Genetic Diversity of Dominant Species Betula pendula in River Valley Forests in the Irtysh River Basin and Sustainable Conservation Measures for the Future
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sampling Method
2.2. DNA Extraction and Microsatellite Analysis
2.3. Genetic Diversity Analysis
2.4. Genetic Structure Analysis
2.5. Haplotype Analysis
3. Results
3.1. Chloroplast Microsatellite Locus Genetic Diversity
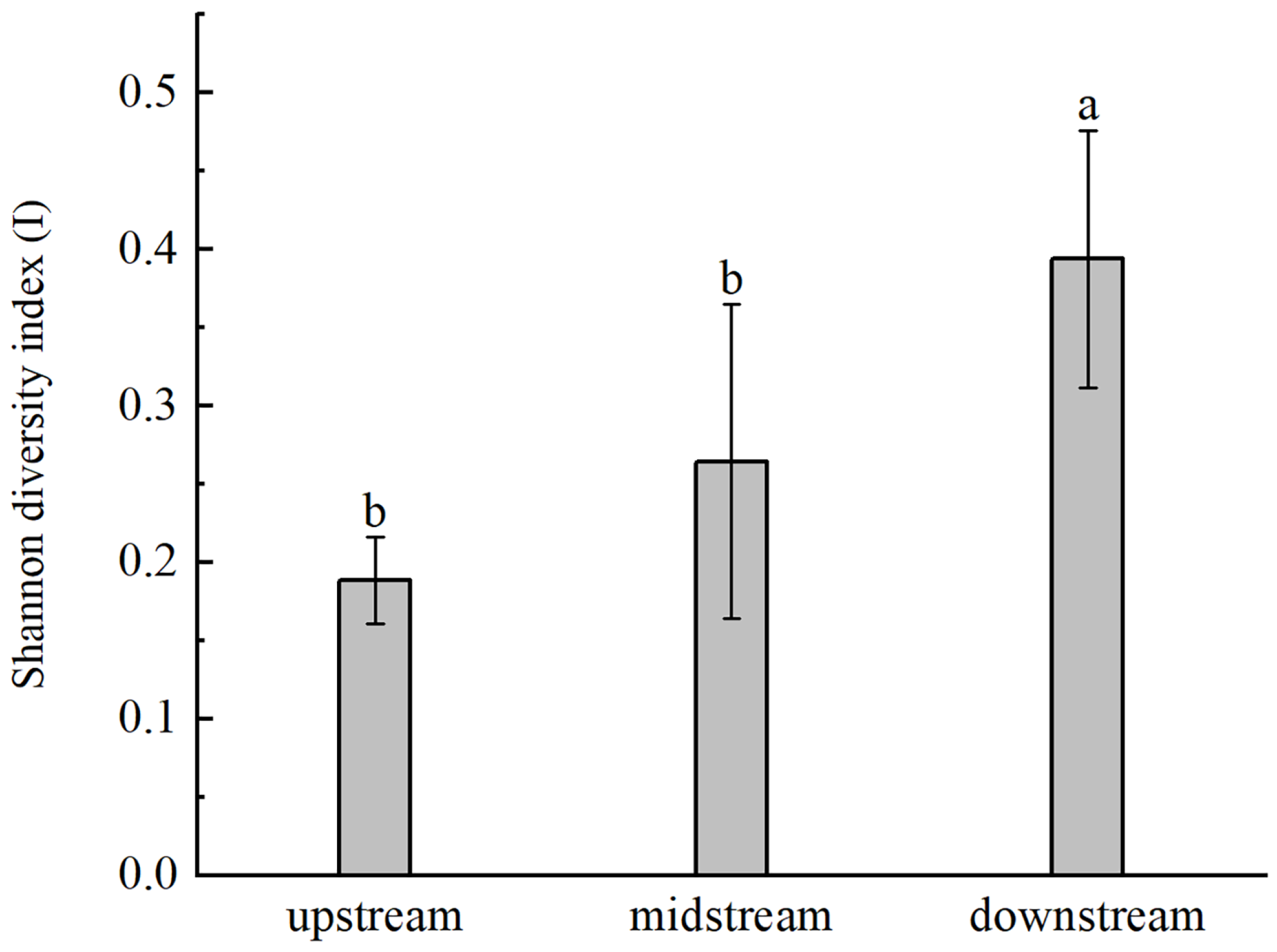
3.2. Genetic Diversity and Genetic Differentiation of B. pendula
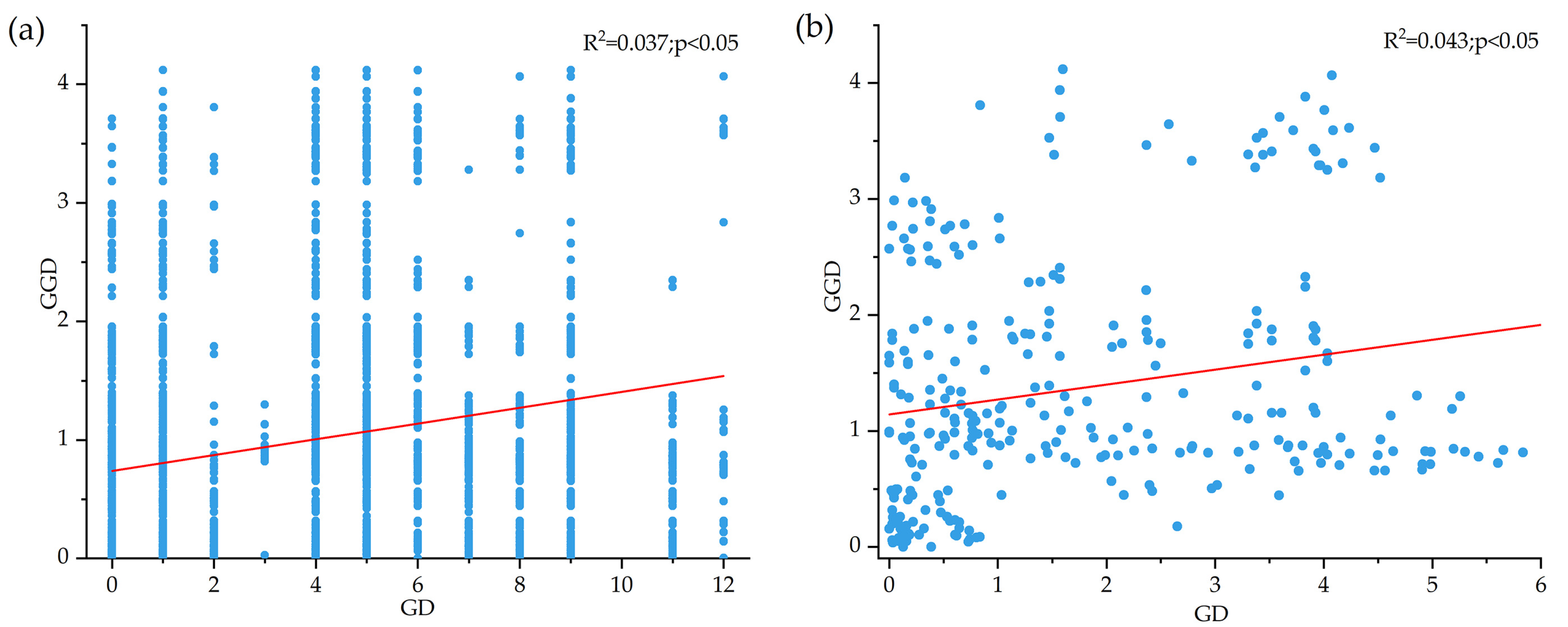
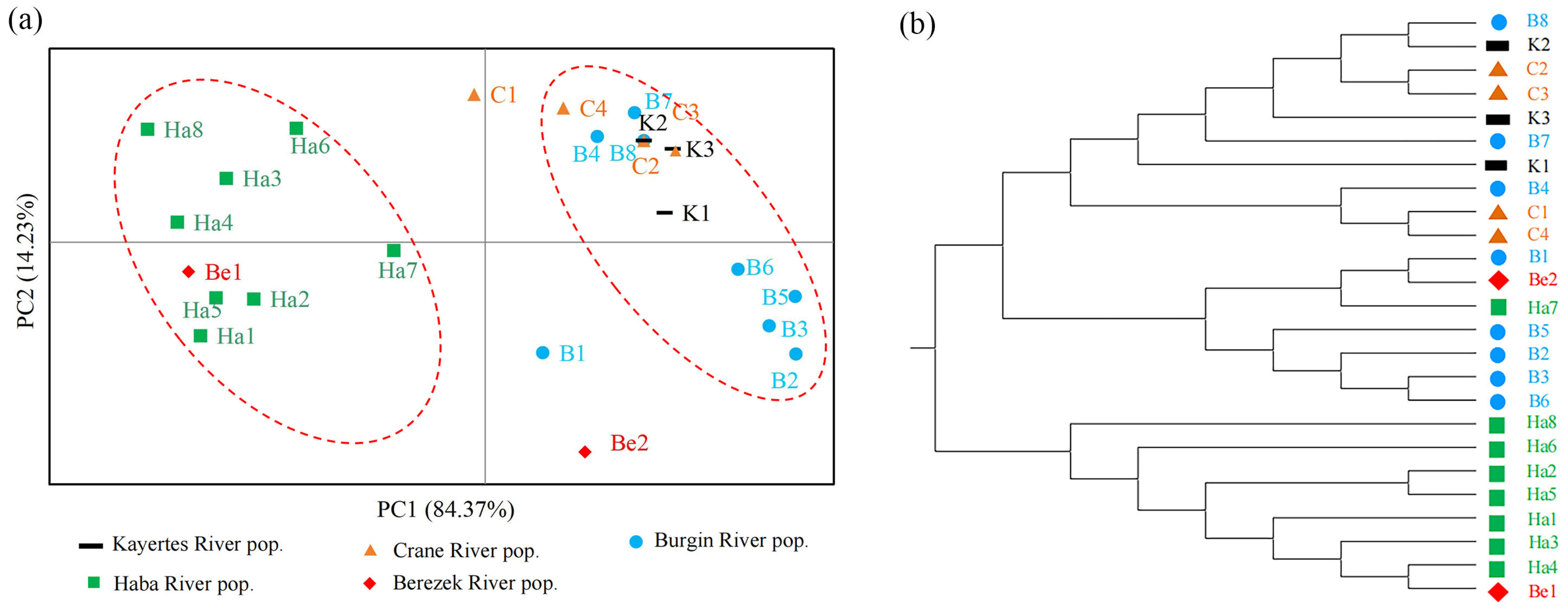
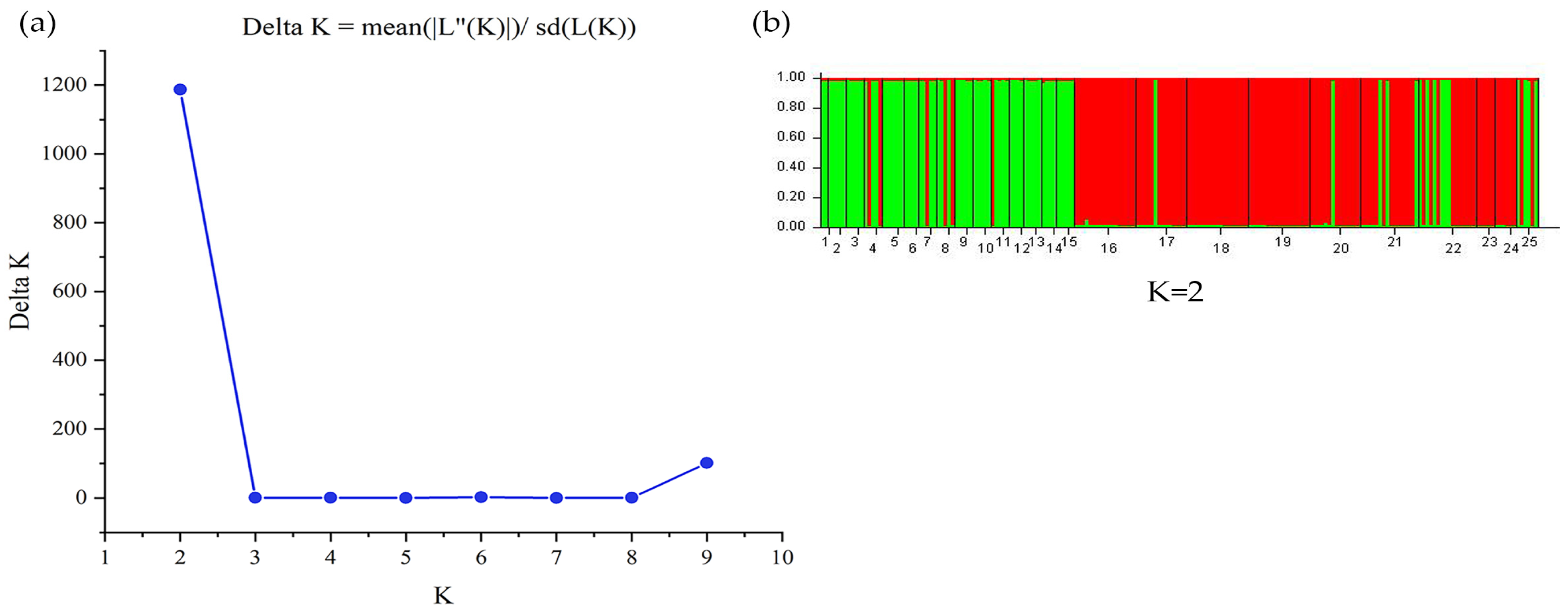
3.3. Genetic Structure of B. pendula
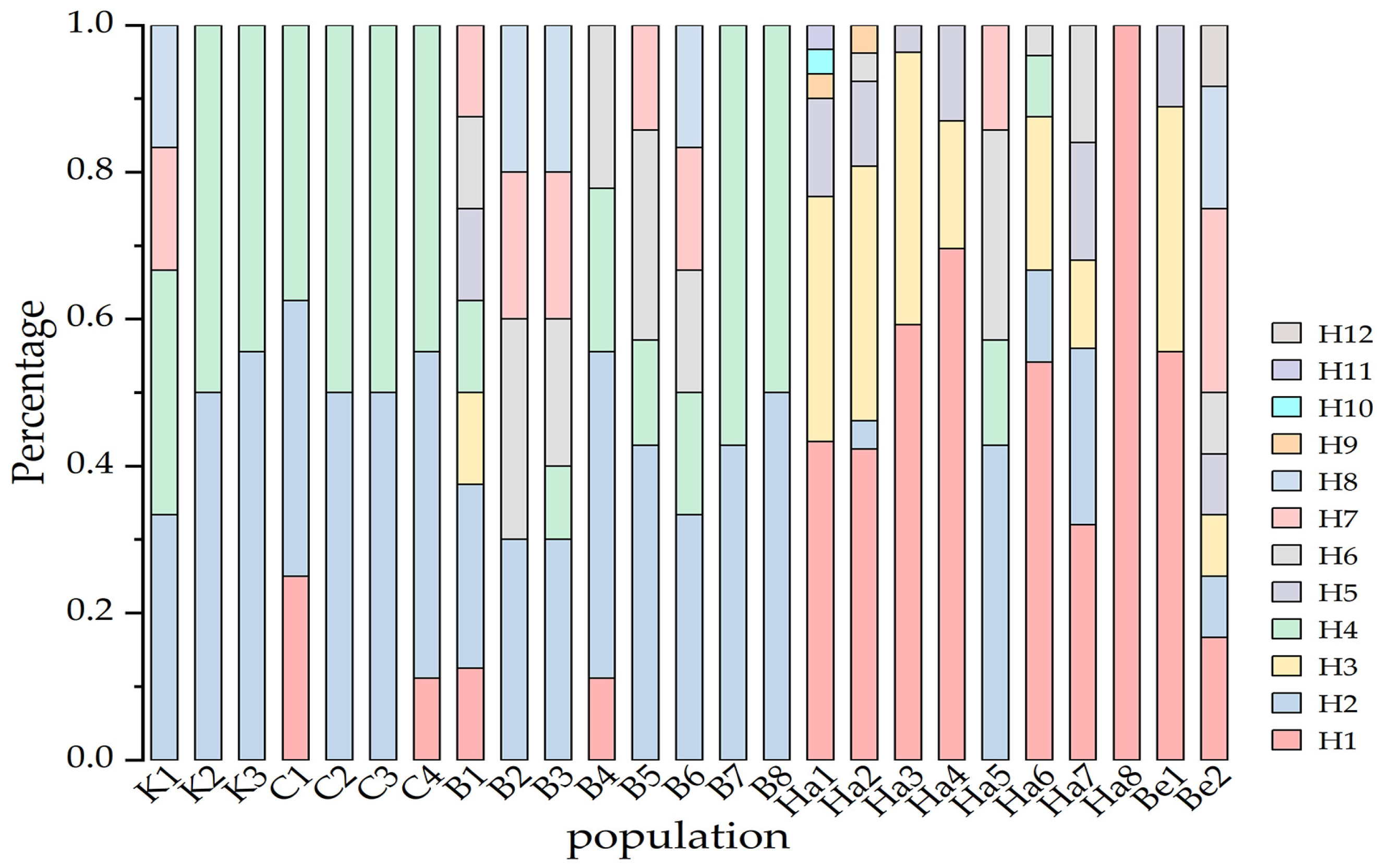
3.4. Geographical Distribution of Chloroplast Haplotype
4. Discussion
4.1. Characteristics of High Genetic Diversity and Low Genetic Differentiation Were Observed in B. pendula of Irtysh River Basin
4.2. Low Genetic Structure Level in B. pendula Population
4.3. The Distribution of B. pendula Haplotype Indicates the Existence of a Refugium in the Irtysh River Basin during the Last Glacial Ice Age
4.4. Protection Strategy of High-Quality Genetic Resources of B. pendula
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Haplotype | Allele Size Combination | |||
|---|---|---|---|---|
| ccmp4 | ccmp5 | BCMS1 | BCMS2 | |
| H1 | 114 | 100 | 167 | 160 |
| H2 | 115 | 100 | 168 | 160 |
| H3 | 114 | 100 | 168 | 160 |
| H4 | 115 | 100 | 167 | 160 |
| H5 | 114 | 101 | 167 | 160 |
| H6 | 115 | 100 | 169 | 160 |
| H7 | 115 | 101 | 168 | 160 |
| H8 | 115 | 101 | 167 | 160 |
| H9 | 114 | 101 | 168 | 160 |
| H10 | 114 | 99 | 167 | 160 |
| H11 | 114 | 99 | 168 | 160 |
| H12 | 115 | 101 | 169 | 160 |
| Hap. | Population | Total | Fre. | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K1 | K2 | K3 | C1 | C2 | C3 | C4 | B1 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | Ha1 | Ha2 | Ha3 | Ha4 | Ha5 | Ha6 | Ha7 | Ha8 | Be1 | Be2 | |||
| H1 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 13 | 11 | 16 | 16 | 0 | 13 | 8 | 5 | 5 | 2 | 94 | 0.377 |
| H2 | 2 | 5 | 5 | 3 | 6 | 4 | 4 | 2 | 3 | 3 | 4 | 3 | 4 | 3 | 5 | 0 | 1 | 0 | 0 | 3 | 3 | 6 | 0 | 0 | 1 | 70 | 0.174 |
| H3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 9 | 10 | 4 | 0 | 5 | 3 | 0 | 3 | 1 | 46 | 0.136 |
| H4 | 2 | 5 | 4 | 3 | 6 | 4 | 4 | 1 | 0 | 1 | 2 | 1 | 2 | 4 | 5 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 47 | 0.116 |
| H5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 1 | 3 | 0 | 0 | 4 | 0 | 1 | 1 | 18 | 0.081 |
| H6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 4 | 0 | 0 | 1 | 21 | 0.048 |
| H7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 13 | 0.035 |
| H8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 9 | 0.023 |
| H9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.0027 |
| H10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0025 |
| H11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0025 |
| H12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0.0025 |
| No. | 4 | 2 | 2 | 3 | 2 | 2 | 3 | 7 | 4 | 5 | 4 | 4 | 5 | 2 | 2 | 6 | 6 | 3 | 3 | 4 | 5 | 5 | 1 | 3 | 7 | ||
References
- Li, Q.; Chen, J. The primary task of watershed-scale comprehensive conservation of Yangtze River Basin: Conservation and sustainable utilization of plant genetic diversity. Biodivers. Sci. 2018, 26, 327–332. [Google Scholar] [CrossRef]
- Koskela, J.; Buck, A.; du Cros, E.T. Climate Change and Forest Genetic Diversity: Implications for Sustainable Forest Management in Europe; Bioversity International: Rome, Italy, 2007; p. 5. [Google Scholar]
- Fady, B.; Aravanopoulos, F.A.; Alizoti, P.; Mátyás, C.; von Wühlisch, G.; Westergren, M.; Belletti, P.; Cvjetkovic, B.; Ducci, F.; Huber, G. Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. For. Ecol. Manag. 2016, 375, 66–75. [Google Scholar] [CrossRef]
- Hughes, F.M.; Rood, S.B. Allocation of river flows for restoration of floodplain forest ecosystems: A review of approaches and their applicability in Europe. Environ. Manag. 2003, 32, 12–33. [Google Scholar] [CrossRef] [PubMed]
- Hughes, F.M.R.; Colston, A.; Mountford, J.O. Restoring riparian ecosystems: The challenge of accommodating variability and designing restoration trajectories. Ecol. Soc. 2005, 10, 12. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Ma, X.; Li, M.; Guo, X.; Yin, M.; Cai, Y.; Yu, X.; Du, N.; Wang, R. Impacts of the yellow River and Qingtongxia dams on genetic diversity of Phragmites australis in Ningxia Plain, China. Aquat. Bot. 2021, 169, 103341. [Google Scholar] [CrossRef]
- Altermatt, F.J. Diversity in riverine metacommunities: A network perspective. Aquat. Ecol. 2013, 47, 365–377. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, Y.; Zhang, S.; Wang, D.; Chen, H. Application of a Linked Hydrodynamic–Groundwater Model for Accurate Groundwater Simulation in Floodplain Areas: A Case Study of Irtysh River, China. Water 2023, 15, 3059. [Google Scholar] [CrossRef]
- Xue, Z.-F.; Liu, T.; Wang, L.; Song, J.-H.; Chen, H.-Y.; Xu, L.; Yuan, Y. Community structure and characteristics of the plain valley forests in the main tributaries of the Ertix River Basin, China. Chin. J. Plant Ecol. 2024, 48. [Google Scholar] [CrossRef]
- Geburek, T.; Hiess, K.; Litschauer, R.; Milasowszky, N. Temporal pollen pattern in temperate trees: Expedience or fate? Oikos 2012, 121, 1603–1612. [Google Scholar] [CrossRef]
- Hynynen, J.; Niemistö, P.; Viherä-Aarnio, A.; Brunner, A.; Hein, S.; Velling, P. Silviculture of birch (Betula pendula Roth and Betula pubescens Ehrh.) in northern Europe. Forestry 2010, 83, 103–119. [Google Scholar] [CrossRef]
- Wagner, S.; Wälder, K.; Ribbens, E.; Zeibig, A. Directionality in fruit dispersal models for anemochorous forest trees. Ecol. Modell. 2004, 179, 487–498. [Google Scholar] [CrossRef]
- Czarnecka, J. Seed dispersal effectiveness in three adjacent plant communities: Xerothermic grassland, brushwood and woodland. Ann. Botan. Fenn. 2005, 42, 161–171. [Google Scholar]
- Belletti, P.; Ferrazzini, D.; Ducci, F.; De Rogatis, A.; Mucciarelli, M. Genetic diversity of Italian populations of Abies alba. Dendrobiology 2017, 77, 147–159. [Google Scholar] [CrossRef]
- Allen, J.A.; Keeland, B. A Guide to Bottomland Hardwood Restoration; US Geological Survey, Biological Resources Division and US Department of Agriculture, Southern Research Station: Asheville, NC, USA, 2001. [Google Scholar]
- Garssen, A.G.; Baattrup-Pedersen, A.; Voesenek, L.A.; Verhoeven, J.T.; Soons, M.B. Riparian plant community responses to increased flooding: A meta-analysis. Global Chang. Biol. 2015, 21, 2881–2890. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Zhang, J.; He, C.; Bao, E.; Duan, A.; Zeng, Y.; Sai, L. Genetic diversity of Populus laurifolia and Populus nigra along Erqis River. For. Res. 2014, 27, 295–301. [Google Scholar] [CrossRef]
- Raymond, L.; Plantegenest, M.; Vialatte, A. Migration and dispersal may drive to high genetic variation and significant genetic mixing: The case of two agriculturally important, continental hoverflies (Episyrphus balteatus and Sphaerophoria scripta). Mol. Ecol. 2013, 22, 5329–5339. [Google Scholar] [CrossRef] [PubMed]
- Gruber, K.; Schöning, C.; Otte, M.; Kinuthia, W.; Hasselmann, M. Distinct subspecies or phenotypic plasticity? Genetic and morphological differentiation of mountain honey bees in East Africa. Ecol. Evol. 2013, 3, 3204–3218. [Google Scholar] [CrossRef] [PubMed]
- Wernberg, T.; Coleman, M.A.; Bennett, S.; Thomsen, M.S.; Tuya, F.; Kelaher, B.P. Genetic diversity and kelp forest vulnerability to climatic stress. Sci. Rep. 2018, 8, 1851. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Jo, Y.S.; Park, C.H.; Friedman, J.M.; Olson, M.S. Population genomic analysis suggests strong influence of river network on spatial distribution of genetic variation in invasive saltcedar across the southwestern United States. Mol. Ecol. 2018, 27, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Sander, N.L.; Pérez-Zavala, F.; Da Silva, C.J.; Arruda, J.C.; Pulido, M.T.; Barelli, M.A.; Rossi, A.B.; Viana, A.P.; Boechat, M.S.; Bacon, C.D. Rivers shape population genetic structure in Mauritia flexuosa (Arecaceae). Ecol. Evol. 2018, 8, 6589–6598. [Google Scholar] [CrossRef]
- Blanchet, S.; Prunier, J.; Paz-Vinas, I.; Saint-Pé, K.; Rey, O.; Raffard, A.; Mathieu-Bégné, E.; Loot, G.; Fourtune, L.; Dubut, V. A river runs through it: The causes, consequences, and management of intraspecific diversity in river networks. Evol. Appl. 2020, 13, 1195–1213. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-X.; Wang, Y.-Q.; Xiao, S.-Y.; Wang, Y.-H.; Liu, J.; Gong, X.; Feng, X.-Y. Rivers have shaped the phylogeography of a narrowly distributed cycad lineage in Southwest China. Conserv. Genet. 2023, 25, 439–453. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, H. Phylogeographic structure of Terminalia franchetii (Combretaceae) in southwest China and its implications for drainage geological history. J. Plant Res. 2011, 124, 63–73. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Zhang, Y.; Kong, Z.C.; Yang, Z.; Yan, Q. Vegetation changes and human activities in the Betula wetland of Habahe in Xinjiang, China since 3600 cal a BP. Acta Ecol. Sin. 2023, 43, 1156–1164. [Google Scholar] [CrossRef]
- Wang, N.; Thomson, M.; Bodles, W.J.; Crawford, R.M.; Hunt, H.V.; Featherstone, A.W.; Pellicer, J.; Buggs, R.J. Genome sequence of dwarf birch (Betula nana) and cross-species RAD markers. Mol. Ecol. 2013, 22, 3098–3111. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zou, Y.-P.; Bai, J.-Y.; Zheng, H.-S. Preparation of total DNA from “recalcitrant plant taxa”. Acta Bot. Sin. 2002, 44, 694–697. Available online: https://www.researchgate.net/publication/258113478_Preparation_of_Total_DNA_from_Recalcitrant_Plant_Taxa (accessed on 12 October 2023).
- Wills, D.M.; Hester, M.L.; Liu, A.; Burke, J.M. Chloroplast SSR polymorphisms in the Compositae and the mode of organellar inheritance in Helianthus annuus. Theor. Appl. Genet. 2005, 110, 941–947. [Google Scholar] [CrossRef]
- Meucci, S.; Schulte, L.; Zimmermann, H.H.; Stoof-Leichsenring, K.R.; Epp, L.; Bronken Eidesen, P.; Herzschuh, U. Holocene chloroplast genetic variation of shrubs (Alnus alnobetula, Betula nana, Salix sp.) at the siberian tundra-taiga ecotone inferred from modern chloroplast genome assembly and sedimentary ancient DNA analyses. Ecol. Evol. 2021, 11, 2173–2193. [Google Scholar] [CrossRef]
- Ismail, N.A.; Rafii, M.; Mahmud, T.; Hanafi, M.; Miah, G. Genetic diversity of torch ginger (Etlingera elatior) germplasm revealed by ISSR and SSR markers. Biomed Res. Int. 2019, 2019, 5904804. [Google Scholar] [CrossRef] [PubMed]
- Thomson, A.M.; Dick, C.W.; Dayanandan, S. A similar phylogeographical structure among sympatric North American birches (Betula) is better explained by introgression than by shared biogeographical history. J. Biogeogr. 2015, 42, 339–350. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Bioinformatics 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Liu, C.; Han, X.; Xiao, C.; Yi, S.; Huang, D.J. Genetic analysis of 32 InDels in four ethnic minorities from Chinese Xinjiang. PLoS ONE 2021, 16, e0250206. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Fan, W.; Gai, H.; Sun, X.; Yang, A.; Zhang, Z.; Ren, M. DataFormater, a software for SSR data formatting to develop population genetics analysis. Mol. Plant Breed 2016, 14, 265–270. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. Structure Harvester: A website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Riahi, L.; Zoghlami, N.; Laucou, V.; Mliki, A.; This, P. Use of chloroplast microsatellite markers as a tool to elucidate polymorphism, classification and origin of Tunisian grapevines. Sci. Hortic. 2011, 130, 781–786. [Google Scholar] [CrossRef]
- Buso, G.; Rangel, P.; Ferreira, M. Analysis of genetic variability of South American wild rice populations (Oryza glumaepatula) with isozymes and RAPD markers. Mol. Ecol. Notes 1998, 7, 107–117. [Google Scholar] [CrossRef]
- De Dato, G.D.; Teani, A.; Mattioni, C.; Aravanopoulos, F.; Avramidou, E.V.; Stojnic, S.; Ganopoulos, I.; Belletti, P.; Ducci, F. Genetic analysis by nuSSR markers of silver Birch (Betula pendula Roth) populations in their Southern European distribution range. Front. Plant Sci. 2020, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Palme, A.; Su, Q.; Palsson, S.; Lascoux, M. Extensive sharing of chloroplast haplotypes among European birches indicates hybridization among Betula pendula, B. pubescens and B. nana. Mol. Ecol. 2004, 13, 167–178. [Google Scholar] [CrossRef]
- Maliouchenko, O.; Palmé, A.E.; Buonamici, A.; Vendramin, G.; Lascoux, M. Comparative phylogeography and population structure of European Betula species, with particular focus on B. pendula and B. pubescens. J. Biogeogr. 2007, 34, 1601–1610. [Google Scholar] [CrossRef]
- Tsuda, Y.; Semerikov, V.; Sebastiani, F.; Vendramin, G.G.; Lascoux, M. Multispecies genetic structure and hybridization in the Betula genus across Eurasia. Mol. Ecol. 2017, 26, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Zeng, J.; Zou, Y.; Bai, J.; Zheng, H. RAPD analysis of genetic variation in natural populations of Betula alnoides from Guangxi, China. Euphytica 2003, 134, 33–41. [Google Scholar] [CrossRef]
- Silvertown, J. The evolutionary maintenance of sexual reproduction: Evidence from the ecological distribution of asexual reproduction in clonal plants. Int. J. Plant Sci. 2008, 169, 157–168. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, L.; Zhang, G.; Guo, H.; Whitt, A.A.; Chen, W.; Han, L.; Ma, C. Effects of grazing intensity on sexual and clonal reproduction in a clonal xerophytic shrub. Pak. J. Bot 2020, 52, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Beck, P.; Caudullo, G.; de Rigo, D.; Tinner, W. Betula Pendula, Betula Pubescens and Other Birches in Europe: Distribution, Habitat, Usage and Threats; Publication Office of the European Union: Luxembourg, 2016; pp. 70–73. [Google Scholar]
- Sannikov, S.; Sannikova, N. Outline of the hydrochory theory for some coniferous species. Dokl. Biol. Sci. 2008, 418, 67. [Google Scholar] [CrossRef] [PubMed]
- Duminil, J.; Fineschi, S.; Hampe, A.; Jordano, P.; Salvini, D.; Vendramin, G.G.; Petit, R.J. Can population genetic structure be predicted from life-history traits? Am. Nat. 2007, 169, 662–672. [Google Scholar] [CrossRef]
- Xie, Y.; Li, Z.; Huang, R.; Xiao, X.; Huang, Y. Genetic diversity of Betula luminifera populations at different elevations in Wuyi Mountain and its association with ecological factors. Front. For. China 2009, 4, 90–95. [Google Scholar] [CrossRef]
- Loveless, M.D.; Hamrick, J.L. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Syst. 1984, 15, 65–95. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114. [Google Scholar] [CrossRef]
- Jay, F.; Manel, S.; Alvarez, N.; Durand, E.Y.; Thuiller, W.; Holderegger, R.; Taberlet, P.; François, O.J. Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol. Ecol. 2012, 21, 2354–2368. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.R.; Lister, A.M.; Barnes, I.; Dalén, L. Refugia revisited: Individualistic responses of species in space and time. Proc. R. Soc. B Biol. Sci. 2010, 277, 661–671. [Google Scholar] [CrossRef]
- Qiu, Y.-X.; Fu, C.-X.; Comes, H.P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Meng, H.H.; Gao, X.Y.; Huang, J.F.; Zhang, M.L. Plant phylogeography in arid Northwest China: Retrospectives and perspectives. J. Syst. Evol. 2015, 53, 33–46. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, B.; Zhang, M.J.P.O. Phylogeography and conservation genetics of the endangered Tugarinovia mongolica (Asteraceae) from Inner Mongolia. Northwest China 2019, 14, e0211696. [Google Scholar] [CrossRef]
- Chen, T.Y.; Lou, A.R. Phylogeography and paleodistribution models of a widespread birch (Betula platyphylla Suk.) across East Asia: Multiple refugia, multidirectional expansion, and heterogeneous genetic pattern. Ecol. Evol. 2019, 9, 7792–7807. [Google Scholar] [CrossRef]
- Li, J.; Zhang, C.; Mipam, T.D.; Zhou, Q.; Chen, S. Effects of Climatic Change on Phylogeography and Ecological Niche of the Endemic Herb Elymus breviaristatus on the Qinghai-Tibet Plateau. Plants 2023, 12, 3326. [Google Scholar] [CrossRef]
- Dubois, H.; Claessens, H.; Ligot, G. Towards silviculture guidelines to produce large-sized silver birch (Betula pendula Roth) logs in Western Europe. Forests 2021, 12, 599. [Google Scholar] [CrossRef]
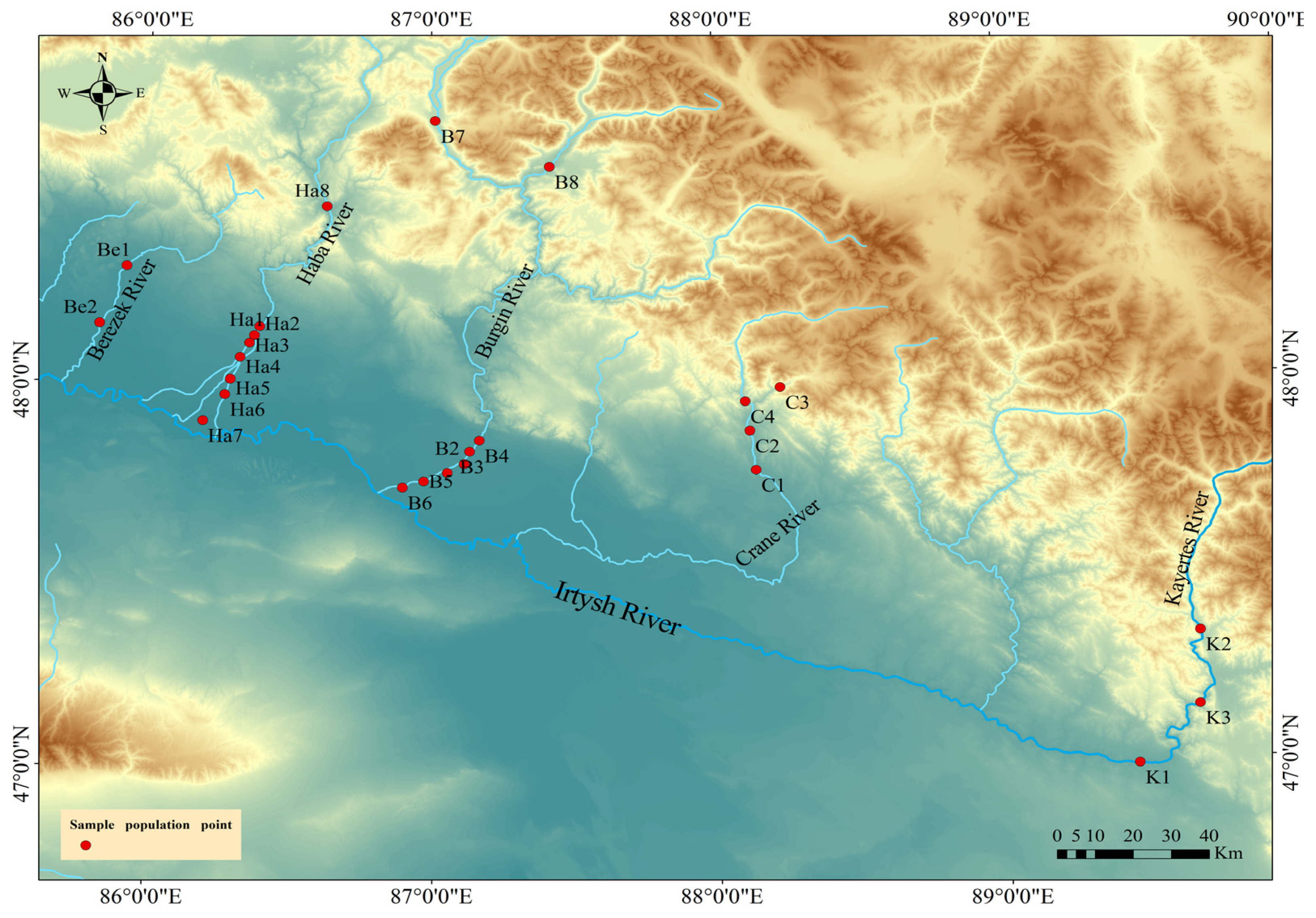
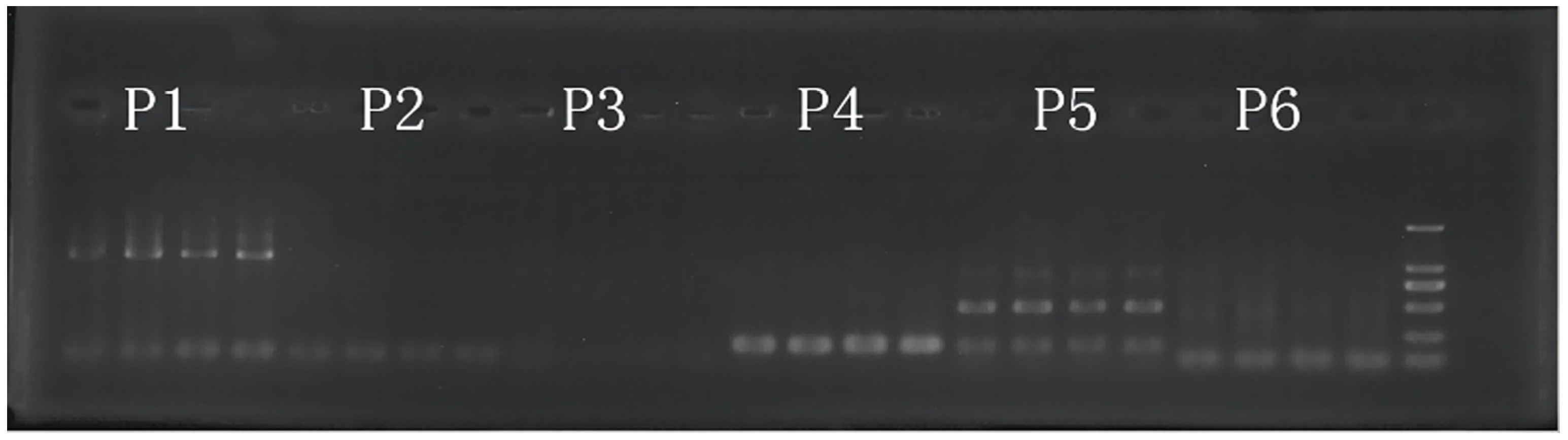


| River | Population | Sample | Longitude (°E) | Latitude (°N) |
|---|---|---|---|---|
| Kayertes River | F1 | 2 | 89°27′ | 46°59′ |
| F2 | 5 | 89°40′ | 47°19′ | |
| F3 | 5 | 89°40′ | 47°8′ | |
| Crane River | K1 | 5 | 88°8′ | 47°46′ |
| K2 | 5 | 88°7′ | 47°52′ | |
| K3 | 5 | 88°13′ | 47°58′ | |
| K4 | 5 | 88°6′ | 47°56′ | |
| Burgin River | B1 | 5 | 87°3′ | 47°45′ |
| B2 | 5 | 87°6′ | 47°47′ | |
| B3 | 5 | 87°8′ | 47°49′ | |
| B4 | 5 | 87°10′ | 47°50′ | |
| B5 | 4 | 86°58′ | 47°44′ | |
| B6 | 5 | 86°53′ | 47°43′ | |
| B7 | 4 | 87°1′ | 48°40′ | |
| B8 | 5 | 87°25′ | 48°33′ | |
| Haba River | Ha1 | 17 | 86°22′ | 48°7′ |
| Ha2 | 14 | 86°23′ | 48°8′ | |
| Ha3 | 17 | 86°21′ | 48°6′ | |
| Ha4 | 17 | 86°19′ | 48°3′ | |
| Ha5 | 14 | 86°17′ | 48°1′ | |
| Ha6 | 16 | 86°16′ | 47°58′ | |
| Ha7 | 16 | 86°11′ | 47°53′ | |
| Ha8 | 5 | 86°37′ | 48°27′ | |
| Berezek River | Be1 | 6 | 85°55′ | 48°18′ |
| Be2 | 6 | 85°49′ | 48°9′ |
| Code | Primer | Repeat Type | Primer Sequence (5′~3′) |
|---|---|---|---|
| P1 | ccmp4 * | (T)11 | 5′-AATGCTGAATCGAYGACCTA-3′ |
| 5′-CCAAAATATTBGGAGGACTCT-3′ | |||
| P2 | ccmp5 * | (T)14 | 5′-TGTTCCAATATCTTCTTGTCATTT-3′ |
| 5′-AGGTTCCATCGGAACAATTAT-3′ | |||
| P3 | ccmp7 * | (A)7 | 5′-CAACATATACCACTGTCAAG-3′ |
| 5′-ACATCATTATTGTATACTCTTTC-3′ | |||
| P4 | BCMS1 † | (T)10 | 5′-GCTCTTTTCGTTAGCGGTTT-3′ |
| 5′-ATTTGAAGCGGGGATACCTT-3′ | |||
| P5 | BCMS2 ‡ | (T)11 | 5′-CCGCTTCAAATTTTAATGAT-3′ |
| 5′-GATGACTTGGGTTTATGTCAA-3′ | |||
| P6 | BCMS3 † | (A)8 | 5′-CGGGCAAAACCAACAAAAT-3′ |
| 5′-GGGTTCGAATCCCTCTCTCT-3′ |
| Parameters of Polymorphism | Primer | |||
|---|---|---|---|---|
| ccmp4 | ccmp5 | BCMS1 | BCMS2 | |
| allele number (Na) | 2 | 3 | 3 | 1 |
| allele loci | 114; 115 | 99; 100; 101 | 167; 168; 169 | 160 |
| effective number of alleles (Ne) | 1.209 | 1.340 | 2.025 | 1.000 |
| Shannon diversity index (I) | 0.186 | 0.305 | 0.735 | 0.000 |
| polymorphism information content (PIC) | 0.3646 | 0.2200 | 0.4253 | 0 |
| Tributary | N | Na | Ne | I | Fst | Nm |
|---|---|---|---|---|---|---|
| Kayertes River | 12 | 1.50 | 1.268 | 0.216 | 0.028 | 8.778 |
| Crane River | 20 | 1.50 | 1.325 | 0.276 | 0.038 | 6.279 |
| Burgin River | 38 | 2.00 | 1.570 | 0.454 | 0.018 | 13.519 |
| Haba River | 116 | 2.25 | 1.324 | 0.362 | 0.055 | 4.255 |
| Berezek River | 12 | 2.00 | 1.716 | 0.546 | 0.308 | 0.563 |
| Irtysh River basin | 198 | 1.85 | 1.440 | 0.371 | 0.315 | 0.544 |
| Kayertes | Crane | Burgin | Haba | Berezek | |
|---|---|---|---|---|---|
| Kayertes | 0.106 | 0.080 | 0.001 | 0.001 | |
| Crane | 0.028 | 0.005 | 0.001 | 0.001 | |
| Burgin | 0.034 | 0.048 | 0.001 | 0.001 | |
| Haba | 0.385 | 0.276 | 0.286 | 0.021 | |
| Berezek | 0.225 | 0.159 | 0.138 | 0.052 |
| Source of Variance | Variance | Percentages of Variance (%) |
|---|---|---|
| Among Pops | 0.201 | 31 |
| Within Pops | 0.437 | 69 |
| Total | 0.638 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Xue, Z.; Liu, T.; Song, J.; Yuan, Y.; Xu, L.; Zhang, Z. Genetic Diversity of Dominant Species Betula pendula in River Valley Forests in the Irtysh River Basin and Sustainable Conservation Measures for the Future. Sustainability 2024, 16, 3217. https://doi.org/10.3390/su16083217
Li Y, Xue Z, Liu T, Song J, Yuan Y, Xu L, Zhang Z. Genetic Diversity of Dominant Species Betula pendula in River Valley Forests in the Irtysh River Basin and Sustainable Conservation Measures for the Future. Sustainability. 2024; 16(8):3217. https://doi.org/10.3390/su16083217
Chicago/Turabian StyleLi, Yanming, Zhifang Xue, Tong Liu, Jihu Song, Ye Yuan, Ling Xu, and Zidong Zhang. 2024. "Genetic Diversity of Dominant Species Betula pendula in River Valley Forests in the Irtysh River Basin and Sustainable Conservation Measures for the Future" Sustainability 16, no. 8: 3217. https://doi.org/10.3390/su16083217
APA StyleLi, Y., Xue, Z., Liu, T., Song, J., Yuan, Y., Xu, L., & Zhang, Z. (2024). Genetic Diversity of Dominant Species Betula pendula in River Valley Forests in the Irtysh River Basin and Sustainable Conservation Measures for the Future. Sustainability, 16(8), 3217. https://doi.org/10.3390/su16083217







