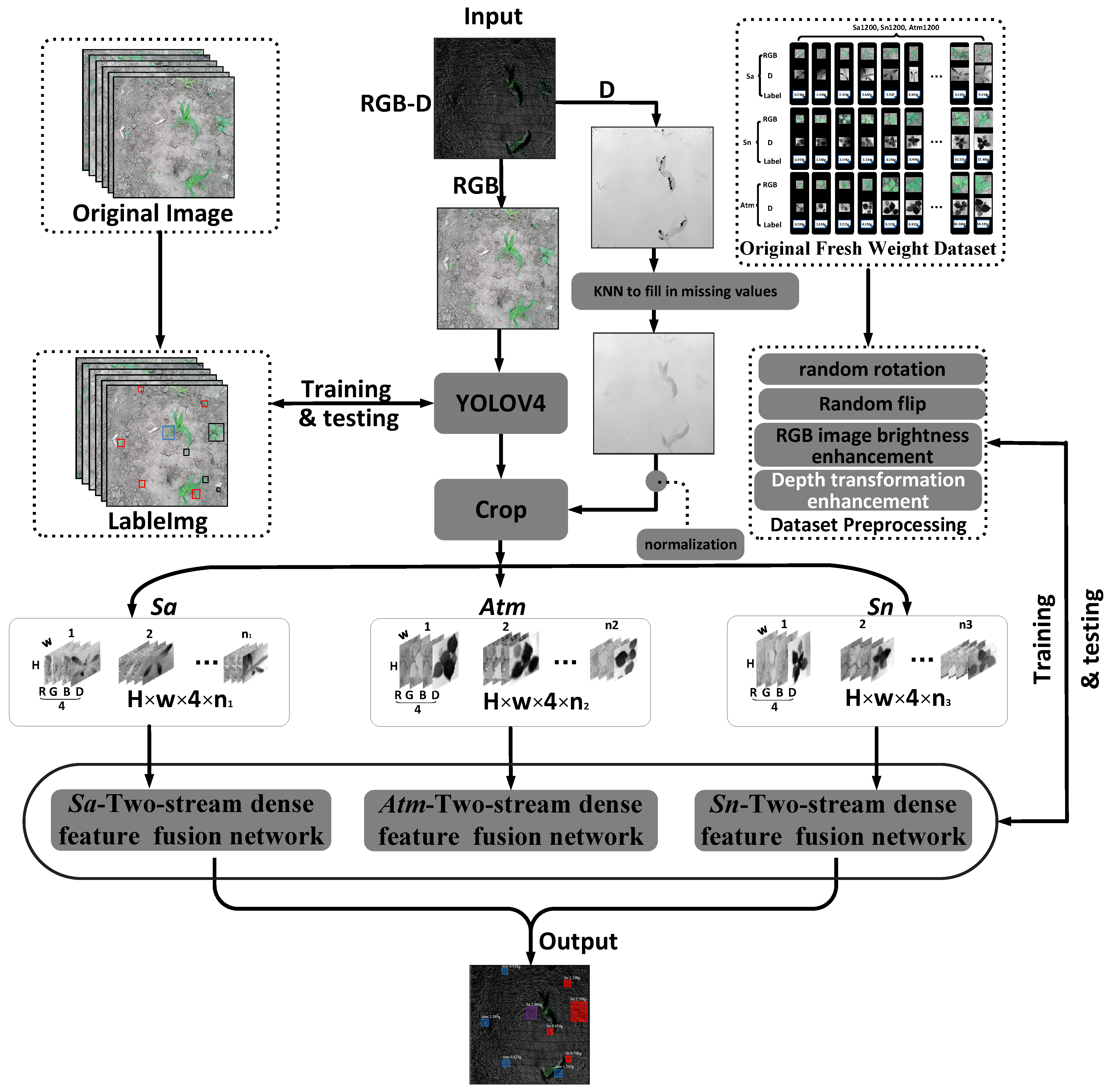
1. Introduction
In the process of weed management, uniform herbicide spraying is currently the most commonly applied weeding method [
1]. However, the large-scale use of herbicides has led to the pollution of the natural environment, increased weed resistance, hidden dangers to food security and biodiversity, and many other agricultural and ecological problems [
2,
3]. This has led to more focused research on precision weed management strategies. In precision weed management, the most commonly used method is to determine the location of weeds through computer vision technology and to apply herbicides to individual weeds [
4,
5]. However, from the perspective of plant physiology, the dosage of herbicides is closely related to the type of weed and its physiological parameters [
6,
7,
8]. Applying a uniform herbicide dosage does not maximize the use of herbicides, and there is therefore still room for optimization. Herbicides in weeds directly act on the cells of the weeds, affecting cell metabolism and transport functions and eventually killing the weeds [
9,
10]. The size of weed cells and tissues directly determines the herbicide dosage [
6,
11]. The aboveground fresh weight is an index that best reflects the size and cell content of plants [
12,
13], and it is suitable for use as a quantitative index to provide a basis for determining herbicide dosages for real-time variable herbicide spraying. Therefore, the development of a vision system that can detect weeds in real time in complex farmland environments and obtain fresh weight data can change the process of determining precise application doses and has important guiding significance for precise weed management.
The use of visual technology is a rapid and effective method for evaluating the fresh weight of plants. Jiang et al. [
14] developed a lettuce weight monitoring system in a plant factory that segmented RGB images and used the number of pixels and the plant weight data to establish a regression equation. Arzani et al. [
15] established a regression relationship between fruit diameter and fresh weight. Reyes et al. [
16] used Mask R-CNN to segment RGB images to obtain plant characteristics and establish a regression equation between fresh weight and characteristics to obtain the fresh weight of lettuce. The experiment was carried out on a hydroponic growth bed. Mortensen et al. [
17] performed 3D point cloud segmentation and obtained the surface area parameters of lettuce for fresh weight prediction. Lee et al. [
18] used the 3D point cloud obtained by Kinect for 3D printing and correlated the weight of the cabbage with the amount of material consumed by the 3D printer. The main method used in the current research is to first extract the plant to be predicted from the background, extract the characteristics of the plant and then establish an association with the fresh weight. However, farmland scenes are complex and changeable; the soil background is uneven, the light fluctuates, the weeds in the field overlap each other, and the types, spatial shapes, and growth positions of the weeds differ. It is therefore difficult to extract weeds from complex backgrounds [
19,
20]. At the same time, the phenotypic information for each plant is extracted as a predictive factor (e.g., measured value [
21], leaf area index [
22], pixel number [
23]) to establish a single linear regression relationship with the fresh weight of the plant; this single-factor approach does not include sufficient information. The fresh weight of weeds is determined not by a single characteristic parameter but by a combination of multiple characteristics. Therefore, it is still difficult to correlate the multidimensional characteristics of weeds with their fresh weight.
Current methods around the extraction of weeds from the background include the use of computer vision techniques and spectral features as the two main directions. This can be effectively distinguished if there is a significant difference in spectral reflectance between the two weeds [
24,
25,
26,
27]. However, the use of spectral cameras is often expensive and demanding (illumination) and it is also difficult to distinguish between weeds with similar spectral features. On the other hand, weed identification using visible light images is mainly based on color features, shape features, or texture features [
28,
29,
30,
31,
32]. Weed detection based on color features uses different weed and crop color thresholds for effective differentiation. However, when faced with similarly colored weeds and crops, it is difficult to distinguish them even with color space conversion [
33], especially for large fields with a relatively large number of weed species, and it is relatively difficult to identify each weed species at a granular level. Identification based on shape and texture is also relatively difficult under conditions of overlapping leaves and similar weed shapes, where shape feature templates are susceptible to interference [
34,
35,
36]. The interspecific similarity of weeds to weeds and the similarity of weeds to crops makes it difficult to perform multi-species weed detection using single-function computer vision methods. In this context, deep learning techniques have performed well in the field of image target detection and recognition [
37,
38,
39,
40]. CNNs can automatically acquire multiple features in visible images that are effective for target object recognition, are robust for multi-species target detection in complex environments, and have been applied to weed recognition research [
41,
42,
43,
44]. The use of CNN technology for weed identification holds good promise.
Weeds are polymorphic, and different types of weeds exhibit different spatial scale information; the same weed may even exist at different spatial scales at different life stages. The use of 2D plane information obtained as RGB images for estimation has limitations, and it is difficult to use this information to accurately describe the spatial stereo information of weeds. As 3D point cloud technology has developed, it has begun to be applied for plant spatial detection. Zhou et al. [
45] used 3D point cloud technology to segment soybean plants. Li et al. [
46] developed a low-cost 3D plant morphological characterization system. Chaivivatrakul et al. [
47] used 3D reconstruction to characterize the morphology of corn plants. Unlike RGB information, 3D point cloud information can provide spatial scale information [
48], and it is obviously more advantageous for describing the spatial structure of weeds. 2D or 3D information essentially obtains phenotypic parameters as a single predictor variable for linear regression. Sapkota et al. [
49] used canopy cover obtained from UAV imagery to build a regression model with ryegrass biomass. However the single linear model is weakly expressive and may ignore other potential information in the imagery that has an impact on above ground biomass. The convolutional neural network model has unique advantages for addressing nonlinear relationships. Such models can recognize the complex and nonlinear relationship between the input and output of the modelling process [
50,
51] and automatically learn implicit characteristic information to directly perform nonlinear regression predictions of fresh weight. At present, it is relatively rare to use convolutional neural networks to directly associate 3D information from weeds with their fresh weight.
In this work, to accurately locate weeds and to predict the fresh weight of weeds with different shapes and positions against a complex farmland background, a combination of 3D point cloud and deep learning techniques is explored.
The contributions of this article are as follows:
A method of data collection and preprocessing for constructing the fresh weight of different kinds of weeds is proposed.
A YOLO-V4 model and a dense fusion network of two-stream features are established for weed detection and fresh weight estimation.
The proposed method is tested and analyzed.
4. Results and Discussion
4.1. Technical Route Results
The main idea of the technical route proposed in this paper is to use YOLO-V4 to locate the target weeds and then send the obtained weed areas to the corresponding two-stream dense feature fusion network by category to predict their fresh weight on the ground.
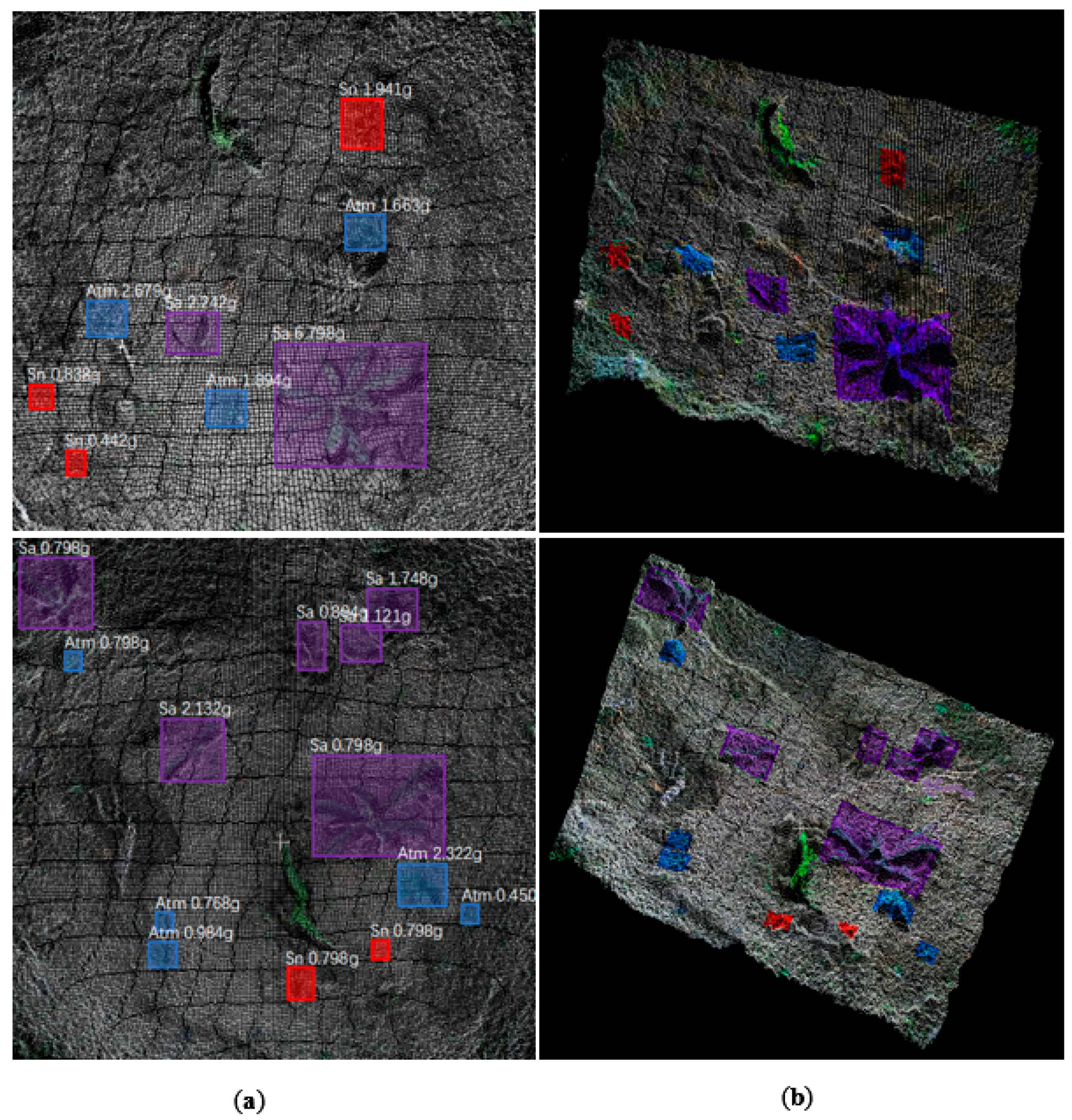
Figure 10 shows the results of the 3D visualization of the aboveground fresh weight detection of weeds (A visualization of the results of the two-stream dense feature fusion network on RGB images can be obtained in
Figure A3 in the
Appendix A). The mAP (IoU value of 0.5) of the model proposed in this paper is 75.34%, and the mIoU is 86.36%. When combining YOLO-V4 with the improved, fastest two-stream dense feature fusion network (AlexNet) model, the prediction speed is 17.8fps. The average relative error of the fresh weight of the weeds in the test set is approximately 4%. This model can provide visual technical support for precision variable-target platforms.
4.2. Comparison of YOLO-V4 with Other Target Detection Algorithms
To find the most suitable convolutional neural network for weed detection, this study compared the YOLO-V4 model with the SSD [
59], YOLO-V5x [
60], M2DNet [
61], and Faster R-CNN [
53] networks. Target detection networks can be divided into two main categories: one-stage target detection networks and two-stage target detection networks. The reason for selecting these four networks for comparison is that YOLO-V4, YOLO-V5x, SSD, and M2DNet are typical representative one-stage networks of different types, and their performance is relatively advanced. The Faster R-CNN network, a typical two-stage network, also exhibits advanced performance. Therefore, this article compares the performance advantages of these four types of networks with regard to the problem of weed detection.
Table 2 shows the mAP scores (mAP is obtained at an IoU value of 0.5), mIoU values, and average detection times of the models.
In the above results, the mAP score of YOLO-V4 is 0.7534, which is higher than the scores of the other four models. This indicates that the combined recall performance and accuracy of YOLO-V4 is better than the other four models. the IoU value of YOLO-V4 is 0.8636, which is higher than the other four models. This indicates that YOLO-V4 is more accurate than the other four models in detecting bounding boxes. the average removal time of YOLO-V4 is 0.033 seconds, which is faster than the other three models. However, the detection speed of YOLO-V4 was slower compared to YOLO-V5x. In our test set1, the minimum pixel size that yolov4 can detect for Sonchus arvensis is 14 × 16, for Abutilon theophrasti Medicus 8 × 10, and for Solanum nigrum 7 × 11. YOLO-V4 is effective for small target weed detection.
4.3. Two-Stream Dense Feature Fusion Network (DenseNet201-Rgbd)
4.3.1. Comparison of Regression Network Results Embedded with the Dense-NiN Module
In describing the model, we mentioned that the Dense-NiN module can be embedded in a typical convolutional neural network. In the embedded VGG19 and AlexNet networks, we add a deep feature fusion layer after each pooling layer to receive the output of the Dense-NiN-Block module. In Inception-V3 and Resnet101, we add the Dense-NiN-Block module before the network convergence layer. The structure of DenseNet201 has been described above. The number of test set2 for each weed species is 300. This study integrates the Resnet101, VGG19, Inception-V3, AlexNet, and DenseNet201 networks of the Dense-NiN module for comparison to select the model with the best fit.
To compare the effects of weed species on the detection results, three weed species,
Abutilon theophrasti Medicus,
Solanum nigrum, and
Sonchus arvensis, were used as training sets to train the convolutional neural network. At the same time, these three weed species were also merged into a single data set to train the model (abbreviated as all). Moreover, to compare RGB-D information and RGB information when using a convolutional network for fresh weight prediction, RGB and RGB-D were used as inputs for network training. The dual-stream dense fusion network architecture proposed in this paper used the RGB-D information for training. The RGB images were used directly with the default network structures of these five networks, and the output module of the original network needed only to be replaced with the output module proposed in this article to achieve a new regression. The RMSEs of the training models are shown in
Figure 11, the
values are shown in
Table 3, and the average times (s) are shown in
Table 4.
The above results show that, in all networks, the accuracy obtained using RGB-D data as the input is higher than that obtained using RGB as the input. This indicates that RBG-D stereo data can indeed provide more information for use in weed fresh weight evaluation. However, the speed usually decreases when RBG-D data are used. This is because the two-stream dense feature fusion network using RGB-D data introduces a denser convolution structure and increases the weight, which causes the speed to drop. In the regression test for the fresh weights of the three weed species, the RMSE values of the dual-stream dense fusion network (DenseNet201-rgbd) are 0.358 for Abutilon theophrasti Medicus, 0.416 for Solanum nigrum, and 0.424 for Sonchus arvensis (Notable among these is the closer detection of RGB-D and RGB for Sonchus arvensis compared to the other two weeds. We provide a specific analysis in session 4.3.3.). The value for all weeds is 0.568, which is higher than those of the other models. The RMSE values of the three aboveground fresh weight prediction models trained using this model are lower than the RMSE value of all weed models trained directly. Therefore, after applying YOLO-V4, a network that can be independently and successfully trained for each weed species can be adaptively selected, and its performance will be better than a trained network using all the weeds as the training set. The of the dual-stream dense fusion network (DenseNet201-rgbd) is also the highest, with a value of 0.9917 for Abutilon theophrasti Medicus, a value of 0.9921 for Solanum nigrum, and a value of 0.9885 for Sonchus arvensis. This network has a good fitting ability. Selecting the corresponding model according to the weed type output by YOLO-V4 does not affect the speed of the model. For example, there are 10 tensors in the output stream of YOLO-V4. Using a different model for each weed type or directly using all the trained weed models requires a calculation time of 10 tensors. The only difference is whether the network is selected according to the weed type. This kind of speed loss is almost negligible.
At the same time, the higher the accuracy of the detection model is, the slower the speed; if higher accuracy is desired, speed must be sacrificed to some extent. When the density of weeds in the environment is high, the accuracy of the model may be reduced, and a faster model can be selected. It is worth noting that the average detection speed of each model is 0.0359 for Solanum nigrum, 0.0378 for Sonchus arvensis, and 0.0390 for Abutilon theophrasti Medicus. We believe that this is due to the size of the weed test set2 image. We calculated the average image size of the three weeds in the test set2. The average size of Solanum nigrum is 104 × 108, the average size of Abutilon theophrasti Medicus is 166 × 175, and the average size of Sonchus arvensis is 150 × 158. The size of the weeds also affects the speed of the network. Therefore, reducing the image size uniformly during the training process of the two-stream dense feature fusion network and reducing the image size by the same proportion during the prediction process could help to improve the efficiency of the model.
On the other hand, we used a non-CNN technique to build a regression model with the canopy area of the weed as the independent variable and the aboveground fresh weight of the weed as the dependent variable. Using a polynomial fit method, Abutilon theophrasti Medicus obtained a minimum RMSE value of 3.632. Solanum nigrum obtained a minimum RMSE value of 3.246. Sonchus arvensis obtained a minimum RMSE value of 2.033. The experiments proved that that using the CNN technique is indeed better than using single factor regression. The method is more advantageous. In a real field environment, the ground is relatively uneven. For example, two identical weeds, one growing at a higher position and the other at a lower position, will have different RGB images even if the height of the camera is 800 mm. If above-ground fresh regression is performed using canopy pixel area, the weed growing in the higher position has a larger canopy pixel area and the weed growing in the lower position has a smaller canopy pixel area. This can lead to such errors, and depth data can help us to resolve such differences effectively.
In practical agricultural applications, the Chinese national standard (GB-T36007-2018) states that field weeding robots should operate at a speed of around 0.4 to 0.5 m per second. Our robots can operate effectively in real time with RGB-D while complying with the Chinese national standard. For us, faster speed is not as effective as more precise accuracy. In the future, robots will inevitably travel at higher speeds, so it is worth considering giving up a certain level of accuracy to use RGB images in the future. It is worth noting that YOLO-V5x is very fast and, although not as accurate as YOLO-V4, is smaller, making it easier for us to deploy to edge computing devices. We still need to evaluate the specific performance of YOLO-V4 and YOLO-V5x on edge computing devices such as the Jet-son AGX Xavier in future work.
4.3.2. The Impact of Different Data Enhancement Methods
To verify the influence of the four data augmentation methods described above in the training model, the control variable method was used to delete one data augmentation method at a time, and the RMSE values were obtained. The results are shown in
Table 5.
According to the experimental results, random rotation and random flipping have limited impacts on the model, but excluding these two methods still reduces the detection accuracy. Removing random rotation increases the average RMSE of the model by 0.052, and removing random flipping increases the average RMSE of the model by up to 0.050. The device cover provides the function of a hood but still allows visible light to pass through. Brightness enhancement can help the model adapt to subtle changes in light. The results show that the result of removing the brightness enhancement transform is 0.115 higher than the RMSE value using the full enhancement method. The depth conversion enhancement function can help the model adapt to uneven ground. Depth enhancement greatly improves the performance of the detection model. If this method is excluded, the RMSE score of the detection model increases by 0.129. Therefore, the depth conversion enhancement method helps to improve the performance of the model.
4.3.3. The Two-Stream Dense Feature Fusion Network (DenseNet201) Is Affected by the Growth Period and Weed Species
To compare the responses of the RGB network and RGB-D network (DenseNet201-rgb and DenseNet201-rgbd) to weeds in different periods, we classified the three weed species by size from small to large according to the quality distribution of the test set. Every fifty adjacent weeds are considered as one stage, and six stages (A, B, C, D, E, and F) stages are considered in the analysis.
Figure 12 shows the actual results for the three weed species.
Comparing the average RMSE value of the RGB data with the average RMSE value of the RGB-D data shows that in stages A and B, the RMSE value for Abutilon theophrasti Medicus increased by 0.113, that for Solanum nigrum decreased by 0.011, and that for Sonchus arvensis decreased by 0.162. The advantage of using RGB-D data is not obvious. In stages C and D, the RMSE for Abutilon theophrasti Medicus increased by 0.209, for Solanum nigrum weeds increased by 0.334, and for Sonchus arvensis increased by 0.111. The RMSE for Abutilon theophrasti Medicus and Solanum nigrum increased significantly, while the increase in the RMSE for Sonchus arvensis was relatively small. In stages E and F, the RMSE for Abutilon theophrasti Medicus weeds increased by 0.650, for Solanum nigrum increased by 0.628, and for Sonchus arvensis increased by 0.249. Compared with those in the first four stages, the RMSE increase for Abutilon theophrasti Medicus and Solanum nigrum was greater, while the increase for Sonchus arvensis was still relatively small. Overall, the RMSE values for Abutilon theophrasti Medicus and Solanum nigrum obtained using RGB images as input gradually increases, and the magnitude of the increase also increases. Although the RMSE value for Sonchus arvensis also exhibits an upward trend, the overall fluctuation is very small. Using RGB-D as the network input, the RMSE values for the predicted values of the weeds in the six stages all fluctuate slightly or even show a downward trend. The results show that in the later stages of weed growth, using RGB-D as the network input provides more stable and accurate results than using RGB as the network input.
In the early weed growth stages, the performances obtained using RGB and RGB-D as inputs are roughly the same. This shows that in the early stage, the regression model is more dependent on the overhead-view area of the plant for regression prediction. At this time, the weeds are very short, so the regression prediction results using RGB and RGB-D are nearly the same. In the subsequent growth stages, as the weeds gradually grow taller, the stems account for a certain percentage of the weight of the weeds, the height of the plants cannot be obtained from the RGB image, and the accuracy of predictions obtained using RGB images begins to decline. The scatter plots of the actual and predicted fresh weights of the weeds show that in the RGB prediction process, at the later stage of growth, the predicted fresh weight value is usually lower than the actual value. Due to a lack of height information, the predicted fresh weight value is too low. Therefore, the RGB-D model exhibits better robustness in the subsequent growth stages of weeds. However, in these six stages, the RMSE values of the results obtained using RGB-D and RGB images for Sonchus arvensis did not change substantially. Given the low height of these weed species, their aboveground fresh weight may depend more on their top-view area. In the early and late stages of growth, the difference between the RMSE values of the RGB and RGB-D predictions is not substantial, but RGB-D still provides a better fitting effect.
4.3.4. Model Analysis
- (1)
Dense connections extract deep features
The Dense-NiN-Block module uses a dense connection structure. The dense structure allows access to all its previous feature maps (including transition layers). Our experiment investigates whether the trained network takes advantage of this opportunity. For each convolutional layer in a block, we calculate the average (absolute) weight assigned to the connection to layer
.
Figure 13 shows the heat map of all four Dense-NiN-Block modules. The average absolute weight replaces the dependence of the convolutional layer on its previous layers. The red dot in position (
,
) indicates that the layer uses the feature map of the previously generated
layer on average.
The figure shows that all layers spread their weights over many inputs within the same block. The feature information from the weed depth data obtained in the early stages of the network is actually used by the deeper convolution filters within the same dense block. The weights of the transition layers also spread their weight across all layers within the preceding dense block, indicating information flow from the first to the last layers of the Dense module through few indirections. Therefore, the NiN module in this study effectively uses the Dense connection method to enhance the use of weed depth information.
- (2)
Model visualization analysis
To explore which information made the greatest contribution to fresh weight prediction as well as the specific impact of depth data, we use Grad-CAM to visualize the network and compare the differences between the RGB-D network and RGB network models (DenseNet201-rgb and DenseNet201-rgbd).
Figure 14 shows the visualization results. Areas with a high thermal value represent the greatest utilization of the feature map of the pixel area.
As shown in
Figure 14, these two networks have learned the channel pixels within the weed area in order to make fresh weight predictions. In the Grad-CAM map output by two-stream dense feature fusion network, the heat value near the middle of the weed area is higher than that at the edges of the weed area. We believe that this phenomenon occurs because the central area of the plant, as the main growth point of the stem, has obviously different height characteristics than the other plant parts. This leads to a large difference between the depth data in this part and in other parts, and this difference can improve the weed fresh weight prediction function of the model; in contrast, the RGB network does not have such advantages. In addition, our model not only considers to the information within the weed outline but also considers the periphery of the weed area (shown in the red circles in the figure). In the actual environment, the ground cannot be flat. Although the camera is set at a distance of 800 mm from the ground, it cannot actually be stabilized at that distance. Due to the unevenness of the ground, the camera position fluctuates around 800 mm above the ground. This results in some weeds being detected in low-lying positions, while some weeds are perceived as being relatively tall. For example, for two weeds of the same quality, the RGB image of the taller weed is larger due to the difference in geographic location, which makes the RGB network prediction value higher. From the perspective of depth data, the depth data value of shorter weeds is higher, and the depth data value of taller weeds is lower. The depth information of the weed outline area does not directly reflect this difference, but the depth data outside the weed outline directly reflects the distance between the camera and the ground. The figure above shows that distance information is also regarded as an important difference feature by the network. At the same time, the information inside and outside the weed outline constitutes the height information for the weed. The high thermal response outside the weed outline area indicates that the network has learned this indirect relationship. Therefore, the value of information outside the range of weed outlines is also used effectively. The RGB network cannot resolve the imaging difference caused by the fluctuation of the distance between the camera and the ground. On the other hand, we explored the results of using only data within the RGB-D weed contour lines. The RMSE results obtained using the network proposed in this paper showed 0.648 for
Abutilon theophrasti Medicus, 0.824 for
Solanum nigrum, and 0.481 for
Sonchus arvensis, all lower than the method used in this paper and more evidence of the importance of ground-to-camera distance information in depth images (areas beyond the weed contour lines). distance information in the depth images (the area beyond the weed contour line).
4.4. The Relationship between IOU and Fresh Weight Prediction
In this study, manual trimming was used to create the data set when training the network. However, when using the two-stream dense feature fusion network model, the output of the YOLO-V4 model was actually accepted. There are certain differences between the two. The specific response to this difference is reflected in the IoU values, so this article compares the RMSE under different IoU values. To specifically reflect the impact of manual trimming and the YOLO-V4 output data on the accuracy of fresh weight prediction. The comparison was performed to reflect the difference in accuracy. The results are shown in
Table 6.
The results above show that the IOU value will have a slight impact on the prediction result. When the IOU value is greater than 50%, the RMSE values for the three weed species using the YOLO-V4 network result as the input prediction value and those using the manual trimming result as the input prediction value are 0.065, 0.034, and 0.060, respectively, and show little difference. However, as the IOU value decreases, the RMSE value gradually increases, and the network prediction accuracy decreases. These results demonstrate that the IOU value affects the accuracy of the two-stream dense feature fusion network. In this article, the IOU threshold for YOLO-V4 is selected as 0.5. Appropriately increasing the IOU threshold can make the network fitting effect more accurate.
4.5. Predictive Effects for Shaded Weeds
In the early stages of corn cultivation, weeds are small and rarely cover each other. During this period, individual weeds are easy to distinguish. However, as the weeds continue to grow, the degree of overlap between them increases, and it becomes more difficult to distinguish them. YOLO-V4 can identify weeds that have a certain degree of overlap, but misidentifications can still occur. Instances of misrecognition can be classified into three situations:
When two weeds cover each other, the network divides them into uniform individuals, as shown by the red bounding box.
When two weeds cover each other, the network identifies only part of the weed but not the whole weed, as shown by the purple bounding boxes.
When two weeds shade each other, the weed cannot be detected, as shown by the black arrow in (a).
Figure 15a shows a situation in which, because of mutual covering, two weeds are identified as one.
Sonchus arvensis is a weed species that relies heavily on its stems to reproduce multiple aboveground parts on the same root that usually overlap considerably and are close together. Therefore, it is easy for mistakes to occur during detection.
Figure 15b,c show the occlusion of
Solanum nigrum and
Abutilon theophrasti Medicus. Unlike
Sonchus arvensis,
Solanum nigrum and
Abutilon theophrasti Medicus have distinct individual characteristics, do not share the same root system, and are usually farther apart. Even if there is occlusion, partial recognition can be achieved, but the abovementioned problems still exist. These problems will affect the accuracy of the subsequent aboveground fresh weight prediction. The first type of error will result in the calculation of the aboveground fresh weight of the provided weed patch data, and the second type of error will cause the prediction value to be too small. However, the purpose of this article is to provide visual support for precise adjustments to herbicide application. Except for the small number of errors of the third type, the detection errors observed in this study would have little effect on variable herbicide application. Therefore, the research in this article still has practical significance.
5. Conclusions
In this study, we propose a new concept for the real-time detection of the aboveground fresh weight of weeds to provide visual support for precision variable herbicide spraying. At the same time, a new model that can detect weeds and predict their fresh weight in real time in the field is developed. The algorithm combines deep learning technology with 3D data. This paper proposes a strategy of using the YOLO-V4 target detection network to obtain the regional weed area and then send the RGB-D data for the weed area into a dual-stream dense feature fusion network regression model to perform a regression on the fresh weight data so that the fresh weight of weeds can be predicted. The error of the model is approximately 4%, and the fastest detection speed is 17.8 fps. To construct a data set for training these two networks, a data collection method that establishes a labelling method is proposed. This method can quickly establish the relationship between the weed RGB-D data and the fresh weight data while avoiding interference with the actual operating environment. When predicting the fresh weight of weeds taller than a certain height, more accurate results are achieved using RGB-D information as the input for the model. The visualization results show that the use of a two-stream dense feature fusion network can better address the imaging differences caused by the uneven land surface and make the predictions more accurate.
In this paper we have only done a preliminary exploration of fresh weight models for three weeds, the richness of weed species is still lacking, and the next step of the workshop is to enrich our weed types. Future work will focus on determining the type and fresh weight of weeds in order to determine the appropriate amount of herbicides to apply in real time, optimizing weeding strategies to reduce the use of herbicides, and applying the model to a variable herbicide-application robot. The approaches used to develop this model can also be extended to the prediction of the fresh weight of crop plants, which could provide support for crop breeding and genetic improvement as well as soil health.