A Novel Deep Learning Method to Identify Single Tree Species in UAV-Based Hyperspectral Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Image Acquisition
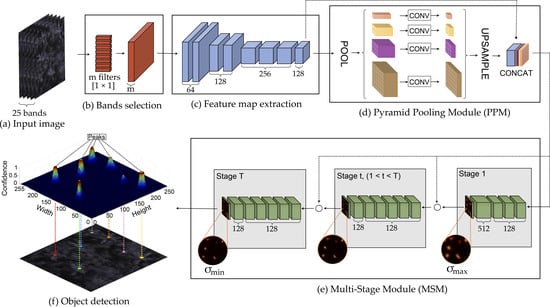
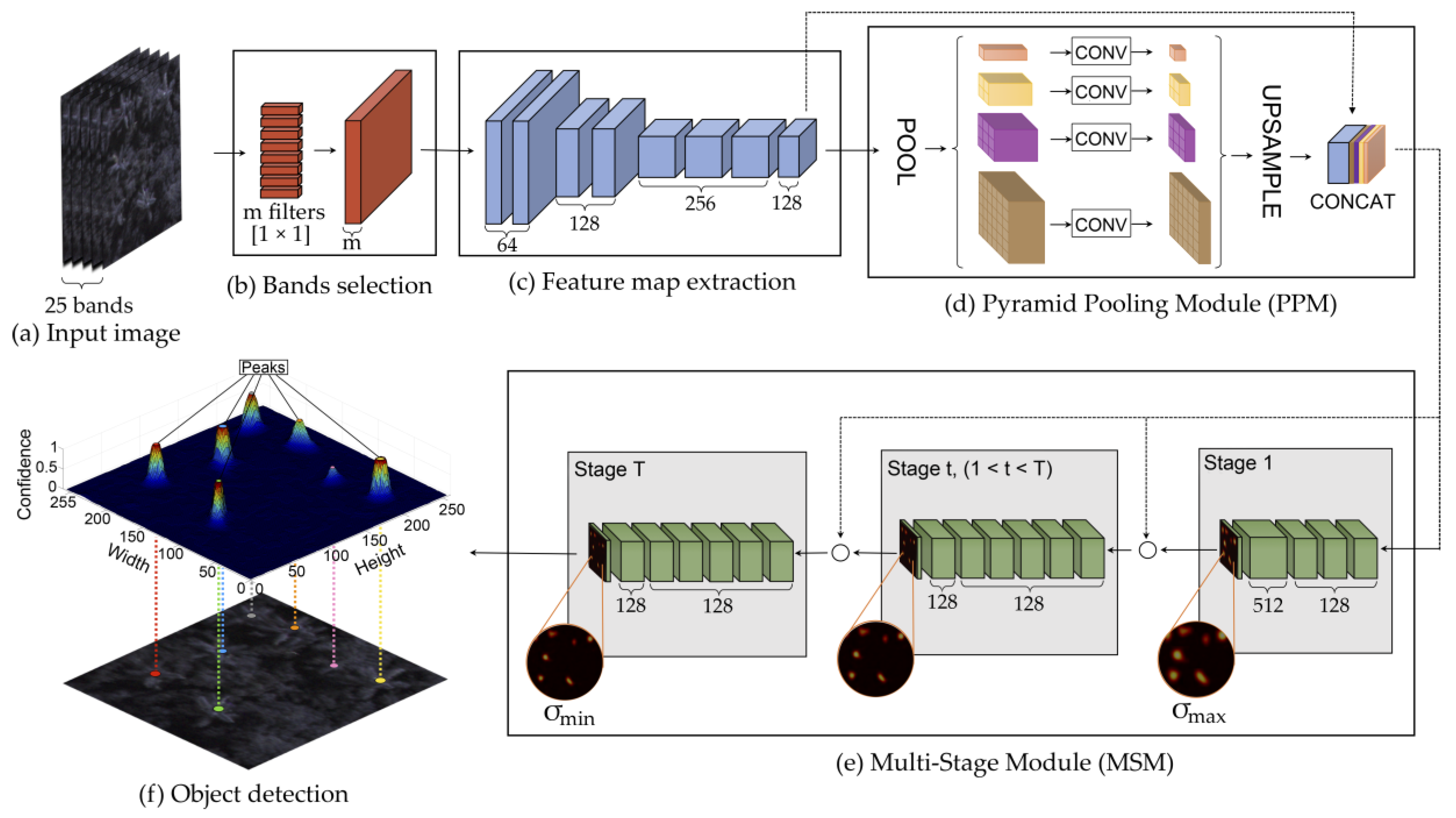
2.3. Proposed Method
2.3.1. Band Learning Machine Module
2.3.2. Feature Map Extraction
2.3.3. Tree Localization
2.4. Experimental Setup
3. Results
3.1. Validation of the Parameters
3.2. Band Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Aasen, H.; Honkavaara, E.; Lucieer, A.; Zarco-Tejada, P.J. Quantitative Remote Sensing at Ultra-High Resolution with UAV Spectroscopy: A Review of Sensor Technology, Measurement Procedures, and Data Correction Workflows. Remote. Sens. 2018, 10, 1091. [Google Scholar] [CrossRef] [Green Version]
- Guimarães, N.; Pádua, L.; Marques, P.; Silva, N.; Peres, E.; Sousa, J.J. Forestry Remote Sensing from Unmanned Aerial Vehicles: A Review Focusing on the Data, Processing and Potentialities. Remote. Sens. 2020, 12, 1046. [Google Scholar] [CrossRef] [Green Version]
- Näsi, R.; Honkavaara, E.; Lyytikäinen-Saarenmaa, P.; Blomqvist, M.; Litkey, P.; Hakala, T.; Viljanen, N.; Kantola, T.; Tanhuanpää, T.; Holopainen, M. Using UAV-Based Photogrammetry and Hyperspectral Imaging for Mapping Bark Beetle Damage at Tree-Level. Remote. Sens. 2015, 7, 15467–15493. [Google Scholar] [CrossRef] [Green Version]
- Saarinen, N.; Vastaranta, M.; Näsi, R.; Rosnell, T.; Hakala, T.; Honkavaara, E.; Wulder, M.A.; Luoma, V.; Tommaselli, A.M.G.; Imai, N.N.; et al. Assessing Biodiversity in Boreal Forests with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote. Sens. 2018, 10, 338. [Google Scholar] [CrossRef] [Green Version]
- Reis, B.P.; Martins, S.V.; Filho, E.I.F.; Sarcinelli, T.S.; Gleriani, J.M.; Marcatti, G.E.; Leite, H.G.; Halassy, M. Management Recommendation Generation for Areas Under Forest Restoration Process through Images Obtained by UAV and LiDAR. Remote. Sens. 2019, 11, 1508. [Google Scholar] [CrossRef] [Green Version]
- Navarro, A.; Young, M.; Allan, B.; Carnell, P.; Macreadie, P.; Ierodiaconou, D. The application of Unmanned Aerial Vehicles (UAVs) to estimate above-ground biomass of mangrove ecosystems. Remote. Sens. Environ. 2020, 242, 111747. [Google Scholar] [CrossRef]
- Casapia, X.T.; Falen, L.; Bartholomeus, H.; Cárdenas, R.; Flores, G.; Herold, M.; Coronado, E.N.H.; Baker, T.R. Identifying and Quantifying the Abundance of Economically Important Palms in Tropical Moist Forest Using UAV Imagery. Remote. Sens. 2019, 12, 9. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, J.; Mu, X.; Li, W.; Yan, G.; Xie, D.; Zhang, W. Quantifying Understory and Overstory Vegetation Cover Using UAV-Based RGB Imagery in Forest Plantation. Remote. Sens. 2020, 12, 298. [Google Scholar] [CrossRef] [Green Version]
- Colgan, M.S.; Baldeck, C.A.; Féret, J.-B.; Asner, G.P. Mapping Savanna Tree Species at Ecosystem Scales Using Support Vector Machine Classification and BRDF Correction on Airborne Hyperspectral and LiDAR Data. Remote Sens. 2012, 4, 3462–3480. [Google Scholar] [CrossRef] [Green Version]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.N.; et al. Individual Tree Detection and Classification with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote. Sens. 2017, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Tuominen, S.; Näsi, R.; Honkavaara, E.; Balazs, A.; Hakala, T.; Viljanen, N.; Pölönen, I.; Saari, H.; Ojanen, H. Assessment of Classifiers and Remote Sensing Features of Hyperspectral Imagery and Stereo-Photogrammetric Point Clouds for Recognition of Tree Species in a Forest Area of High Species Diversity. Remote. Sens. 2018, 10, 714. [Google Scholar] [CrossRef] [Green Version]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote. Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Chen, Y.; Lu, D.; Li, G.; Chen, E. Classification of Land Cover, Forest, and Tree Species Classes with ZiYuan-3 Multispectral and Stereo Data. Remote. Sens. 2019, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of machine-learning classification in remote sensing: An applied review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef] [Green Version]
- Osco, L.P.; Ramos, A.P.M.; Pereira, D.R.; Moriya, É.; Imai, N.N.; Matsubara, E.; Estrabis, N.; De Souza, M.; Marcato, J.; Goncalves, W.N.; et al. Predicting Canopy Nitrogen Content in Citrus-Trees Using Random Forest Algorithm Associated to Spectral Vegetation Indices from UAV-Imagery. Remote. Sens. 2019, 11, 2925. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.D.; Yokoya, N.; Bui, D.T.; Yoshino, K.; Friess, D.A. Remote Sensing Approaches for Monitoring Mangrove Species, Structure, and Biomass: Opportunities and Challenges. Remote. Sens. 2019, 11, 230. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, G.T.; Imai, N.N.; Tommaselli, A.M.G.; De Moraes, M.V.A.; Honkavaara, E. Evaluation of Hyperspectral Multitemporal Information to Improve Tree Species Identification in the Highly Diverse Atlantic Forest. Remote. Sens. 2020, 12, 244. [Google Scholar] [CrossRef] [Green Version]
- Marrs, J.; Ni-Meister, W. Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data. Remote. Sens. 2019, 11, 819. [Google Scholar] [CrossRef] [Green Version]
- Imangholiloo, M.; Saarinen, N.; Markelin, L.; Rosnell, T.; Näsi, R.; Hakala, T.; Honkavaara, E.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Characterizing Seedling Stands Using Leaf-Off and Leaf-On Photogrammetric Point Clouds and Hyperspectral Imagery Acquired from Unmanned Aerial Vehicle. Forests 2019, 10, 415. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Leng, W.; Liu, K.; Liu, L.; He, Z.; Zhu, Y. Object-Based Mangrove Species Classification Using Unmanned Aerial Vehicle Hyperspectral Images and Digital Surface Models. Remote. Sens. 2018, 10, 89. [Google Scholar] [CrossRef] [Green Version]
- Näsi, R.; Honkavaara, E.; Blomqvist, M.; Lyytikäinen-Saarenmaa, P.; Hakala, T.; Viljanen, N.; Kantola, T.; Holopainen, M. Remote sensing of bark beetle damage in urban forests at individual tree level using a novel hyperspectral camera from UAV and aircraft. Urban For. Urban Green. 2018, 30, 72–83. [Google Scholar] [CrossRef]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree Species Classification of Drone Hyperspectral and RGB Imagery with Deep Learning Convolutional Neural Networks. Remote. Sens. 2020, 12, 1070. [Google Scholar] [CrossRef] [Green Version]
- Sothe, C.; Almeida, C.M.D.; Schimalski, M.B.; Rosa, L.E.C.L.; Castro, J.D.B.; Feitosa, R.Q.; Dalponte, M.; Lima, C.L.; Liesenberg, V.; Miyoshi, G.T.; et al. Comparative performance of convolutional neural network, weighted and conventional support vector machine and random forest for classifying tree species using hyperspectral and photogrammetric data. GIScience Remote. Sens. 2020, 57, 369–394. [Google Scholar] [CrossRef]
- Safonova, A.; Tabik, S.; Alcaraz-Segura, D.; Rubtsov, A.; Maglinets, Y.; Herrera, F. Detection of Fir Trees (Abies sibirica) Damaged by the Bark Beetle in Unmanned Aerial Vehicle Images with Deep Learning. Remote. Sens. 2019, 11, 643. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Fu, H.; Yu, L.; Cracknell, A. Deep Learning Based Oil Palm Tree Detection and Counting for High-Resolution Remote Sensing Images. Remote Sens. 2017, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote. Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Khamparia, A.; Singh, K. A systematic review on deep learning architectures and applications. Expert Syst. 2019, 36, e12400. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.B.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. arXiv 2017, arXiv:1708.02002. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.B.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. arXiv 2015, arXiv:1506.01497. [Google Scholar]
- Dos Santos, A.A.; Marcato Junior, J.; Araújo, M.S.; Di Martini, D.R.; Tetila, E.C.; Siqueira, H.L.; Aoki, C.; Eltner, A.; Matsubara, E.T.; Pistori, H.; et al. Assessment of CNN-Based Methods for Individual Tree Detection on Images Captured by RGB Cameras Attached to UAVs. Sensors 2019, 19, 3595. [Google Scholar] [CrossRef] [Green Version]
- Lobo Torres, D.; Feitosa, R.; Nigri Happ, P.; Cue La Rosa, L.; Junior, J.; Martins, J.; Bressan, P.; Gonçalves, W.; Liesenberg, V. Applying Fully Convolutional Architectures for Semantic Segmentation of a Single Tree Species in Urban Environment on High Resolution UAV Optical Imagery. Sensors 2020, 20, 563. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Sylvain, J.-D.; Drolet, G.; Brown, N. Mapping dead forest cover using a deep convolutional neural network and digital aerial photography. ISPRS J. Photogramm. Remote. Sens. 2019, 156, 14–26. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual tree-crown detection in RGB imagery using self-supervised deep learning neural networks. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban Tree Species Classification Using a WorldView-2/3 and LiDAR Data Fusion Approach and Deep Learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote. Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Hennessy, A.; Clarke, K.; Lewis, M. Hyperspectral Classification of Plants: A Review of Waveband Selection Generalisability. Remote. Sens. 2020, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Alshehhi, R.; Marpu, P.R.; Woon, W.L.; Mura, M.D. Simultaneous extraction of roads and buildings in remote sensing imagery with convolutional neural networks. ISPRS J. Photogramm. Remote. Sens. 2017, 130, 139–149. [Google Scholar] [CrossRef]
- Audebert, N.; Saux, B.L.; Lefevre, S. Deep Learning for Classification of Hyperspectral Data: A Comparative Review. IEEE Geosci. Remote Sens. Mag. 2019, 7, 159–173. [Google Scholar] [CrossRef] [Green Version]
- Bioucas-Dias, J.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.M.; Chanussot, J. Hyperspectral Remote Sensing Data Analysis and Future Challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef] [Green Version]
- Richards, J.A.; Jia, X. Remote Sensing Digital Image Analysis: An Introduction, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2005; ISBN 3-540-25128-6. [Google Scholar]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data. Remote. Sens. 2018, 10, 1218. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Song, B.; Zhang, S.; Liu, X. A Novel Principal Component Analysis Method for the Reconstruction of Leaf Reflectance Spectra and Retrieval of Leaf Biochemical Contents. Remote. Sens. 2017, 9, 1113. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.A.; Wichern, D.W. Applied Multivariate Statistical Analysis; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2007; ISBN 978-0-13-187715-3. [Google Scholar]
- Özcan, A.H.; Hisar, D.; Sayar, Y.; Ünsalan, C. Tree crown detection and delineation in satellite images using probabilistic voting. Remote Sens. Lett. 2017, 8, 761–770. [Google Scholar] [CrossRef]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of Citrus Trees from Unmanned Aerial Vehicle Imagery Using Convolutional Neural Networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef] [Green Version]
- Ampatzidis, Y.; Partel, V. UAV-Based High Throughput Phenotyping in Citrus Utilizing Multispectral Imaging and Artificial Intelligence. Remote. Sens. 2019, 11, 410. [Google Scholar] [CrossRef] [Green Version]
- Osco, L.P.; Arruda, M.D.S.D.; Junior, J.M.; Da Silva, N.B.; Ramos, A.P.M.; Moryia, É.A.S.; Imai, N.N.; Pereira, D.R.; Creste, J.E.; Matsubara, E.T.; et al. A convolutional neural network approach for counting and geolocating citrus-trees in UAV multispectral imagery. ISPRS J. Photogramm. Remote. Sens. 2020, 160, 97–106. [Google Scholar] [CrossRef]
- Brasil Descreto s/n de 16 de julho de 2002. 2002. Available online: http://www.planalto.gov.br/ccivil_03/dnn/2002/Dnn9609.htm (accessed on 15 October 2016).
- Brasil Descreto s/n de 14 de maio de 2004. 2004. Available online: http://www.planalto.gov.br/CCIVIL_03/_Ato2004-2006/2004/Decreto/_quadro.htm (accessed on 15 October 2016).
- Berveglieri, A.; Tommaselli, A.M.G.; Imai, N.N.; Ribeiro, E.A.W.; Guimarães, R.B.; Honkavaara, E. Identification of Successional Stages and Cover Changes of Tropical Forest Based on Digital Surface Model Analysis. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2016, 9, 5385–5397. [Google Scholar] [CrossRef]
- Berveglieri, A.; Imai, N.N.; Tommaselli, A.M.G.; Casagrande, B.; Honkavaara, E. Successional stages and their evolution in tropical forests using multi-temporal photogrammetric surface models and superpixels. ISPRS J. Photogramm. Remote. Sens. 2018, 146, 548–558. [Google Scholar] [CrossRef]
- Giombini, M.I.; Bravo, S.P.; Sica, Y.V.; Tosto, D.S. Early genetic consequences of defaunation in a large-seeded vertebrate-dispersed palm (Syagrus romanzoffiana). Heredity 2017, 118, 568–577. [Google Scholar] [CrossRef]
- Elias, G.; Colares, R.; Rocha Antunes, A.; Padilha, P.; Tucker Lima, J.; Santos, R. Palm (Arecaceae) Communities in the Brazilian Atlantic Forest: A Phytosociological Study. Floresta e Ambiente 2019, 26. [Google Scholar] [CrossRef]
- Da Silva, F.R.; Begnini, R.M.; Lopes, B.C.; Castellani, T.T. Seed dispersal and predation in the palm Syagrus romanzoffiana on two islands with different faunal richness, southern Brazil. Stud. Neotrop. Fauna Environ. 2011, 46, 163–171. [Google Scholar] [CrossRef]
- Brasil, D.F. Espécies Nativas da Flora Brasileira de Valor Econômico Atual ou Potencial: Plantas para o Futuro-Região Centro-Oeste. 2011. Available online: https://www.alice.cnptia.embrapa.br/handle/doc/1073295 (accessed on 3 March 2020).
- Lorenzi, H. Árvores Brasileiras, 1st ed.; Instituto Plantarum de Estudos da Flora: Nova Odessa, Brazil, 1992; Volume 1, ISBN 85-86714-14-3. [Google Scholar]
- Mendes, C.; Ribeiro, M.; Galetti, M. Patch size, shape and edge distance influence seed predation on a palm species in the Atlantic forest. Ecography 2015, 39. [Google Scholar] [CrossRef]
- Sica, Y.; Bravo, S.P.; Giombini, M. Spatial Pattern of Pindó Palm (Syagrus romanzoffiana) Recruitment in Argentinian Atlantic Forest: The Importance of Tapir and Effects of Defaunation. Biotropica 2014, 46. [Google Scholar] [CrossRef]
- Honkavaara, E.; Saari, H.; Kaivosoja, J.; Pölönen, I.; Hakala, T.; Litkey, P.; Mäkynen, J.; Pesonen, L. Processing and Assessment of Spectrometric, Stereoscopic Imagery Collected Using a Lightweight UAV Spectral Camera for Precision Agriculture. Remote Sens. 2013, 5, 5006–5039. [Google Scholar] [CrossRef] [Green Version]
- Honkavaara, E.; Rosnell, T.; Oliveira, R.; Tommaselli, A. Band registration of tuneable frame format hyperspectral UAV imagers in complex scenes. ISPRS J. Photogramm. Remote. Sens. 2017, 134, 96–109. [Google Scholar] [CrossRef]
- Honkavaara, E.; Khoramshahi, E. Radiometric Correction of Close-Range Spectral Image Blocks Captured Using an Unmanned Aerial Vehicle with a Radiometric Block Adjustment. Remote. Sens. 2018, 10, 256. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.M.; Milton, E.J. The use of the empirical line method to calibrate remotely sensed data to reflectance. Int. J. Remote Sens. 1999, 20, 2653–2662. [Google Scholar] [CrossRef]
- Miyoshi, G.T.; Imai, N.N.; Tommaselli, A.M.G.; Honkavaara, E.; Näsi, R.; Moriya, É.A.S. Radiometric block adjustment of hyperspectral image blocks in the Brazilian environment. Int. J. Remote Sens. 2018, 39, 4910–4930. [Google Scholar] [CrossRef] [Green Version]
- Aich, S.; Stavness, I. Improving Object Counting with Heatmap Regulation. arXiv 2018, arXiv:1803.05494. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar]
- Story, M.; Congalton, R.G. Accuracy assessment: A user’s perspective. Photogramm. Eng. Remote Sens. 1986, 52, 397–399. [Google Scholar]
- Jensen, J.R. Remote Sensing of the Environment: An Earth Resource Perspective; Prentice Hall Series in Geographic Information Science; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2007; ISBN 978-0-13-188950-7. [Google Scholar]
- Clark, M.L.; Roberts, D.A. Species-Level Differences in Hyperspectral Metrics among Tropical Rainforest Trees as Determined by a Tree-Based Classifier. Remote Sens. 2012, 4, 1820–1855. [Google Scholar] [CrossRef] [Green Version]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote. Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Natesan, S.; Armenakis, C.; Vepakomma, U. RESNET-Based tree species classification using UAV images. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019. [Google Scholar] [CrossRef] [Green Version]








| Band | λ/FWHM | Band | λ/FWHM | Band | λ/FWHM | Band | λ/FWHM | Band | λ/FWHM |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 506.22/12.44 | 6 | 580.16/15.95 | 11 | 650.96/14.44 | 16 | 700.28/18.94 | 21 | 750.16/17.97 |
| 2 | 519.94/17.38 | 7 | 591.90/16.61 | 12 | 659.72/16.83 | 17 | 710.06/19.70 | 22 | 769.89/18.72 |
| 3 | 535.09/16.84 | 8 | 609.00/15.08 | 13 | 669.75/19.80 | 18 | 720.17/19.31 | 23 | 780.49/17.36 |
| 4 | 550.39/16.53 | 9 | 620.22/16.26 | 14 | 679.84/20.45 | 19 | 729.57/19.01 | 24 | 790.30/17.39 |
| 5 | 565.10/17.26 | 10 | 628.73/15.30 | 15 | 690.28/18.87 | 20 | 740.42/17.98 | 25 | 819.66/17.84 |
| 2016 | 2017 | 2018 | Total | |
|---|---|---|---|---|
| Training | 106 | 174 | 175 | 455 |
| Validation | 81 | 112 | 112 | 305 |
| Test | 79 | 116 | 116 | 311 |
| Method | Precision | Recall | F-Measure |
|---|---|---|---|
| Baseline + 25 bands | 0.898 | 0.881 | 0.889 |
| Baseline + PCA (5 bands) | 0.979 | 0.871 | 0.921 |
| Proposed method | 0.973 | 0.945 | 0.959 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyoshi, G.T.; Arruda, M.d.S.; Osco, L.P.; Marcato Junior, J.; Gonçalves, D.N.; Imai, N.N.; Tommaselli, A.M.G.; Honkavaara, E.; Gonçalves, W.N. A Novel Deep Learning Method to Identify Single Tree Species in UAV-Based Hyperspectral Images. Remote Sens. 2020, 12, 1294. https://doi.org/10.3390/rs12081294
Miyoshi GT, Arruda MdS, Osco LP, Marcato Junior J, Gonçalves DN, Imai NN, Tommaselli AMG, Honkavaara E, Gonçalves WN. A Novel Deep Learning Method to Identify Single Tree Species in UAV-Based Hyperspectral Images. Remote Sensing. 2020; 12(8):1294. https://doi.org/10.3390/rs12081294
Chicago/Turabian StyleMiyoshi, Gabriela Takahashi, Mauro dos Santos Arruda, Lucas Prado Osco, José Marcato Junior, Diogo Nunes Gonçalves, Nilton Nobuhiro Imai, Antonio Maria Garcia Tommaselli, Eija Honkavaara, and Wesley Nunes Gonçalves. 2020. "A Novel Deep Learning Method to Identify Single Tree Species in UAV-Based Hyperspectral Images" Remote Sensing 12, no. 8: 1294. https://doi.org/10.3390/rs12081294
APA StyleMiyoshi, G. T., Arruda, M. d. S., Osco, L. P., Marcato Junior, J., Gonçalves, D. N., Imai, N. N., Tommaselli, A. M. G., Honkavaara, E., & Gonçalves, W. N. (2020). A Novel Deep Learning Method to Identify Single Tree Species in UAV-Based Hyperspectral Images. Remote Sensing, 12(8), 1294. https://doi.org/10.3390/rs12081294