Home Sweet Home: Setting the Best Thriving Conditions for the Ad Hoc Engineered Microbial Consortium in the Zero Mile System
Abstract
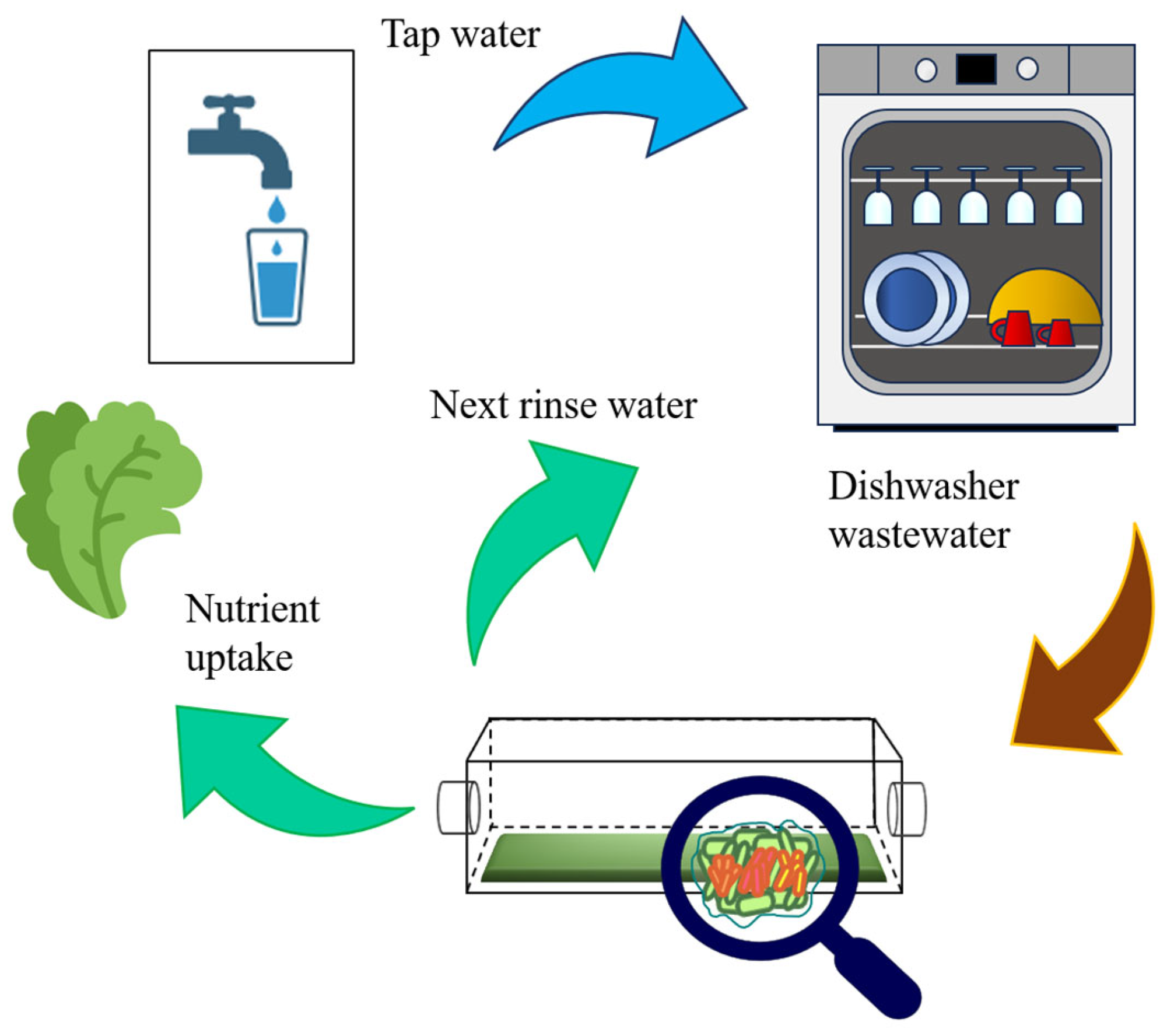
:1. Introduction
2. Materials and Methods
2.1. Trichormus variabilis
2.2. Assembling the Microbial Consortium
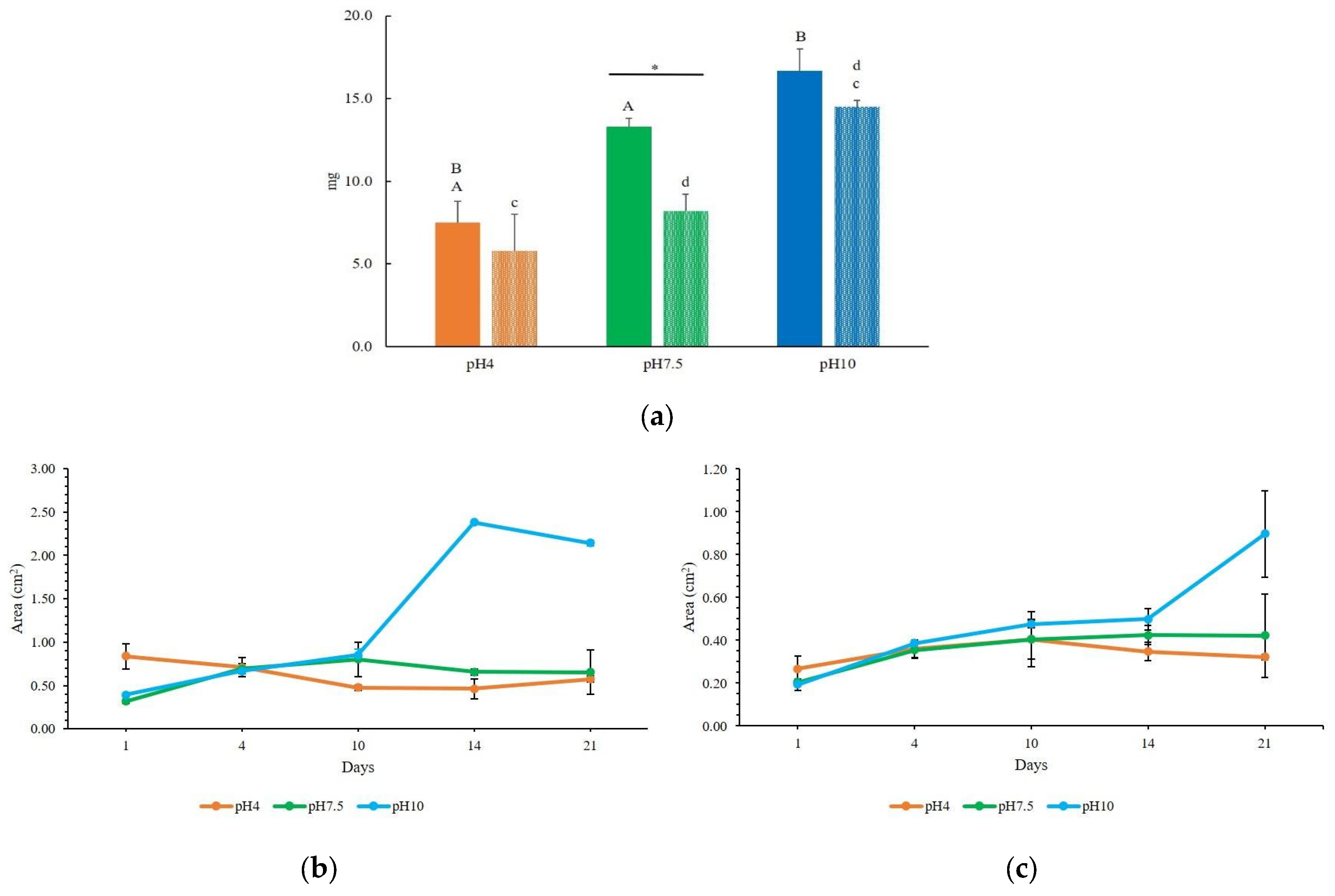
2.3. Operational Conditions of the Consortium: Effect of Light, pH and Organic Load
2.4. Dishwasher Wastewater
2.5. Cultivation-Based Approach: Isolation of Dishwasher Wastewater Bacteria on Solid Media and Identification by Sanger Sequencing
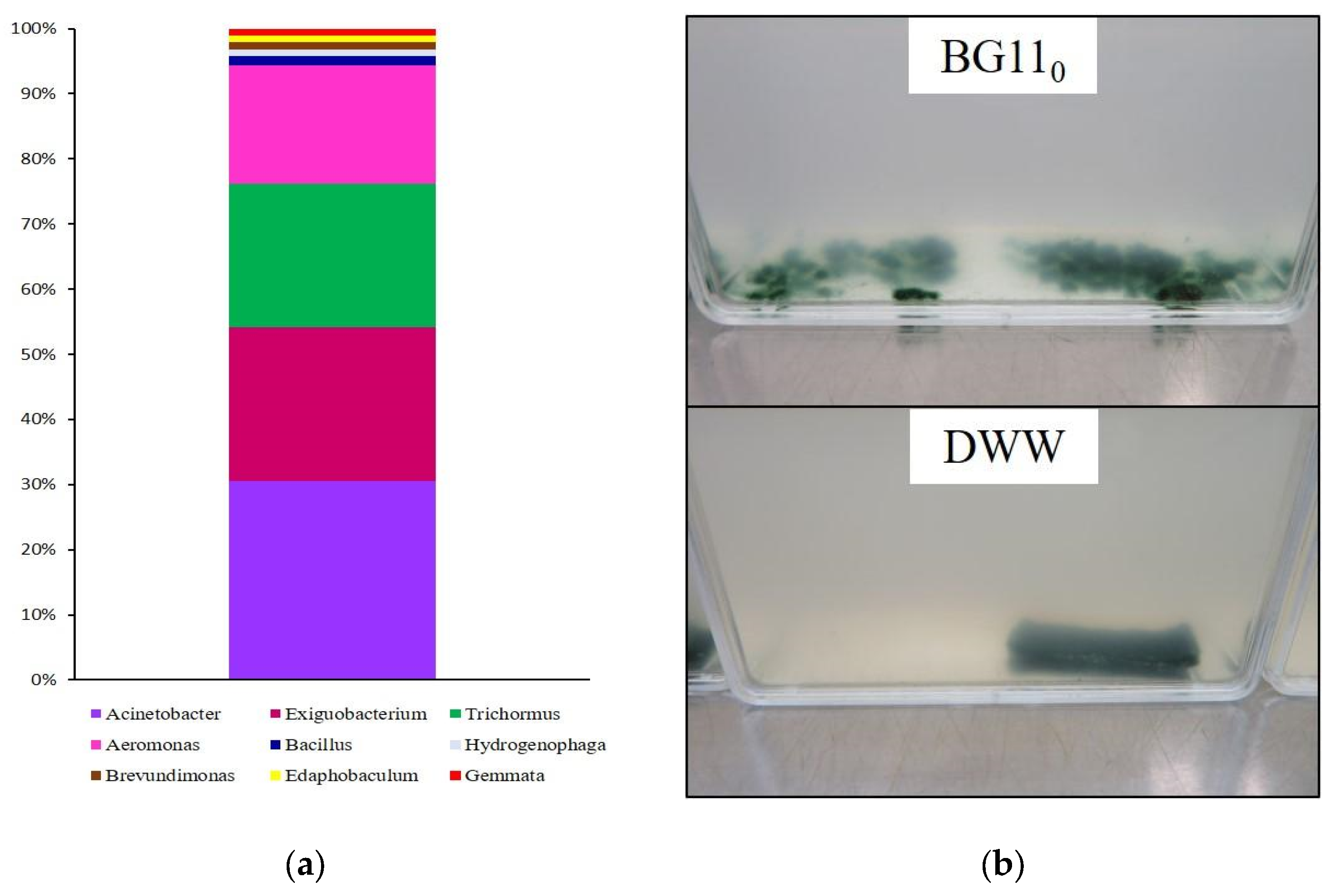
2.6. DNA Metabarcoding Approach: Taxonomic Composition of Dishwasher Wastewater Colonisers and Consortium Components
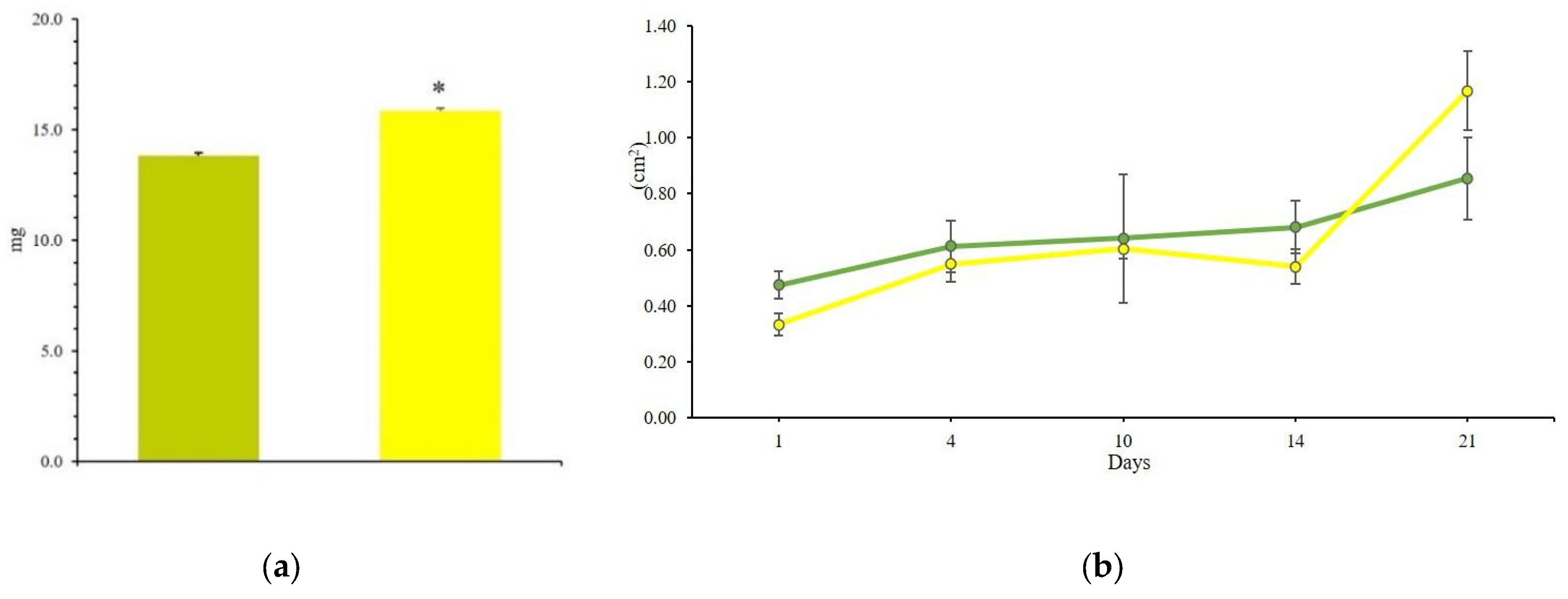
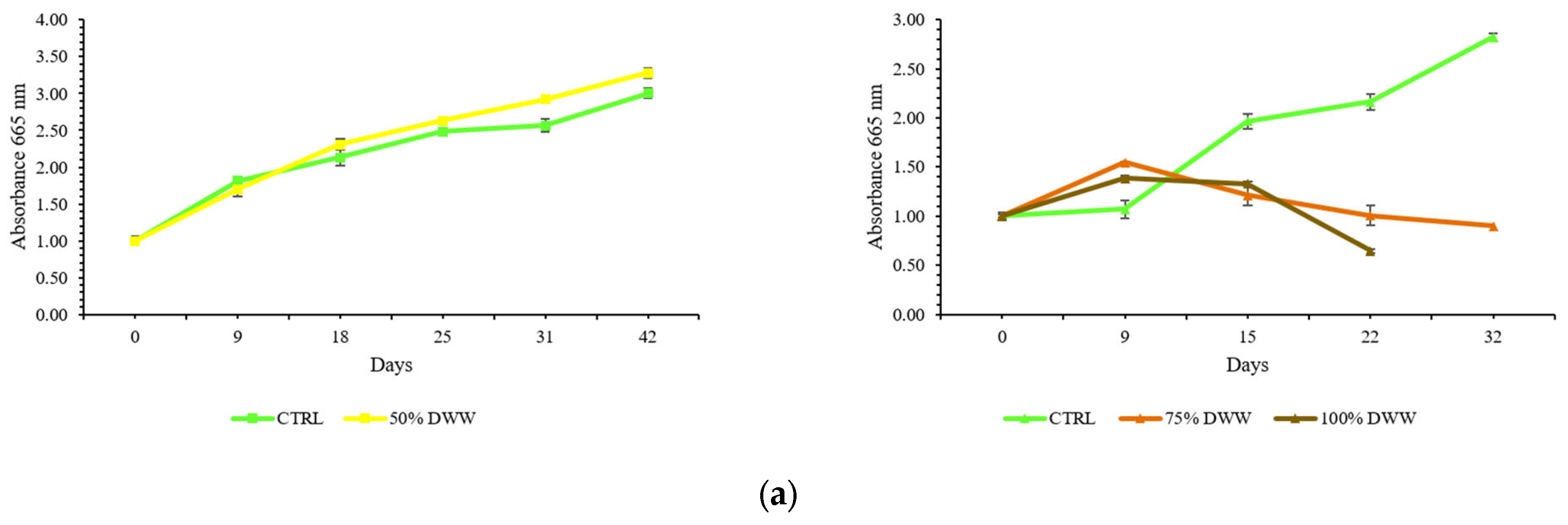
2.7. Testing the Consortium with Three Dilutions of Dishwasher Wastewater
2.8. Statistical Analysis
3. Results and Discussion
3.1. Consortium Assemblage
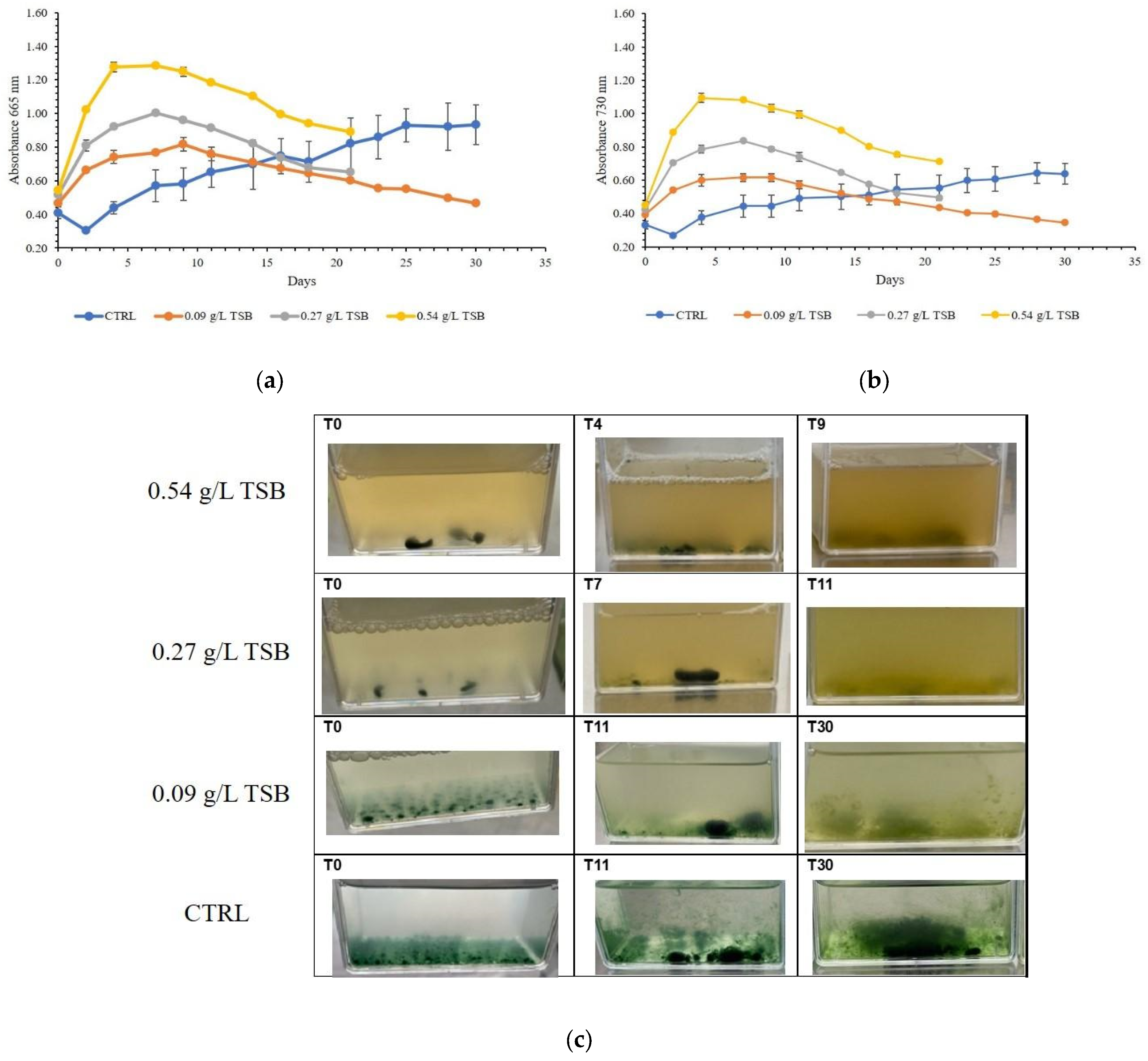
3.2. Setting the Operational Conditions of the Zero Mile for Consortium Thriving
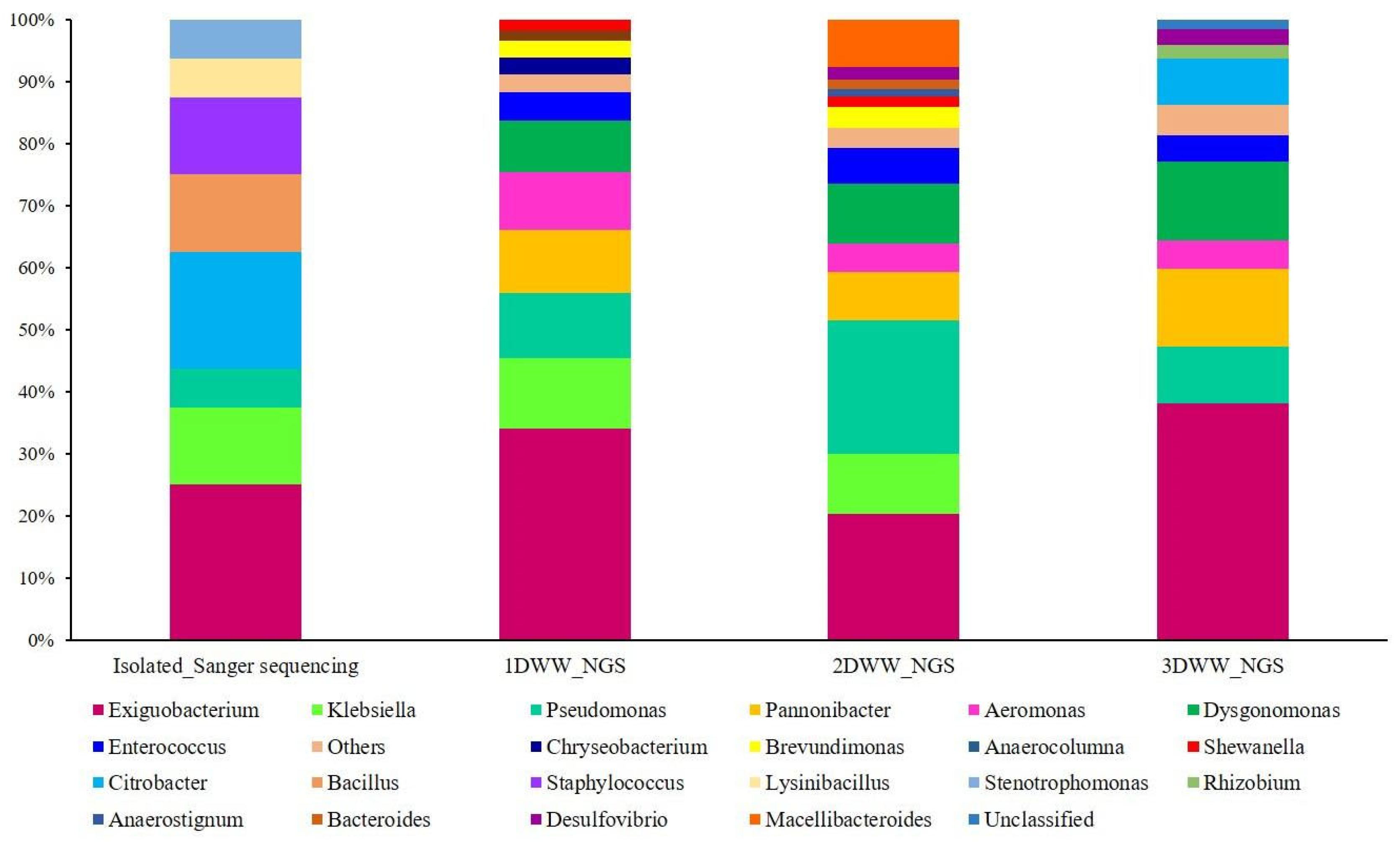
3.3. Dishwasher Wastewater Microbial Colonisers
3.4. Design Implications
- Modularity and independence of the biofiltering, watering and cultivation sub-systems;
- Control of the biofilter light conditions, both through passive strategies (positioning and photo-adaptive materials) and active strategies (direct light from artificial sources);
- Separation of the plant cultivation sub-system in building structure and vegetable support;
- Flexibility of the plant cultivation sub-system in order to host different cultivation typologies;
- Accessibility, maintainability and mobility of all components and subsystems;
- Up-scaling of the system focusing on the mesoscale and urban scale, also looking at the key issues of socialisation and living space regeneration.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schroeder, P.; Anggraeni, K.; Weber, U. The Relevance of Circular Economy Practices to the Sustainable Development Goals. J. Ind. Ecol. 2019, 23, 77–95. [Google Scholar] [CrossRef]
- UNESCO. The United Nations World Water Development Report, 2017: Wastewater: The Untapped Resource; UNESCO: Paris, France, 2017; Available online: https://unesdoc.unesco.org/ark:/48223/pf0000247153 (accessed on 4 August 2021).
- UNESCO. The United Nations World Water Development Report 2023: Partnerships and Cooperation for Water; UNESCO: Paris, France, 2023; Available online: https://unesdoc.unesco.org/ark:/48223/pf0000384655 (accessed on 6 September 2023).
- Raschid-Sally, L.; Jayakody, P. Drivers and Characteristics of Wastewater Agriculture in Developing Countries: Results from a Global Assessment; International Water Management Institute: Colombo, Sri Lanka, 2008; Volume 127. [Google Scholar]
- Cisneros, B.J. 18—Water Recycling and Reuse: An Overview. In Water Reclamation and Sustainability; Ahuja, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 431–454. [Google Scholar]
- Posadas, E.; García-Encina, P.-A.; Soltau, A.; Domínguez, A.; Díaz, I.; Muñoz, R. Carbon and nutrient removal from centrates and domestic wastewater using algal–bacterial biofilm bioreactors. Bioresour. Technol. 2013, 139, 50–58. [Google Scholar] [CrossRef]
- Costa, F.; Nebuloni, A. The Jetsons’ Kitchen—A Zero-Mile System for Waste Water Recycling and Cultivation; FrancoAngeli: Milan, Italy, 2021; pp. 1–134. Available online: https://www.francoangeli.it/Libro/The-Jetsons%27-Kitchen?Id=27055 (accessed on 6 September 2023).
- Congestri, R.; Savio, S.; Farrotti, S.; Amati, A.; Krasojevic, K.; Perini, N.; Costa, F.; Migliore, L. Developing a microbial consortium for removing nutrients in dishwasher wastewater: Towards a biofilter for its up-cycling. Water Sci. Technol. 2020, 82, 1142–1154. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.L.; Pires, J.C.M.; Simões, M. A review on the use of microalgal consortia for wastewater treatment. Algal Res. 2017, 24, 403–415. [Google Scholar] [CrossRef]
- Fito, J.; Alemu, K. Microalgae–bacteria consortium treatment technology for municipal wastewater management. Nanotechnol. Environ. Eng. 2019, 4, 4. [Google Scholar] [CrossRef]
- Subashchandrabose, S.R.; Ramakrishnan, B.; Megharaj, M.; Venkateswarlu, K.; Naidu, R. Consortia of cyanobacteria/microalgae and bacteria: Biotechnological potential. Biotechnol. Adv. 2011, 29, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, R.; Guieysse, B. Algal–bacterial processes for the treatment of hazardous contaminants: A review. Water Res. 2006, 40, 2799–2815. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Y.; Wu, C.; Muylaert, K.; Vyverman, W.; Yu, H.-Q.; Muñoz, R.; Rittmann, B. Advanced nutrient removal from surface water by a consortium of attached microalgae and bacteria: A review. Bioresour. Technol. 2017, 241, 1127–1137. [Google Scholar] [CrossRef]
- Alabiso, A.; Frasca, S.; Cantelmo, V.; D’andrea, M.M.; Braglia, R.; Scuderi, F.; Costa, F.; Savio, S.; Congestri, R.; Migliore, L. A dirty job: Dishwasher wastewater reuse and upcycle through an ad hoc engineered microbial consortium. npj Clean Water 2023, 6, 66. [Google Scholar] [CrossRef]
- Perera, I.A.; Abinandan, S.; Subashchandrabose, S.R.; Venkateswarlu, K.; Naidu, R.; Megharaj, M. Advances in the technologies for studying consortia of bacteria and cyanobacteria/microalgae in wastewaters. Crit. Rev. Biotechnol. 2019, 39, 709–731. [Google Scholar] [CrossRef]
- Lazaro, C.Z.; Varesche, M.B.A.; Silva, E.L. Effect of inoculum concentration, pH, light intensity and lighting regime on hydrogen production by phototrophic microbial consortium. Renew. Energy 2015, 75, 1–7. [Google Scholar] [CrossRef]
- Stappers, P.J.; Sleeswijk Visser, F.; Keller, I. The role of prototypes and frameworks in research through design. In The Routledge Companion to Design Research; Rodgers, P.A., Yee, J., Eds.; Routledge: London, UK, 2014; pp. 167–174. [Google Scholar]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Z.; Liao, Q.; Zhu, X.; Li, J.; Lee, D.-J. Effect of culture conditions on the kinetics of hydrogen production by photosynthetic bacteria in batch culture. Int. J. Hydrogen Energy 2011, 36, 14004–14013. [Google Scholar] [CrossRef]
- Di Pippo, F.; Ellwood, N.T.W.; Guzzon, A.; Siliato, L.; Micheletti, E.; De Philippis, R.; Albertano, P.B. Effect of light and temperature on biomass, photosynthesis and capsular polysaccharides in cultured phototrophic biofilms. J. Appl. Phycol. 2012, 24, 211–220. [Google Scholar] [CrossRef]
- Abedi, S.; Astaraei, F.R.; Ghobadian, B.; Tavakoli, O.; Jalili, H.; Chivasa, S.; Greenwell, H.C. Bioenergy production using Trichormus variabilis—A review. Biofuels Bioprod. Biorefin. 2019, 13, 1365–1382. [Google Scholar] [CrossRef]
- Hill, W.R. Effects of light. In Algal Ecology—Freshwater Benthic Ecosystems; Jan Stevenson, R., Bothwell, M.L., Lowe, R.L., Eds.; Academic Press: San Diego, CA, USA, 1996; pp. 121–148. [Google Scholar]
- Jia, H.; Yuan, Q. Removal of nitrogen from wastewater using microalgae and microalgae–bacteria consortia. Cogent Environ. Sci. 2016, 2, 1275089. [Google Scholar] [CrossRef]
- Bertinelli, A.; Hamill, J.; Mahadevan, M.; Miles, F. Serious injuries from dishwasher powder ingestions in small children. J. Paediatr. Child Health 2006, 42, 129–133. [Google Scholar] [CrossRef]
- Johansson, E.; Wernersson, E.S.; Hakanson, H. Effects on the Survival of Enterococcus faecium in Dishwater. Foodserv. Res. Int. 2004, 15, 118–128. [Google Scholar] [CrossRef]
- Magg, H.; Waeschenbach, G. Application of Enzymes in Automatic-Dishwashing Detergents. In Enzymes in Detergency; van Ee, J.H., Misset, O., Bass, E.J., Eds.; Marcel Dekker: New York, NY, USA, 1997; Volume 69, pp. 231–250. [Google Scholar]
- Giraldez-Ruiz, N.; Mateo, P.; Bonilla, I.; Fernandez-Pinas, F. The relationship between intracellular pH, growth characteristics and calcium in the cyanobacterium Anabaena sp. strain PCC7120 exposed to low pH. New Phytol. 1997, 137, 599–605. [Google Scholar] [CrossRef]
- Isengard, H.-D. Water content, one of the most important properties of food. Food Control 2001, 12, 395–400. [Google Scholar] [CrossRef]
- Zupančič, J.; Turk, M.; Črnigoj, M.; Avguštin, J.A.; Gunde-Cimerman, N. The dishwasher rubber seal acts as a reservoir of bacteria in the home environment. BMC Microbiol. 2019, 19, 300. [Google Scholar] [CrossRef]
- Raghupathi, P.K.; Zupančič, J.; Brejnrod, A.D.; Jacquiod, S.; Houf, K.; Burmølle, M.; Gunde-Cimerman, N.; Sørensen, S.J. Microbial Diversity and Putative Opportunistic Pathogens in Dishwasher Biofilm Communities. Appl. Environ. Microbiol. 2018, 84, e02755-17. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Kathariou, S.; Tiedje, J.M. The Exiguobacterium genus: Biodiversity and biogeography. Extremophiles 2009, 13, 541–555. [Google Scholar] [CrossRef]
- White, R.A.; Grassa, C.J.; Suttle, C.A. Draft Genome Sequence of Exiguobacterium pavilionensis Strain RW-2, with Wide Thermal, Salinity, and pH Tolerance, Isolated from Modern Freshwater Microbialites. Genome Announc. 2013, 1, e00597-13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, W.; Guo, Y.; Zhang, Z.; Shi, W.; Cui, F.; Lens, P.N.; Tay, J.H. Microalgal-bacterial consortia: From interspecies interactions to biotechnological applications. Renew. Sustain. Energy Rev. 2020, 118, 109563. [Google Scholar] [CrossRef]
- Srisawat, P.; Higuchi-Takeuchi, M.; Numata, K. Microbial autotrophic biorefineries: Perspectives for biopolymer production. Polym. J. 2022, 54, 1139–1151. [Google Scholar] [CrossRef]
- Karana, E.; Barati, B.; Giaccardi, E. Living Artefacts: Conceptualizing Livingness as a Material Quality in Everyday Artefacts. Int. J. Des. 2020, 14, 37–53. [Google Scholar]
- Garrido-Baserba, M.; Vinardell, S.; Molinos-Senante, M.; Rosso, D.; Poch, M. The Economics of Wastewater Treatment Decentralization: A Techno-economic Evaluation. Environ. Sci. Technol. 2018, 52, 8965–8976. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Baserba, M.; Barnosell, I.; Molinos-Senante, M.; Sedlak, D.L.; Rabaey, K.; Schraa, O.; Verdaguer, M.; Rosso, D.; Poch, M. The third route: A techno-economic evaluation of extreme water and wastewater decentralization. Water Res. 2022, 218, 118408. [Google Scholar] [CrossRef] [PubMed]
- Garrido Baserba, M.; Sedlak, D.; Barnosell, I.; Molinos-Senante, M.; Schraa, O.; Rosso, D.; Verdaguer, M.; Poch, M. Using water and wastewater decentralization to enhance the resiliency and sustainability of cities. Res. Sq. 2023. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alabiso, A.; Frasca, S.; Bartolini, M.; Congestri, R.; D’Andrea, M.M.; Buratti, G.; Costa, F.; Meraviglia, M.; Nebuloni, A.; Migliore, L. Home Sweet Home: Setting the Best Thriving Conditions for the Ad Hoc Engineered Microbial Consortium in the Zero Mile System. Sustainability 2024, 16, 2227. https://doi.org/10.3390/su16062227
Alabiso A, Frasca S, Bartolini M, Congestri R, D’Andrea MM, Buratti G, Costa F, Meraviglia M, Nebuloni A, Migliore L. Home Sweet Home: Setting the Best Thriving Conditions for the Ad Hoc Engineered Microbial Consortium in the Zero Mile System. Sustainability. 2024; 16(6):2227. https://doi.org/10.3390/su16062227
Chicago/Turabian StyleAlabiso, Annamaria, Sara Frasca, Matteo Bartolini, Roberta Congestri, Marco Maria D’Andrea, Giorgio Buratti, Fiammetta Costa, Matteo Meraviglia, Attilio Nebuloni, and Luciana Migliore. 2024. "Home Sweet Home: Setting the Best Thriving Conditions for the Ad Hoc Engineered Microbial Consortium in the Zero Mile System" Sustainability 16, no. 6: 2227. https://doi.org/10.3390/su16062227





