Mapping Phragmites australis Aboveground Biomass in the Momoge Wetland Ramsar Site Based on Sentinel-1/2 Images
Abstract
:1. Introduction
2. Materials and Methods
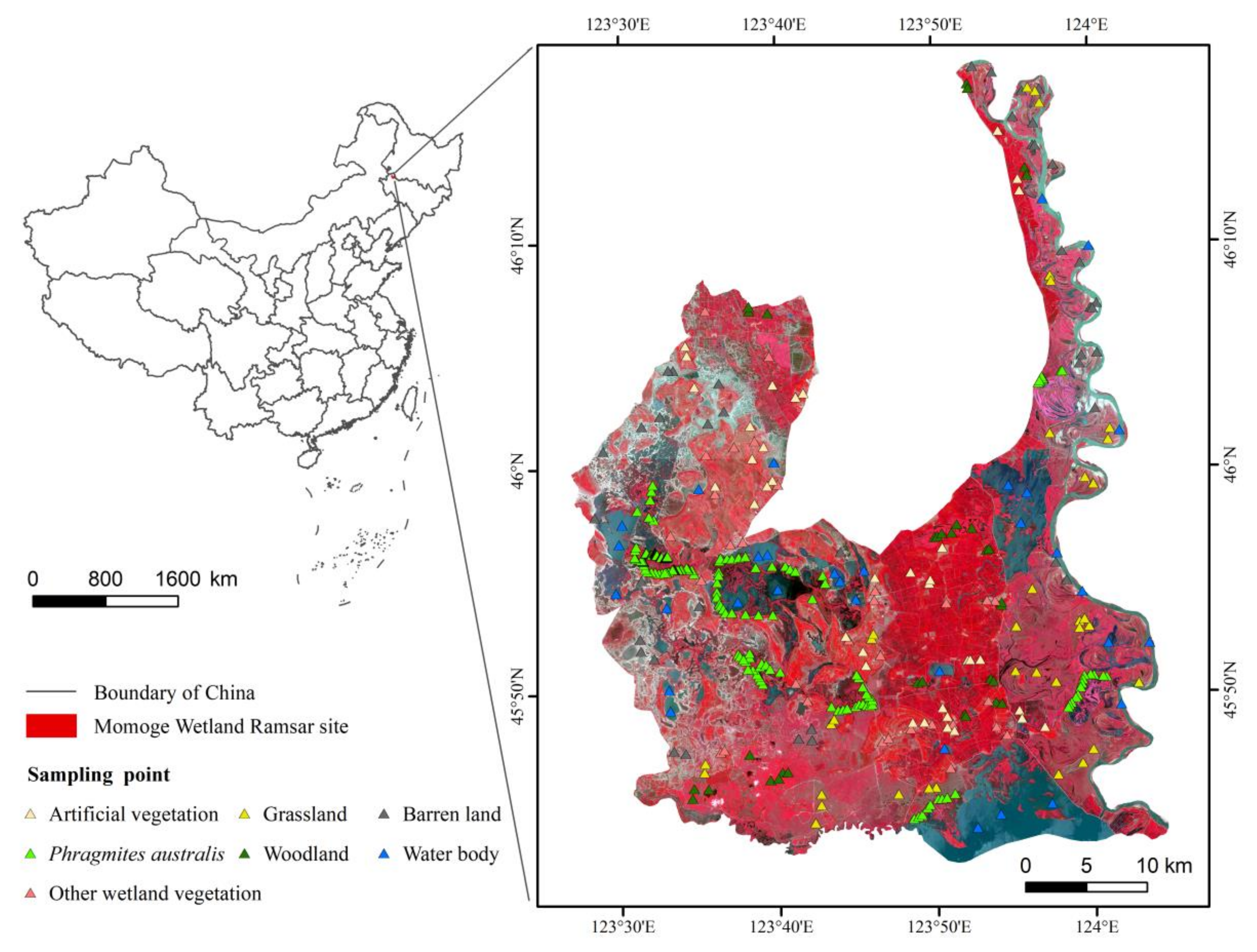
2.1. Study Area
2.2. Datasets
2.2.1. Sentinel-1/2 Image Selection and Processing
2.2.2. Field Survey Dataset
2.2.3. Biomass Sampling of P. australis
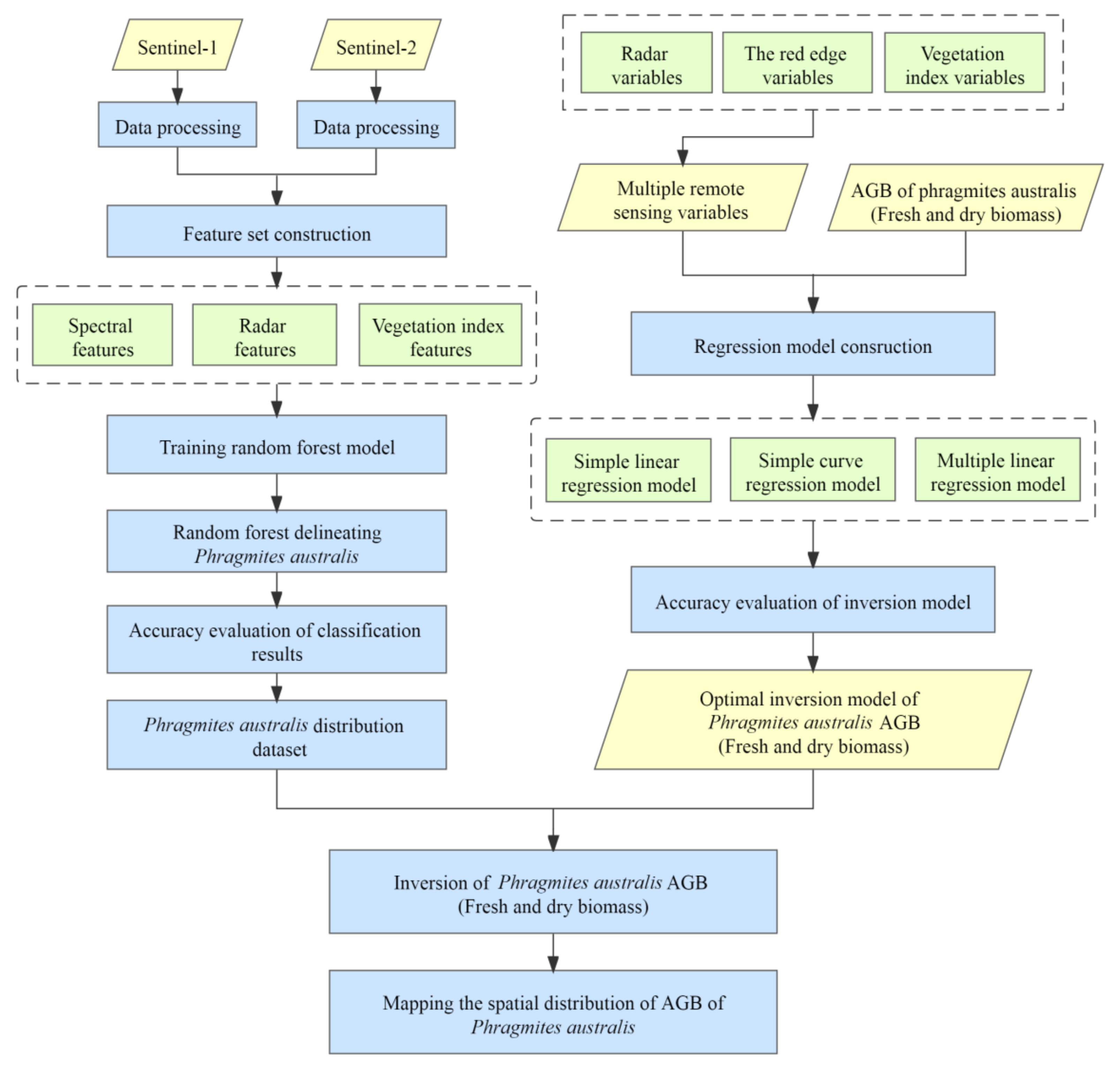
2.3. Methods
2.3.1. Classification System and Classification Feature Sets
2.3.2. Training the Random Forest Model
2.3.3. Selecting Remote Sensing Variables for Predicting P. australis AGB
2.3.4. AGB Inversion Regression Model
2.3.5. Precision Validation
3. Results
3.1. Classification Accuracy and Spatial Pattern of P. australis
3.2. Optimal Regression Model for Predicting P. australis AGB
3.2.1. Sensitivity of Different Remote Sensing Variables to P. australis AGB
3.2.2. Optimal Regression Model and Accuracy Evaluation for Predicting P. australis AGB
3.3. Spatial Estimates of P. australis AGB
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Simple Linear Regression Model | R2 | F | P | |||||
|---|---|---|---|---|---|---|---|---|
| Fresh AGB | Dry AGB | Fresh AGB | Dry AGB | Fresh AGB | Dry AGB | Fresh AGB | Dry AGB | |
| DVI | Y = 94.601X + 32.786 | Y = 58.795X + 3.855 | 0.168 | 0.287 | 19.748 | 39.456 | 0.000 | 0.000 |
| EVI | Y = −25474.1X + 12841.298 | Y = -11140X + 5614.933 | 0.099 | 0.084 | 10.762 | 8.971 | 0.001 | 0.003 |
| IRECI | Y = 988.958X + 48.703 | Y = 688.448X + 11.128 | 0.148 | 0.317 | 16.960 | 45.440 | 0.000 | 0.000 |
| MCARI | Y = −496.785X + 159.414 | Y = -319.359X + 84.169 | 0.184 | 0.345 | 22.101 | 51.646 | 0.000 | 0.000 |
| MSAVI | Y = 77.709X + 25.610 | Y = 54.533X − 5.275 | 0.099 | 0.215 | 10.725 | 26.881 | 0.001 | 0.000 |
| NDVI | Y = 62.920X + 45.448 | Y = 47.494X + 6.611 | 0.084 | 0.213 | 9.033 | 26.534 | 0.003 | 0.000 |
| SAVI | Y = 48.968X + 38.738 | Y = 33.942X + 4.326 | 0.105 | 0.223 | 11.456 | 28.096 | 0.001 | 0.000 |
| B7 | Y = 92.998X + 35.745 | Y = 63.534X + 2.731 | 0.167 | 0.241 | 19.631 | 31.084 | 0.000 | 0.000 |
| B6 | Y = 109.958X + 31.961 | Y = 75.392X + 0.017 | 0.154 | 0.321 | 17.829 | 46.252 | 0.000 | 0.000 |
| B5 | Y = 583.525X − 46.448 | Y = 334.526X − 39.109 | 0.132 | 0.268 | 14.938 | 35.877 | 0.000 | 0.000 |
| VH | Y = 309.324X + 26.062 | 0.066 | 6.905 | 0.01 | ||||
| Curve Regression Model | R2 | F | P | |
|---|---|---|---|---|
| Fresh AGB | Fresh AGB | Fresh AGB | Fresh AGB | |
| DVI | Y = 1013.351X − 2062.661X2 + 1434.618X3 | 0.192 | 7.603 | 0.000 |
| IRECI | Y = 3060.607X − 86,367.302X2 + 999,864.866X3 + 37.16 | 0.403 | 43.44 | 0.000 |
| MCARI | Y = −1385.306X + 2895.829X2 + 225.919 | 0.134 | 7.535 | 0.001 |
| MSAVI | Y = 722.027X − 1308.796X2 + 805.478X3 − 66.292 | 0.115 | 4.174 | 0.008 |
| NDVI | Y = 686.102X − 1525.248X2 + 1109.235X3 − 24.43 | 0.112 | 4.024 | 0.01 |
| SAVI | Y = 412.465X − 599.9X2 + 293.628X3 − 21.856 | 0.125 | 4.592 | 0.005 |
| B7 | Y = 247.359X − 447.164X2 + 372.205X3 + 22.05 | 0.172 | 6.642 | 0.000 |
| B6 | Y = 250.181X − 490.966X2 + 478.936X3 + 22.842 | 0.158 | 6.016 | 0.001 |
| B5 | Y = 87.605X + 1134.670X2 + 7.059 | 0.185 | 11.016 | 0.000 |
| Curve Regression Model | R2 | F | P | |
| Dry AGB | Dry AGB | Dry AGB | Dry AGB | |
| DVI | Y = 454.874X − 815.043X2 + 525.493X3 − 55.01 | 0.301 | 13.788 | 0.000 |
| IRECI | Y = 540.258X + 13,446.345X2 − 206,133.785X3 + 9.802 | 0.432 | 53.047 | 0.000 |
| MCARI | Y = 170.852X − 1597.673X2 + 47.478 | 0.246 | 15.807 | 0.000 |
| MSAVI | Y = 181.920X − 303.982X2 + 205.433X3 − 18.921 | 0.226 | 9.333 | 0.000 |
| NDVI | Y = 290.012X − 590.474X2 + 427.825X3 − 20.792 | 0.231 | 9.598 | 0.000 |
| SAVI | Y = 160.815X − 207.596X2 + 101.023X3 − 17.116 | 0.233 | 9.737 | 0.000 |
| B7 | Y = -99.743X + 423.332X2 − 328.471X3 + 20.538 | 0.355 | 17.621 | 0.000 |
| B6 | Y = −251.365X + 874.941X2 − 721.383X3 + 36.285 | 0.332 | 15.869 | 0.000 |
| B5 | Y = 1002.791X − 1528.997X2 − 111.212 | 0.277 | 18.54 | 0.000 |
| VH | Y = 1831.173X − 30,031.630X2 + 146,088.610X3 + 5.927 | 0.1 | 3.566 | 0.017 |
References
- Mitsch, W.J.; Gosselink, J.G. Wetlands; John Wiley & Sons Inc.: New York, NY, USA, 1986. [Google Scholar]
- Mitsch, W.J.; Gosselink, J.G. Wetlands, 2nd ed.; Van Nostrand Reinhold: New York, NY, USA, 1993. [Google Scholar]
- Duman, T.; Schäfer, K.V.R. Partitioning net ecosystem carbon exchange of native and invasive plant communities by vegetation cover in an urban tidal wetland in the New Jersey Meadowlands (USA). Ecol. Eng. 2018, 114, 16–24. [Google Scholar] [CrossRef]
- Mitsch, W.J.; Gosselink, J.G. Wetlands, 3rd ed.; John Wiley & Son, Inc.: New York, NY, USA, 2000. [Google Scholar]
- Guo, N.; Liu, J. Overview of plant biomass research. Subtrop. Plant Sci. 2011, 40, 83–88. [Google Scholar]
- Mao, D.; Luo, L.; Wang, Z.; Wilson, M.C.; Zeng, Y.; Wu, B.; Wu, J. Conversions between natural wetlands and farmland in China: A multiscale geospatial analysis. Sci. Total Environ. 2018, 634, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, X.; Zhou, Y. Progress in estimating methods of wetland vegetation biomass. Geogr. Geo-Inf. Sci. 2004, 5, 104–109, 113. [Google Scholar]
- Shao, C.; Chen, Z.; Dong, H. Study on the growth and biomass of Phragmites communis in liaohe Estuarine wetland. J. Liaoning Univ. (Nat. Sci. Ed.) 1995, 1, 89–94. [Google Scholar]
- Liu, L.; Han, M.; Liu, Y. Spatial distribution of wetland vegetation biomass and its influencing factors in the Yellow River Delta Nature Reserve. Acta Ecol. Sin. 2017, 37, 4346–4355. [Google Scholar]
- Wang, J.; Mao, D.; Du, H. Study on forest and swamp mapping of Hani Wetland using Sentinel 1/2 satellite imagery. Wetl. Sci. Manag. 2021, 17, 2–7+12. [Google Scholar]
- Gaurav, K.; Kiran, K.S. Mapping and Monitoring the Selected Wetlands of Punjab, India, Using Geospatial Techniques. J. Indian Soc. Remote Sens. 2020, 48, 615–625. [Google Scholar]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: A review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, A. High density biomass estimation for wetland vegetation using WorldView-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Jensen, D.; Cavanaugh, K.; Simard, M.; Christensen, A.; Rovai, A. Aboveground biomass distributions and vegetation composition changes in Louisiana’s Wax Lake Delta. Estuar. Coast. Shelf Sci. 2021, 250, 107–139. [Google Scholar] [CrossRef]
- Yang, Q.; Yuan, Q.; Li, T. Mapping PM 2.5 concentration at high resolution using a cascade random forest based downscaling model: Evaluation and application. J. Clean. Prod. 2020, 277, 1–12. [Google Scholar] [CrossRef]
- Gou, F.; Zhao, C.; Yang, J. Spatial pattern of aboveground biomass and its response to water and salinity in sugan Lake wetland. Acta Ecol. Sin. 2021, 19, 1–11. [Google Scholar]
- Yu, H.; Wu, Y.; Jin, Y. Retrieval of aboveground biomass from MODIS SWIR data and its spatio-temporal variation in arid region. Remote Sens. Technol. Appl. 2017, 32, 524–530. [Google Scholar]
- Dinh, H.T.M. Potential value of combining ALOS PALSAR and Landsat-derived tree cover data for forest biomass retrieval in Madagascar. Remote Sens. Environ. 2018, 213, 206–214. [Google Scholar]
- Ramon, T.; Paul, S.; Dirk, G. GMES Sentinel- 1 mission. Remote Sens. Environ. 2012, 120, 9–24. [Google Scholar]
- Su, W.; Hou, N.; Li, Q. Retrieval of leaf area index of maize canopy based on Sentinel-2 remote sensing images. Trans. Chin. Soc. Agric. Mach. 2018, 49, 151–156. [Google Scholar]
- Oliver, C.; Maurizio, S. Exploring combinations of multi-temporal and multi-frequency radar backscatter observations to estimate above-ground biomass of tropical forest. Remote Sens. Environ. 2019, 232, 111313. [Google Scholar]
- Oliver, S.; Andrea, N.; Osama, Y. Dimensionality Reduction and Feature Selection for Object-Based Land Cover Classification based on Sentinel-1 and Sentinel-2 Time Series Using Google Earth Engine. Remote Sens. 2019, 12, 76. [Google Scholar]
- Jelének, J.; Kopačková-Strnadová, V. Synergic use of Sentinel-1 and Sentinel-2 data for automatic detection of earthquake-triggered landscape changes: A case study of the 2016 Kaikoura earthquake (Mw 7.8), New Zealand. Remote Sens. Environ. 2021, 265. [Google Scholar] [CrossRef]
- Investigators at Free University of Bozen-Bolzano Report Findings in Remote Sensing. Exploiting Time Series of Sentinel-1 and Sentinel-2 Imagery To Detect Meadow Phenology In Mountain Regions. Remote Sensing. 2019, 11, 542. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, C.; Heng, W. Retrieving Surface Soil Moisture over Wheat-Covered Areas Using Data from Sentinel-1 and Sentinel-2. Water 2021, 13, 1981. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Wu, W. Prediction of Soil Organic Carbon under Different Land Use Types Using Sentinel-1/-2 Data in a Small Watershed. Remote Sens. 2021, 13, 1229. [Google Scholar] [CrossRef]
- Xing, X.; Yang, X.; Xu, B. Estimation of aboveground biomass using remote sensing based on random forest algorithm. J. Geo-Inf. Sci. 2021, 23, 1312–1324. [Google Scholar]
- O’Shea, R.E. Advancing cyanobacteria biomass estimation from hyperspectral observations: Demonstrations with HICO and PRISMA imagery. Remote Sens. Environ. 2021, 266, 112693. [Google Scholar] [CrossRef]
- Tewes, A. Assimilation of Sentinel-2 Estimated LAI into a Crop Model: Influence of Timing and Frequency of Acquisitions on Simulation of Water Stress and Biomass Production of Winter Wheat. Agronomy 2020, 10, 1813. [Google Scholar] [CrossRef]
- Yuan, Y.; Yuan, L.; Sheng, H. Influence of Spectral Bandwidth and Position on Chlorophyll Content Retrieval at Leaf and Canopy Levels. J. Indian Soc. Remote Sens. 2016, 44, 583–593. [Google Scholar]
- Schmid, T.; Koch, M.; Gumuzzio, J.; Mather, P.M. A spectral library for a semi-arid wetland and its application to studies of wetland degradation using hyperspectral and multispectral data. Int. J. Remote Sens. 2004, 25, 2485–2496. [Google Scholar] [CrossRef]
- Li, C.; Zhou, L.; Xu, W. Estimating Aboveground Biomass Using Sentinel-2 MSI Data and Ensemble Algorithms for Grassland in the Shengjin Lake Wetland, China. Remote Sens. 2021, 13, 1595. [Google Scholar] [CrossRef]
- The Secretariat of the Convention on Wetlands. The List of Wetlands of International Importance. 2020. Available online: https://www.ramsar.org/sites/default/files/documents/library/sitelist.pdf (accessed on 15 October 2021).
- Jiang, H.; He, C.; Luo, W. Hydrological restoration and water resource management of siberian crane (Grus leucogeranus) stopover wetlands. Water 2018, 10, 1714. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zuo, Y.; Ren, B. Study on insect diversity in momoge national nature reserve. J. Northeast. Norm. Univ. (Nat. Sci.) 2011, 43, 112–116. [Google Scholar]
- Li, S.; An, Y.; Wang, X. Species composition and quantitative characteristics of plant communities in Momoge Wetland under different surface water levels. Wetl. Sci. 2015, 13, 466–471. [Google Scholar]
- Wu, B.; Qian, J.; Zeng, Y. Land Cover Atlas of the People’s Republic of China (1:10 Million); China Cartographic Publishing House: Beijing, China, 2017. [Google Scholar]
- Yan, H.; Zhu, W.; Mao, D. Remote sensing analysis of anthropogenic stress in internationally important wetlands of Yangtze river delta. China Environ. Sci. 2020, 40, 3605–3615. [Google Scholar]
- Hu, B.; Xu, Y.; Huang, X. Improving Urban Land Cover Classification with Combined Use of Sentinel-2 and Sentinel-1 Imagery. ISPRS Int. J. Geo-Inf. 2021, 10, 533. [Google Scholar] [CrossRef]
- Ghorbanian, A.; Zaghian, S.; Asiyabi, R.M. Mangrove Ecosystem Mapping Using Sentinel-1 and Sentinel-2 Satellite Images and Random Forest Algorithm in Google Earth Engine. Remote Sens. 2021, 13, 2565. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, C.; Luo, W. Urban impervious surface extraction based on multi-features and random forest. IEEE Access 2020, 8, 226609–226623. [Google Scholar] [CrossRef]
- Yao, M. Random Forests and Its Application to the Classification of Remote Sensing Image; Huaqiao University: Quanzhou, China, 2014. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Chen, Q.; Wang, G. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2014, 13, 1–43. [Google Scholar] [CrossRef]
- Zhang, G.; Ganguly, S.; Nemani, R.R. Estimation of forest aboveground biomass in California using canopy height and leaf area index estimated from satellite data. Remote Sens. Environ. 2014, 151, 44–56. [Google Scholar] [CrossRef]
- Mao, D.; Wang, Z.; Li, L. Spatiotemporal dynamics of grassland aboveground net primary productivity and its association with climatic pattern and changes in Northern China. Ecol. Indic. 2014, 41, 40–48. [Google Scholar] [CrossRef]
- Yang, C.; Liu, J.; Huang, H. Correlation analysis between tropical forest vegetation biomass and remote sensing geo-data. Geogr. Res. 2005, 24, 473–479. [Google Scholar]
- Zhang, J.; Pan, G. Comparison and application of multiple linear regression and BP neural network prediction model. J. Kunming Univ. Technol. Nat. Sci. Ed. 2013, 38, 61–67. [Google Scholar]
- Li, J.; Shu, X.; Chen, S. Remote sensing monitoring model of poyang Lake wetland vegetation biomass based on LandSAT-TM data. J. Guangzhou Univ. (Nat. Sci. Ed.) 2005, 6, 494–498. [Google Scholar]
- Xu, J. Mathematical Methods in Modern Geography; Higher Education Press: Beijing, China, 2002; pp. 47–60. [Google Scholar]
- Li, X.; Yeh, A.; Liu, K. Inventory of mangrove wetlands in the pearl river estuary of China using remote sensing. J. Geogr. Sci. 2006, 16, 155–164. [Google Scholar] [CrossRef]
- Lei, X.; Yang, B.; Jiang, W. Changes of vegetation pattern and its influencing factors in east dongting wetland. Geogr. Res. 2012, 31, 461–470. [Google Scholar]
- Xiang, D.; Wu, Y.; Zhang, R. Classification of coastal wetland vegetation using remote sensing. Shanxi Meteorol. 2011, 2, 19–24. [Google Scholar]
- Shan, J.; Fang, W.; Lu, M. Local detrended fluctuation analysis for spectral red-edge parameters extraction. Nonlinear Dyn. 2018, 93, 995–1008. [Google Scholar]
- Yumiko, K.; Brenda, T.; Marilyn, D. Evaluation of red and red-edge reflectance-based vegetation indices for rice biomass and grain yield prediction models in paddy fields. Precis. Agric. 2016, 17, 507–530. [Google Scholar]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef] [Green Version]
- Filella, I.; Penuelas, J. The red edge position and shape as indicators of plant chlorophyll content, biomass and hydric status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Hansen, P.; Schjoerring, J. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Song, K.; Zhang, B.; Li, F. Correlation analysis of hyperspectral reflectance with soybean leaf area and aboveground fresh biomass. Trans. Chin. Soc. Agric. Eng. 2005, 21, 36–40. [Google Scholar]
- Frampton, W.; Dash, J.; Watmough, G. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, X.; Ying, G. Biomass model of typical steppe region based on VEGETATION index: A case study of Xilinhot City, Inner Mongolia. Chin. J. Plant Ecol. 2007, 31, 23–31. [Google Scholar]
- Zhang, M.; Ustin, S.; Rejmankova, E. Monitoring Pacific coast salt marshes using remote sensing. Ecol. Appl. 1997, 7, 1039–1053. [Google Scholar] [CrossRef]
- Shi, Z.; Ma, Y.; Wang, Y. Advances in land use/cover classification using remote sensing images. Chin. Agric. Sci. Bull. 2012, 28, 273–278. [Google Scholar]
- Swatantran, A.; Dubayah, R.; Roberts, D. Mapping biomass and stress in the Sierra Nevada using lidar and hyperspectral data fusion. Remote Sens. Environ. 2011, 115, 2917–2930. [Google Scholar] [CrossRef] [Green Version]
- Megan, W.; Lang, E.; Kasischke, S.; Prince, S.D.; Pittman, K.W. Assessment of C-band synthetic aperture radar data for mapping and monitoring Coastal Plain forested wetlands in the Mid-Atlantic Region, U.S.A. Remote Sens. Environ. 2007, 112, 4120–4130. [Google Scholar]
- Wang, G.S.; Wang, N.; Guo, W.L. Modelling Forest Aboveground Biomass Based on GF-3 Dual-Polarized and WorldView-3 Data: A Case Study in Datong National Wetland Park, China. Math. Probl. Eng. 2021, 2021, 9925940. [Google Scholar] [CrossRef]
- Bucha, T.; Papčo, J.; Sačkov, I.; Pajtík, J. Woody Above-Ground Biomass Estimation on Abandoned Agriculture Land Using Sentinel-1 and Sentinel-2 Data. Remote Sens. 2021, 13, 2488. [Google Scholar] [CrossRef]
- Ren, L.; Wang, M.; Li, C. Impacts of human activities on river runoff in the northern area of China. J. Hydrol. 2002, 261, 204–217. [Google Scholar] [CrossRef]
- Xu, Z.; He, Y.; Yan, B.; Ren, H. Effects of nutrient and water level changes on wetland plants. Chin. J. Ecol. 2006, 25, 87–92. [Google Scholar]
- Lawson, T.; Vialet-Chabrand, S. Speedy stomata, photosynthesis and plant water use efficiency. New Phytol. 2019, 221, 93–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorai, M.; Laajili, W.; Santiago, L. Rapid recovery of photosynthesis and water relations following soil drying and rewatering is related to the adaptation of desert shrub Ephedra alata subsp. alenda (Ephedraceae) to arid environments. Environ. Exp. Bot. 2015, 109, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Wang, X.; Tong, S. Study on plant community species diversity in momoge Wetland restoration area. Wetl. Sci. 2021, 19, 458–464. [Google Scholar]
- Sun, X.; Chen, Y.; Zhuo, N.; Cui, Y. Effects of salinity and concomitant species on growth of Phragmites australis populations at different levels of genetic diversity. Sci. Total Environ. 2021, 780, 146516. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, W.; Ji, R.; Zhao, Y.; Feng, R. Dynamic Response of Phragmites australis and Suaeda salsa to Climate Change in the Liaohe Delta Wetland. J. Meteorol. Res. 2021, 35, 157–171. [Google Scholar] [CrossRef]




| Acquisition Date: 15 July 2020 | Acquisition Date: 23 July 2020 | ||
|---|---|---|---|
| Sentinel-2 Band | Spatial Resolution (m) | Sentinel-1 Band | Spatial Resolution (m) |
| B2 Blue | 10 | VV polarization backscattering coefficient | 10 |
| B3 Green | 10 | ||
| B4 Red | 10 | ||
| B5 VRE1 | 20 | ||
| B6 VRE2 | 20 | ||
| B7 VRE3 | 20 | VH polarization backscattering coefficient | 10 |
| B8 NIR | 10 | ||
| B8a VRE | 20 | ||
| B11 SWIR1 | 20 | ||
| B12 SWIR2 | 20 | ||
| Variable Types | Name of Remote Sensing Variables | Description or Calculation Formula |
|---|---|---|
| Radar characteristics | VV | VV polarization backscattering coefficient |
| VH | VH polarization backscattering coefficient | |
| Red edge band characteristics | B5 (VRE-1) | Sentinel-2 Vegetation Red Edge band 1 |
| B6 (VRE-2) | Sentinel-2 Vegetation Red Edge band 2 | |
| B7 (VRE-3) | Sentinel-2 Vegetation Red Edge band 3 | |
| Vegetation index characteristics | DVI | NIR-Red |
| EVI | 2.5(NIR − Red)/(NIR + 6Red − 7.5Blue + 1) | |
| NDVI | (NIR − Red)/(NIR + Red) | |
| SAVI | ((NIR − Red)/(NIR + Red + L))(1 + L) | |
| MSAVI | ||
| MCARI | ((VRE1 − Red) − 0.2(VRE1 − Green))(VRE1/NIR) | |
| IRECI | (VRE3-Red)/(VRE1/VRE2) |
| P. australis | Water Body | Barren Land | Wood Land | Grassland | Other Wetland Vegetation | Artificial Vegetation | Total | |
|---|---|---|---|---|---|---|---|---|
| P. australis | 4173 | 19 | 0 | 3 | 0 | 59 | 237 | 4491 |
| Water body | 13 | 11,808 | 0 | 0 | 0 | 0 | 0 | 11,821 |
| Barren land | 0 | 0 | 2900 | 0 | 25 | 0 | 216 | 3141 |
| Woodland | 26 | 0 | 0 | 2137 | 11 | 159 | 11 | 2344 |
| Grassland | 0 | 0 | 2 | 0 | 4409 | 436 | 12 | 4859 |
| Other wetland vegetation | 0 | 0 | 3 | 143 | 1396 | 3800 | 14 | 5436 |
| Artificial vegetation | 312 | 0 | 133 | 572 | 0 | 38 | 2169 | 3224 |
| Total | 4524 | 11,827 | 3038 | 2855 | 5841 | 4572 | 2659 | 35,316 |
| Producer Accuracy% | 92.24 | 99.84 | 95.46 | 74.85 | 75.48 | 84.86 | 81.57 | |
| User Accuracy% | 92.92 | 99.89 | 92.36 | 91.17 | 90.76 | 71.39 | 67.28 |
| Multiple Linear Regression Model | R2 | F | P | |
|---|---|---|---|---|
| Fresh weight of AGB | Y = 856.114 + 379.777(B5) + 199.002(DVI) + 726.696(B7) − 785.183(B6) − 1514.958(IRECI) −208.821(MCARI) − 206.846(SAVI) − 1754.943(EVI) + 374.596(MSAVI) − 146.105(NDVI) −209.012(VH) | 0.692 | 7.438 | 0.000 |
| Dry weight of AGB | Y = −314.773 + 404.26(B7) − 446.934(B6) + 140.101(IRECI) + 29.898(DVI) + 212.375(B5) +106.868(MCARI) − 56.928(SAVI) + 88.964(MSAVI) − 16.539(NDVI) + 530.148(EVI) − 73.929(VH) | 0.754 | 9.252 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Mao, D.; Zhang, D.; Wang, Z.; Du, B.; Yan, H.; Qiu, Z.; Feng, K.; Wang, J.; Jia, M. Mapping Phragmites australis Aboveground Biomass in the Momoge Wetland Ramsar Site Based on Sentinel-1/2 Images. Remote Sens. 2022, 14, 694. https://doi.org/10.3390/rs14030694
Zhao Y, Mao D, Zhang D, Wang Z, Du B, Yan H, Qiu Z, Feng K, Wang J, Jia M. Mapping Phragmites australis Aboveground Biomass in the Momoge Wetland Ramsar Site Based on Sentinel-1/2 Images. Remote Sensing. 2022; 14(3):694. https://doi.org/10.3390/rs14030694
Chicago/Turabian StyleZhao, Yuxin, Dehua Mao, Dongyou Zhang, Zongming Wang, Baojia Du, Hengqi Yan, Zhiqiang Qiu, Kaidong Feng, Jingfa Wang, and Mingming Jia. 2022. "Mapping Phragmites australis Aboveground Biomass in the Momoge Wetland Ramsar Site Based on Sentinel-1/2 Images" Remote Sensing 14, no. 3: 694. https://doi.org/10.3390/rs14030694








