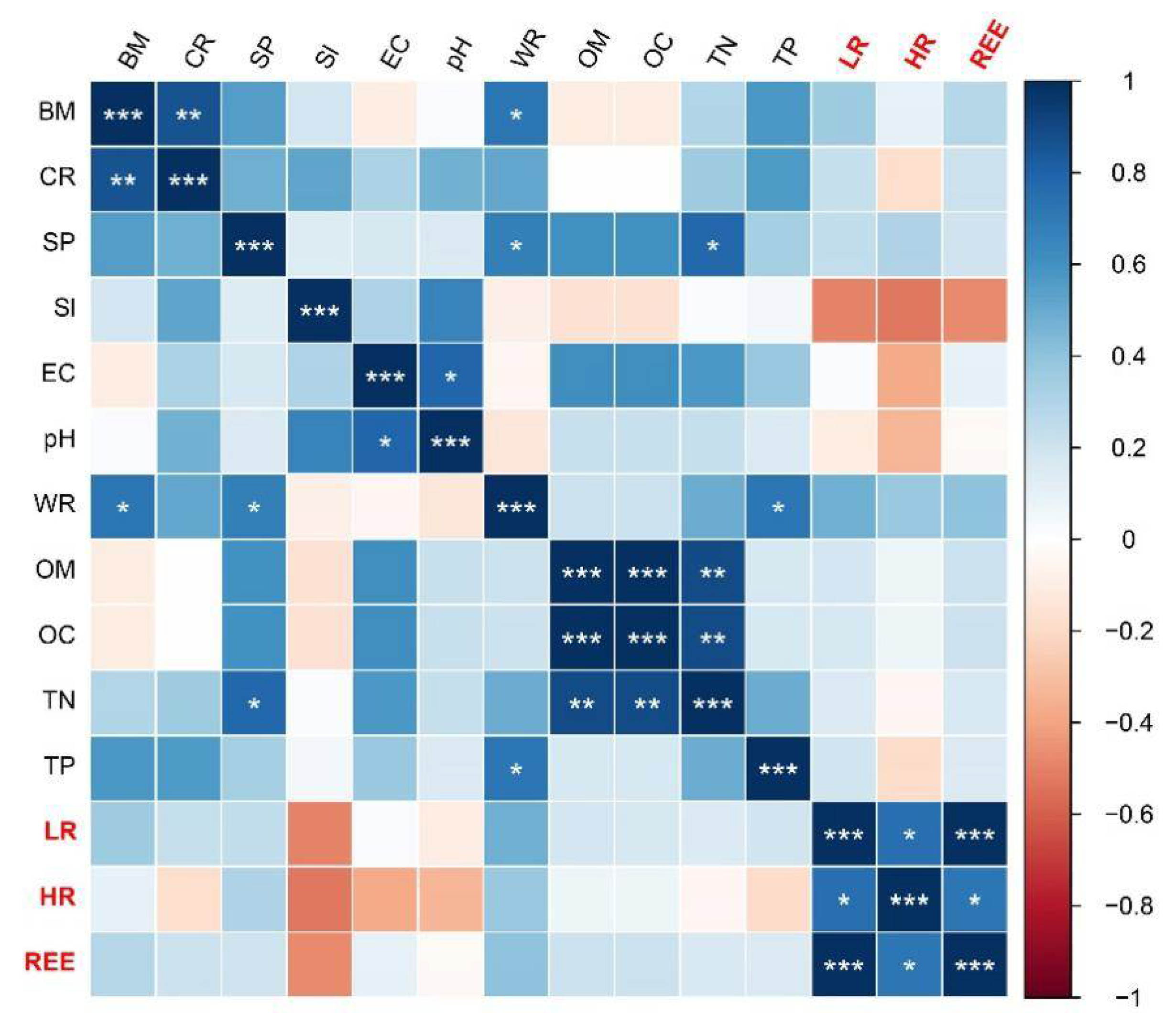
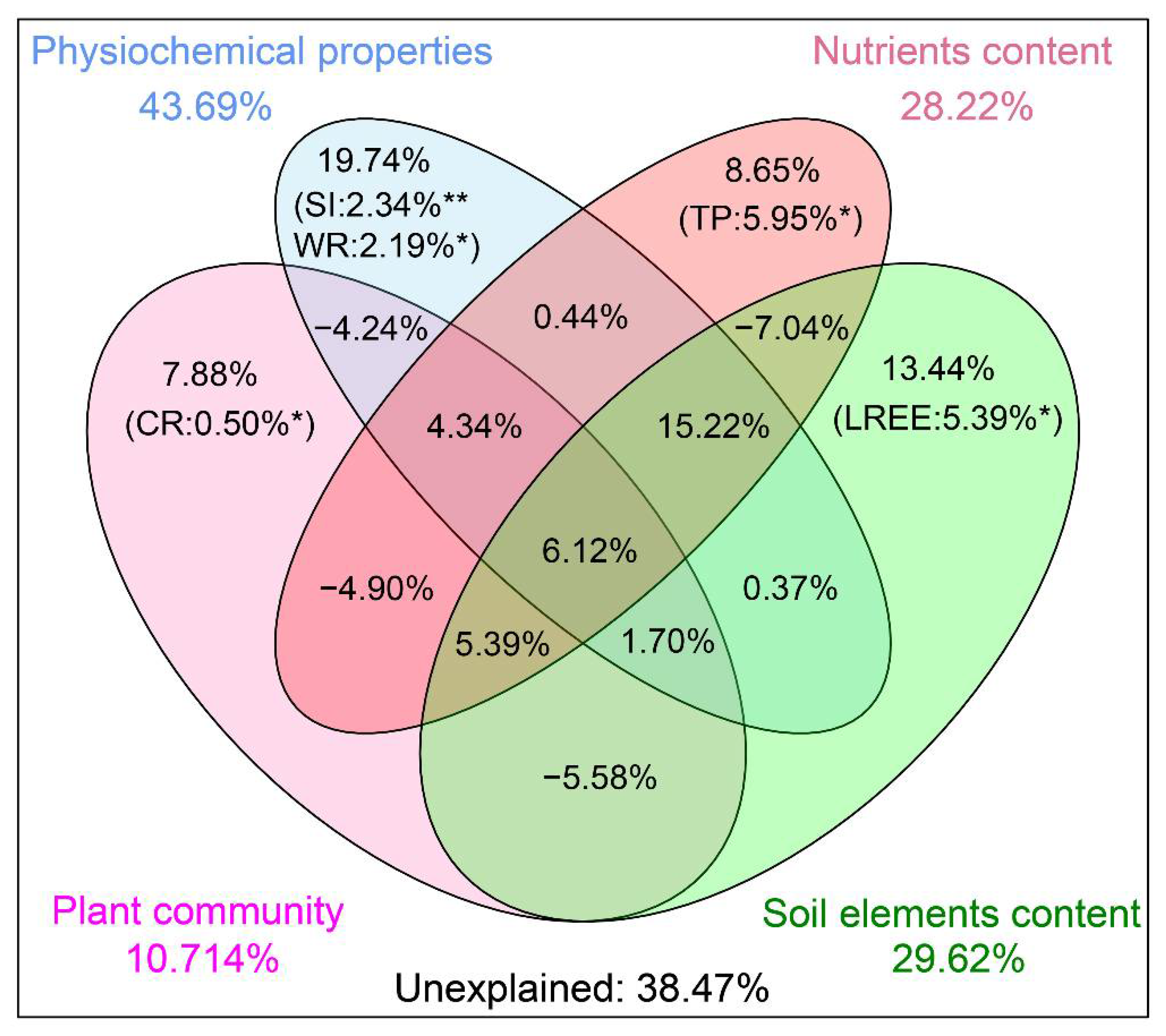
3.4. Environmental Factors Affecting Microbial Communities
4. Discussion
4.1. Effects of Exogenous REEs on Soil Physical and Chemical Properties
4.2. Effects of Exogenous REE on Soil Microbial Community
4.3. Environmental Factors Shape the Soil Microbial Community Together
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Castor, S.B.; Hedrick, J.B. Rare Earth Elements. Ind. Miner. Rocks 2006, 7, 769–792. [Google Scholar] [CrossRef]
- Ramos, S.J.; Dinali, G.S.; Oliveira, C.; Martins, G.C.; Moreira, C.G.; Siqueira, J.O.; Guilherme, L.R.G. Rare Earth Elements in the Soil Environment. Curr. Pollut. Rep. 2016, 2, 28–50. [Google Scholar] [CrossRef]
- Gwenzi, W.; Mangori, L.; Danha, C.; Chaukura, N.; Dunjana, N.; Sanganyado, E. Sources, behaviour, and environmental and human health risks of high-technology rare earth elements as emerging contaminants. Sci. Total Environ. 2018, 636, 299–313. [Google Scholar] [CrossRef] [PubMed]
- USGS U.S. Geological Survey, Mineral Commodity Summaries, January 2022. Available online: https://www.usgs.gov/centers/national-minerals-information-center/rare-earths-statistics-and-information (accessed on 10 October 2022).
- Alonso, E.; Sherman, A.M.; Wallington, T.J.; Everson, M.P.; Field, F.R.; Roth, R.; Kirchain, R.E. Evaluating Rare Earth Element Availability: A Case with Revolutionary Demand from Clean Technologies. Environ. Sci. Technol. 2012, 46, 3406–3414. [Google Scholar] [CrossRef]
- Adeel, M.; Lee, J.Y.; Zain, M.; Rizwan, M.; Nawab, A.; Ahmad, M.; Shafiq, M.; Yi, H.; Jilani, G.; Javed, R.; et al. Cryptic footprints of rare earth elements on natural resources and living organisms. Environ. Int. 2019, 127, 785–800. [Google Scholar] [CrossRef]
- Wang, L.; Liang, T. Geochemical fractions of rare earth elements in soil around a mine tailing in Baotou, China. Sci. Rep. 2015, 5, 12483. [Google Scholar] [CrossRef]
- Wang, L.; Liang, T. Anomalous abundance and redistribution patterns of rare earth elements in soils of a mining area in Inner Mongolia, China. Environ. Sci. Pollut. Res. 2016, 23, 11330–11338. [Google Scholar] [CrossRef]
- Thomas, P.J.; Carpenter, D.; Boutin, C.; Allison, J.E. Rare earth elements (REEs): Effects on germination and growth of selected crop and native plant species. Chemosphere 2013, 96, 57–66. [Google Scholar] [CrossRef]
- Li, J.; Verweij, R.A.; van Gestel, C.A. Lanthanum toxicity to five different species of soil invertebrates in relation to availability in soil. Chemosphere 2018, 193, 412–420. [Google Scholar] [CrossRef]
- Tang, W.; Wang, G.; Zhang, S.; Li, T.; Xu, X.; Deng, O.; Luo, L.; He, Y.; Zhou, W. Physiochemical responses of earthworms (Eisenia fetida) under exposure to lanthanum and cerium alone or in combination in artificial and contaminated soils. Environ. Pollut. 2021, 296, 118766. [Google Scholar] [CrossRef]
- Pagano, G.; Thomas, P.J.; Di Nunzio, A.; Trifuoggi, M. Human exposures to rare earth elements: Present knowledge and research prospects. Environ. Res. 2019, 171, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Li, Y.; Li, H.; Yu, J.; Ye, B.; Liang, T. Rare earth elements in human hair from a mining area of China. Ecotoxicol. Environ. Saf. 2013, 96, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, H.; Zheng, S.; Miao, Y.; Yin, S.; Li, P.; Bian, Y. Direct Quantification of Rare Earth Elements Concentrations in Urine of Workers Manufacturing Cerium, Lanthanum Oxide Ultrafine and Nanoparticles by a Developed and Validated ICP-MS. Int. J. Environ. Res. Public Health 2016, 13, 350. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, S.; Trujillo-Reyes, J.; Hernandez-Viezcas, J.A.; White, J.C.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Cerium Biomagnification in a Terrestrial Food Chain: Influence of Particle Size and Growth Stage. Environ. Sci. Technol. 2016, 50, 6782–6792. [Google Scholar] [CrossRef]
- Hawthorne, J.; Roche, R.D.L.T.; Xing, B.; Newman, L.A.; Ma, X.; Majumdar, S.; Gardea-Torresdey, J.; White, J.C. Particle-Size Dependent Accumulation and Trophic Transfer of Cerium Oxide through a Terrestrial Food Chain. Environ. Sci. Technol. 2014, 48, 13102–13109. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Van Der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setala, H.; van der Putten, W.H.; Wall, D.H. Ecological Linkages Between Aboveground and Belowground Biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- Schloter, M.; Dilly, O.; Munch, J. Indicators for evaluating soil quality. Agric. Ecosyst. Environ. 2003, 98, 255–262. [Google Scholar] [CrossRef]
- Li, S.; Zhao, B.; Jin, M.; Hu, L.; Zhong, H.; He, Z. A comprehensive survey on the horizontal and vertical distribution of heavy metals and microorganisms in soils of a Pb/Zn smelter. J. Hazard. Mater. 2020, 400, 123255. [Google Scholar] [CrossRef]
- Jiao, S.; Liu, Z.; Lin, Y.; Yang, J.; Chen, W.; Wei, G. Bacterial communities in oil contaminated soils: Biogeography and co-occurrence patterns. Soil Biol. Biochem. 2016, 98, 64–73. [Google Scholar] [CrossRef]
- Bajaj, S.; Singh, D.K. Biodegradation of persistent organic pollutants in soil, water and pristine sites by cold-adapted microorganisms: Mini review. Int. Biodeterior. Biodegradation 2015, 100, 98–105. [Google Scholar] [CrossRef]
- Gonzalez, V.; Vignati, D.A.; Leyval, C.; Giamberini, L. Environmental fate and ecotoxicity of lanthanides: Are they a uniform group beyond chemistry? Environ. Int. 2014, 71, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Dushyantha, N.; Batapola, N.; Ilankoon, I.M.S.K.; Rohitha, S.; Premasiri, R.; Abeysinghe, B.; Ratnayake, N.; Dissanayake, K. The story of rare earth elements (REEs): Occurrences, global distribution, genesis, geology, mineralogy and global production. Ore Geol. Rev. 2020, 122, 103521. [Google Scholar] [CrossRef]
- Guan, H.; Li, J.; Mou, Y.; Gao, W.; Zhang, C. Spatial Heterogeneity of Rare Earth Elements Accumulation in the Soil Surrounding Bayan Obo Mining Area. Rare Earth. 2021, 42, 27–35. [Google Scholar] [CrossRef]
- Xu, C.; Campbell, I.H.; Kynicky, J.; Allen, C.M.; Chen, Y.; Huang, Z.; Qi, L. Comparison of the Daluxiang and Maoniuping carbonatitic REE deposits with Bayan Obo REE deposit, China. Lithos 2008, 106, 12–24. [Google Scholar] [CrossRef]
- Wang, L.; Liang, T. Accumulation and fractionation of rare earth elements in atmospheric particulates around a mine tailing in Baotou, China. Atmospheric Environ. 2014, 88, 23–29. [Google Scholar] [CrossRef]
- Wang, L.; Liang, T.; Zhang, Q.; Li, K. Rare earth element components in atmospheric particulates in the Bayan Obo mine region. Environ. Res. 2014, 131, 64–70. [Google Scholar] [CrossRef]
- Guan, H.; Jinxia, L.W.; Jun, M. Community Composition and Seasonal Variation of Ground-Dwelling Arthropods Communities in Desert Steppe around Bayan Obo Mine. J. Inn. Mong. Univ. 2022, 53, 77–85. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000; pp. 355–356. [Google Scholar]
- Guo, H.; Nasir, M.; Lv, J.; Dai, Y.; Gao, J. Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotoxicol. Environ. Saf. 2017, 144, 300–306. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Chao, Y.; Liu, W.; Chen, Y.; Chen, W.; Zhao, L.; Ding, Q.; Wang, S.; Tang, Y.-T.; Zhang, T.; Qiu, R.-L. Structure, Variation, and Co-occurrence of Soil Microbial Communities in Abandoned Sites of a Rare Earth Elements Mine. Environ. Sci. Technol. 2016, 50, 11481–11490. [Google Scholar] [CrossRef]
- Liu, J.; Liu, W.; Zhang, Y.; Chen, C.; Wu, W.; Zhang, T.C. Microbial communities in rare earth mining soil after in-situ leaching mining. Sci. Total Environ. 2020, 755, 142521. [Google Scholar] [CrossRef]
- Jelusic, M.; Lestan, D. Effect of EDTA washing of metal polluted garden soils. Part I: Toxicity hazards and impact on soil properties. Sci. Total Environ. 2014, 475, 132–141. [Google Scholar] [CrossRef]
- Wang, X.F. Concentration and Fractionation of Rare Eanh Elements in Soils SuHDunding RareEanh Ore Area. Rock Miner. Anal. 2019, 38, 137–146. [Google Scholar] [CrossRef]
- Liu, J.; He, X.-X.; Lin, X.-R.; Chen, W.-C.; Zhou, Q.-X.; Shu, W.-S.; Huang, L.-N. Ecological Effects of Combined Pollution Associated with E-Waste Recycling on the Composition and Diversity of Soil Microbial Communities. Environ. Sci. Technol. 2015, 49, 6438–6447. [Google Scholar] [CrossRef]
- Luo, C.; Deng, Y.; Liang, J.; Zhu, S.; Wei, Z.; Guo, X.; Luo, X. Exogenous rare earth element-yttrium deteriorated soil microbial community structure. J. Rare Earths 2018, 36, 430–439. [Google Scholar] [CrossRef]
- Wei, Z.; Hao, Z.; Li, X.; Guan, Z.; Cai, Y.; Liao, X. The effects of phytoremediation on soil bacterial communities in an abandoned mine site of rare earth elements. Sci. Total Environ. 2019, 670, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, F.; Huang, Y.; Zhou, M.; Gao, J.; Yan, T.; Sheng, H.; An, L. Sphingomonas sp. Cra20 Increases Plant Growth Rate and Alters Rhizosphere Microbial Community Structure of Arabidopsis thaliana Under Drought Stress. Front. Microbiol. 2019, 10, 1221. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Waqas, M.; Asaf, S.; Kamran, M.; Shahzad, R.; Bilal, S.; Khan, M.A.; Kang, S.-M.; Kim, Y.-H.; Yun, B.-W.; et al. Plant growth-promoting endophyte Sphingomonas sp. LK11 alleviates salinity stress in Solanum pimpinellifolium. Environ. Exp. Bot. 2016, 133, 58–69. [Google Scholar] [CrossRef]
- Magnani, D.; Solioz, M. How Bacteria Handle Copper. In Molecular Microbiology of Heavy Metals; Springer: New York, NY, USA, 2007; pp. 259–285. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Chen, A.; Shi, Q.; Ouyang, Y.; Chen, Y. Effect of Ce3+ on membrane permeability of Escherichia coli cell. J. Rare Earths 2012, 30, 947–951. [Google Scholar] [CrossRef]
- Peng, L.; Weiying, Z.; Xi, L.; Yi, L. Structural Basis for the Biological Effects of Pr(III) Ions: Alteration of Cell Membrane Permeability. Biol. Trace Element Res. 2007, 120, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Azarbad, H.; Niklińska, M.; van Gestel, C.A.; van Straalen, N.M.; Röling, W.F.; Laskowski, R. Microbial community structure and functioning along metal pollution gradients. Environ. Toxicol. Chem. 2013, 32, 1992–2002. [Google Scholar] [CrossRef] [PubMed]
- Bardgett, R.D.; Wardle, D.A.; Yeates, G.W. Linking above-ground and below-ground interactions: How plant responses to foliar herbivory influence soil organisms. Soil Biol. Biochem. 1998, 30, 1867–1878. [Google Scholar] [CrossRef]
- Das, S.K.; Varma, A. Role of Enzymes in Maintaining Soil Health. In Soil Enzymology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 25–42. [Google Scholar] [CrossRef]
- Ma, J.; Qin, J.; Ma, H.; Zhou, Y.; Shen, Y.; Xie, Y.; Xu, D. Soil characteristic changes and quality evaluation of degraded desert steppe in arid windy sandy areas. PeerJ 2022, 10, e13100. [Google Scholar] [CrossRef]
- Na, X.; Yu, H.; Wang, P.; Zhu, W.; Niu, Y.; Huang, J. Vegetation biomass and soil moisture coregulate bacterial community succession under altered precipitation regimes in a desert steppe in northwestern China. Soil Biol. Biochem. 2019, 136, 107520. [Google Scholar] [CrossRef]
- Dion, P. Microbiology of Extreme Soils; Springer: New York, NY, USA, 2008. [Google Scholar]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Graham, D.W.; Sreekrishnan, T.; Ahammad, S.Z. Exploring the impacts of physicochemical characteristics and heavy metals fractions on bacterial communities in four rivers. J. Environ. Manag. 2023, 325, 116453. [Google Scholar] [CrossRef]
- He, Y.; Wang, H.; Liu, Y.; Zuo, M.; Wang, X.; Shen, Y.; Li, G.; Hu, Y.; Zhao, M.; Yang, Y.; et al. Distribution and Variation of Soil Bacterial Community of Two Lead-Zinc Tailings in Qinling Mountains. Geomicrobiol. J. 2022, 39, 1–11. [Google Scholar] [CrossRef]


| Sites | Plant Community Properties | Soil Physicochemical Properties | Soil Nutrients Contents | Soil Elements Content | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM (g) | CR (%) | SP | pH | EC (μs/cm) | SI (%) | WR (%) | OM(g/kg) | OC(g/kg) | TN (g/kg) | TP (g/kg) | LREE (μg/g) | HREE (μg/g) | REE (μg/g) | HM (μg/g) | |
| S1 | 53.14 ± 10.22 a | 25.00 ± 1.75 bc | 7.17 ± 0.76 ab | 8.07 ± 0.24 c | 1.45 ± 0.08 b | 52.6 ± 7.21 ab | 4.34 ± 0.52 a | 29.41 ± 2.62 ab | 17.06 ± 1.52 ab | 1.92 ± 0.08 ab | 0.05 ± 0.02 a | 1032.98 ± 506.28 a | 31.99 ± 8.24 a | 1064.93 ± 514.5 a | 418.80 ± 32.24 a |
| S2 | 43.83 ± 5.23 ab | 27.17 ± 3.57 ab | 7.17 ± 0.8 ab | 8.39 ± 0.15 ab | 1.67 ± 0.13 ab | 48.72 ± 3.72 bc | 4.47 ± 0.3 a | 26.86 ± 1.24 ab | 15.58 ± 0.71 ab | 1.76 ± 0.06 bc | 0.04 ± 0.01 b | 889.17 ± 105.18 a | 25.58 ± 1.75 b | 914.77 ± 106.5 a | 291.77 ± 6.05 b |
| S3 | 36.13 ± 4.01 bc | 24.08 ± 0.63 bcd | 4.25 ± 0.43 d | 8.28 ± 0.02 abc | 1.65 ± 0.02 ab | 48.16 ± 0.77 bc | 2.71 ± 1.07 bcd | 24.13 ± 0.68 b | 14.00 ± 0.4 b | 1.57 ± 0.03 c | 0.03 ± 0 b | 486.75 ± 119.05 b | 20.94 ± 1.43 bc | 507.70 ± 120.4 b | 285.93 ± 12.1 b |
| S4 | 16.29 ± 4.88 d | 17.00 ± 3.68 ef | 4.42 ± 0.95 d | 8.24 ± 0.09 abc | 1.67 ± 0.04 ab | 44.42 ± 1.04 c | 2.11 ± 0.19 d | 28.43 ± 1.44 ab | 16.49 ± 0.84 ab | 1.8 ± 0.07 bc | 0.03 ± 0.01 b | 506.8 ± 80.85 b | 21.67 ± 1.85 bc | 528.47 ± 82.4 b | 248.77 ± 12.7 b |
| S5 | 15.11 ± 3.12 d | 14.58 ± 1.44 f | 5.83 ± 0.52 bc | 7.95 ± 0.22 bc | 1.65 ± 0.03 ab | 45.57 ± 2.97 c | 2.48 ± 0.17 cd | 31.45 ± 3.87 a | 18.24 ± 2.25 a | 1.98 ± 0.27 ab | 0.03 ± 0 b | 326.52 ± 21.65 b | 20.28 ± 1.74 bc | 346.80 ± 23.37 b | 277.13 ± 9.63 b |
| S6 | 21.08 ± 3.47 d | 19.42 ± 2.24 de | 5.00 ± 0.75 cd | 8.18 ± 0.4 bc | 1.6 ± 0.14 ab | 49.00 ± 0.51 bc | 2.33 ± 0.09 cd | 30.91 ± 4.75 a | 17.93 ± 2.76 a | 1.98 ± 0.26 ab | 0.03 ± 0 b | 338.92 ± 15.54 b | 19.64 ± 0.24 c | 358.53 ± 15.75 b | 299.27 ± 33.86 b |
| S7 | 21.99 ± 3.45 d | 20.83 ± 1.76 cde | 7.08 ± 0.8 ab | 8.46 ± 0.14 a | 1.81 ± 0.41 a | 56.85 ± 1.37 a | 2.9 ± 0.14 bcd | 28.57 ± 1.42 ab | 16.57 ± 0.83 ab | 1.99 ± 0.12 ab | 0.03 ± 0 b | 298.01 ± 42.53 b | 19.13 ± 1.39 c | 317.13 ± 42.28 b | 259.63 ± 4.25 b |
| S8 | 34.09 ± 5.56 c | 27.5 ± 2.38 ab | 5.42 ± 0.14 cd | 8.75 ± 0.1 a | 1.79 ± 0.06 a | 59.23 ± 3.17 a | 3.07 ± 0.36 bc | 27.3 ± 4.19 ab | 15.84 ± 2.43 ab | 1.87 ± 0.25 abc | 0.03 ± 0 b | 241.25 ± 60.92 b | 16.63 ± 2.12 c | 257.90 ± 62.91 b | 287.57 ± 14.12 b |
| S9 | 46.51 ± 2.71 a | 30.00 ± 4.16 a | 7.58 ± 1.04 a | 8.70 ± 0.02 a | 1.85 ± 0.05 a | 59.12 ± 5.56 a | 3.31 ± 0.12 b | 29.9 ± 2.41 a | 17.34 ± 1.4 a | 2.12 ± 0.03 a | 0.03 ± 0 b | 200.91 ± 29.43 b | 16.13 ± 1.5 c | 217.05 ± 30.94 b | 292.73 ± 15.01 b |
| Taxa | REE | TP | pH | EC | WR | SI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | r | p | |
| Actinobacteriota | −0.480 | 0.003 | −0.039 | 0.823 | 0.392 | 0.018 | 0.107 | 0.536 | −0.234 | 0.170 | −0.209 | 0.222 |
| Verrucomicrobiota | 0.338 | 0.044 | 0.299 | 0.077 | −0.533 | 0.001 | −0.407 | 0.014 | 0.053 | 0.757 | −0.407 | 0.014 |
| Planctomycetota | 0.364 | 0.029 | 0.358 | 0.032 | −0.197 | 0.250 | −0.173 | 0.313 | 0.198 | 0.248 | −0.176 | 0.305 |
| Gemmatimonadetes | 0.576 | 0.000 | 0.495 | 0.020 | −0.422 | 0.010 | −0.432 | 0.009 | 0.220 | 0.198 | −0.421 | 0.011 |
| Abditibacteriota | 0.530 | 0.001 | 0.397 | 0.017 | −0.271 | 0.109 | −0.235 | 0.167 | 0.193 | 0.258 | −0.259 | 0.128 |
| YC-ZSS-LKJ147 | 0.555 | 0.000 | 0.600 | 0.000 | −0.193 | 0.259 | −0.499 | 0.002 | 0.568 | 0.000 | 0.014 | 0.934 |
| Subgroup_10 | 0.589 | 0.000 | 0.589 | 0.000 | −0.234 | 0.169 | −0.384 | 0.021 | 0.363 | 0.030 | −0.274 | 0.106 |
| Sphingomonas | 0.671 | 0.000 | 0.618 | 0.000 | −0.129 | 0.452 | −0.226 | 0.184 | 0.430 | 0.009 | −0.337 | 0.044 |
| Roseisolibacter | 0.704 | 0.000 | 0.585 | 0.000 | −0.426 | 0.010 | −0.408 | 0.013 | 0.358 | 0.032 | −0.500 | 0.002 |
| Adhaeribacter | 0.333 | 0.047 | 0.232 | 0.172 | 0.550 | 0.750 | 0.304 | 0.071 | 0.147 | 0.147 | −0.254 | 0.135 |
| Abditibacterium | 0.530 | 0.001 | 0.397 | 0.017 | −0.271 | 0.244 | −0.235 | 0.167 | 0.258 | 0.258 | −0.259 | 0.128 |
| Pseudomonas | −0.349 | 0.037 | −0.324 | 0.054 | 0.285 | 0.092 | 0.265 | 0.118 | −0.134 | 0.435 | 0.038 | 0.826 |
| Candidatus_Udaeobacter | 0.311 | 0.065 | 0.273 | 0.107 | −0.520 | 0.001 | −0.398 | 0.016 | 0.046 | 0.788 | −0.403 | 0.015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, H.; Mu, Y.; Song, R.; Lan, Y.; Du, X.; Li, J.; Chi, W.; Sang, W. Soil Microbial Communities in Desert Grassland around Rare Earth Mine: Diversity, Variation, and Response Patterns. Sustainability 2022, 14, 15629. https://doi.org/10.3390/su142315629
Guan H, Mu Y, Song R, Lan Y, Du X, Li J, Chi W, Sang W. Soil Microbial Communities in Desert Grassland around Rare Earth Mine: Diversity, Variation, and Response Patterns. Sustainability. 2022; 14(23):15629. https://doi.org/10.3390/su142315629
Chicago/Turabian StyleGuan, Haibo, Yanjun Mu, Rutao Song, Yuecen Lan, Xiongfeng Du, Jinxia Li, Wenfeng Chi, and Weiguo Sang. 2022. "Soil Microbial Communities in Desert Grassland around Rare Earth Mine: Diversity, Variation, and Response Patterns" Sustainability 14, no. 23: 15629. https://doi.org/10.3390/su142315629
APA StyleGuan, H., Mu, Y., Song, R., Lan, Y., Du, X., Li, J., Chi, W., & Sang, W. (2022). Soil Microbial Communities in Desert Grassland around Rare Earth Mine: Diversity, Variation, and Response Patterns. Sustainability, 14(23), 15629. https://doi.org/10.3390/su142315629







